Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora)
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Identification
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Genetic Distances, Operated Taxonomic Unite Delimitation, and Phylogenetic Analyses
2.4. Secondary Structure Prediction
3. Results
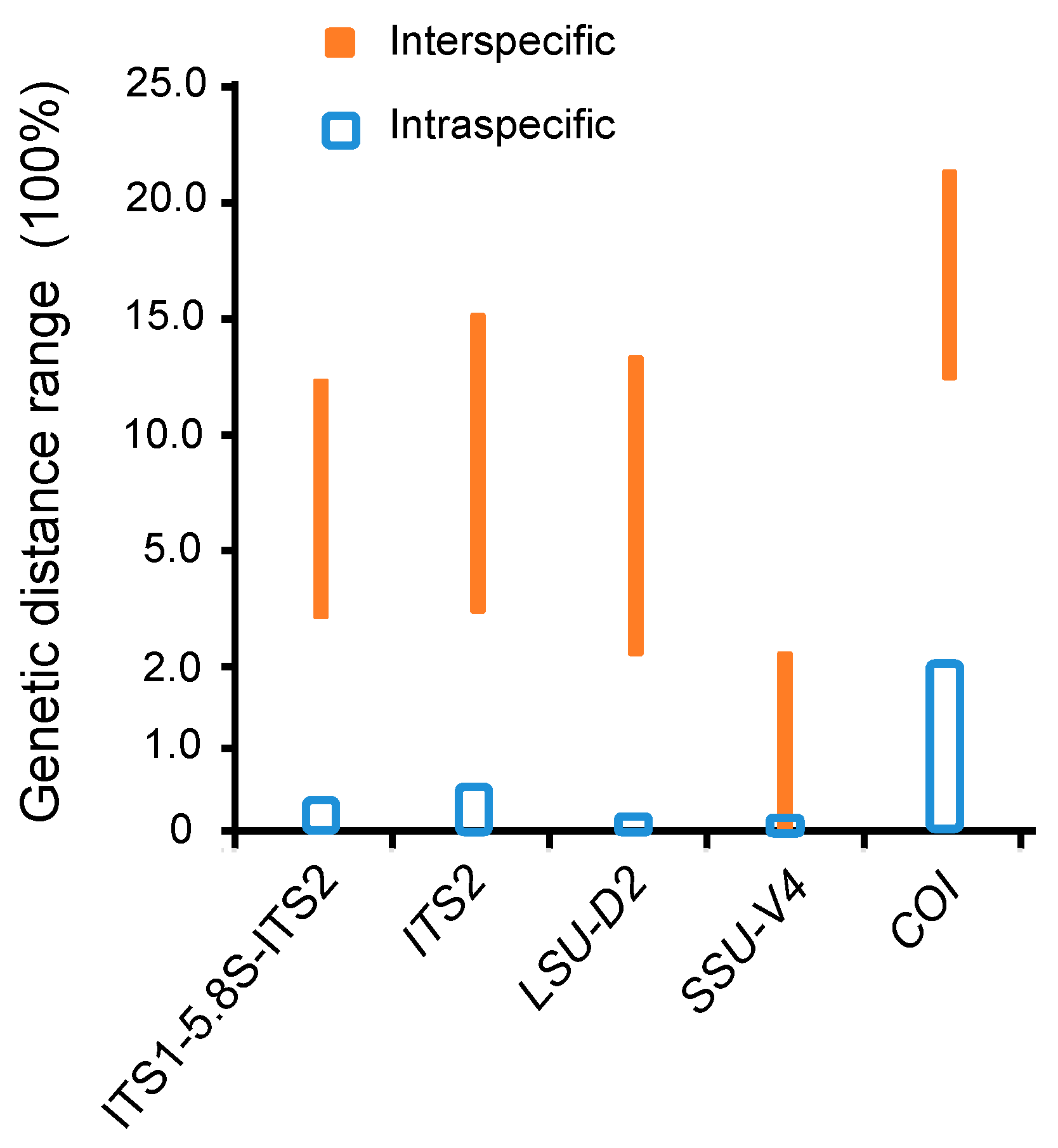
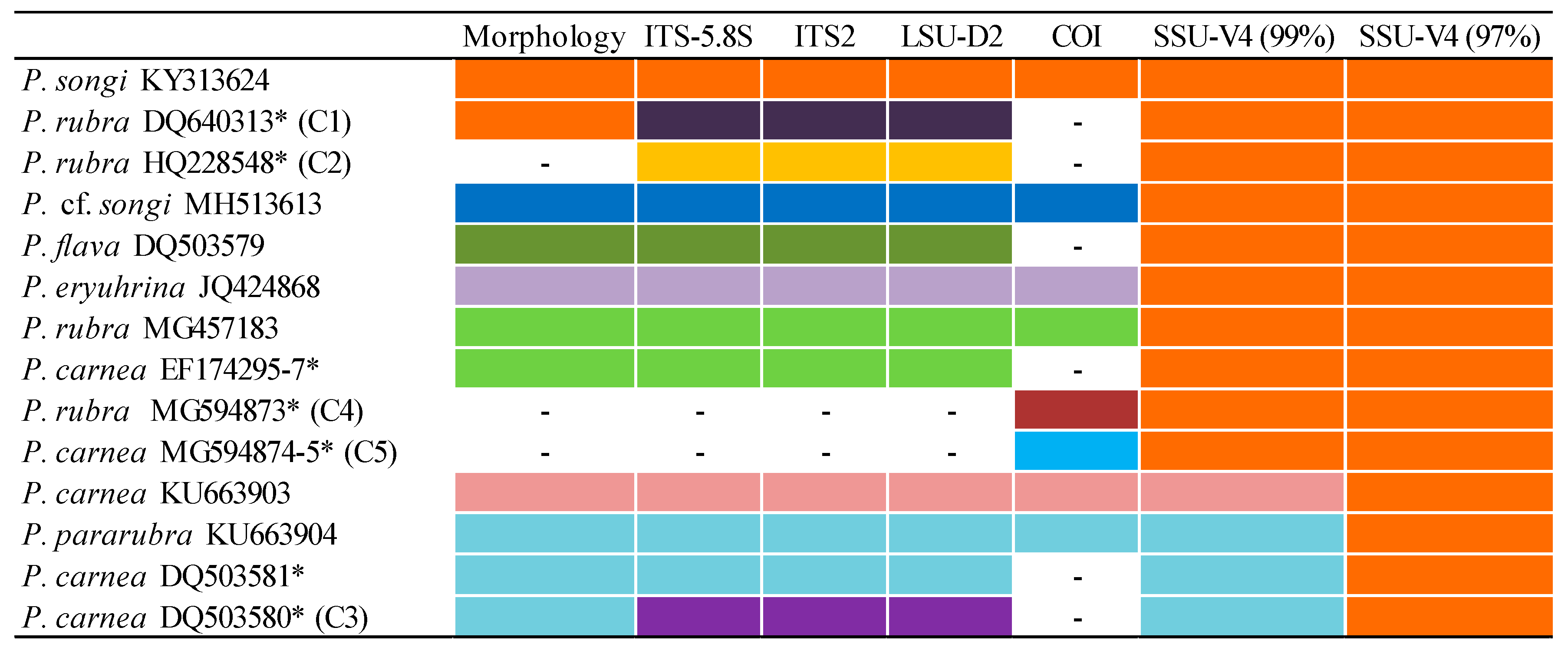
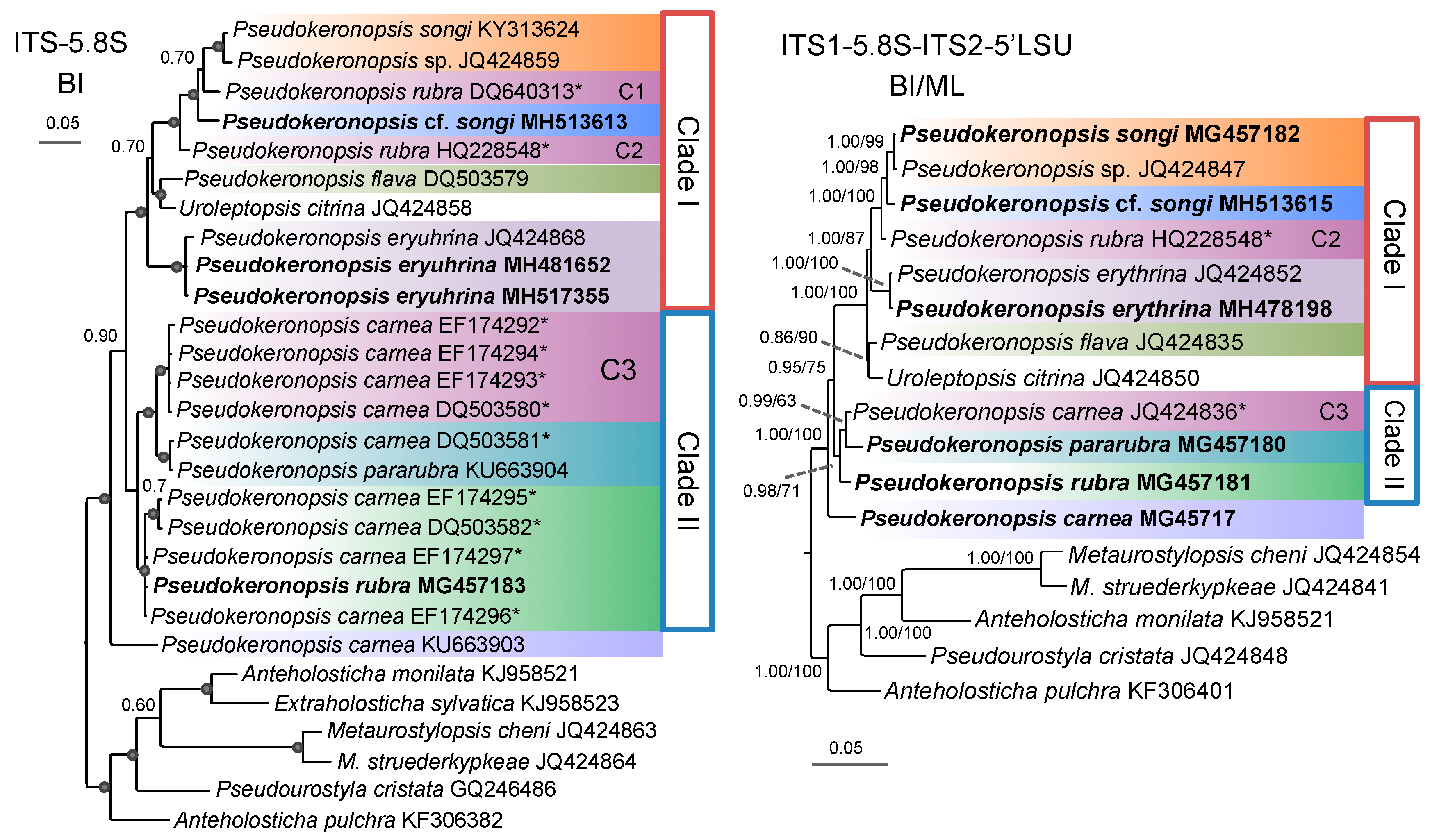
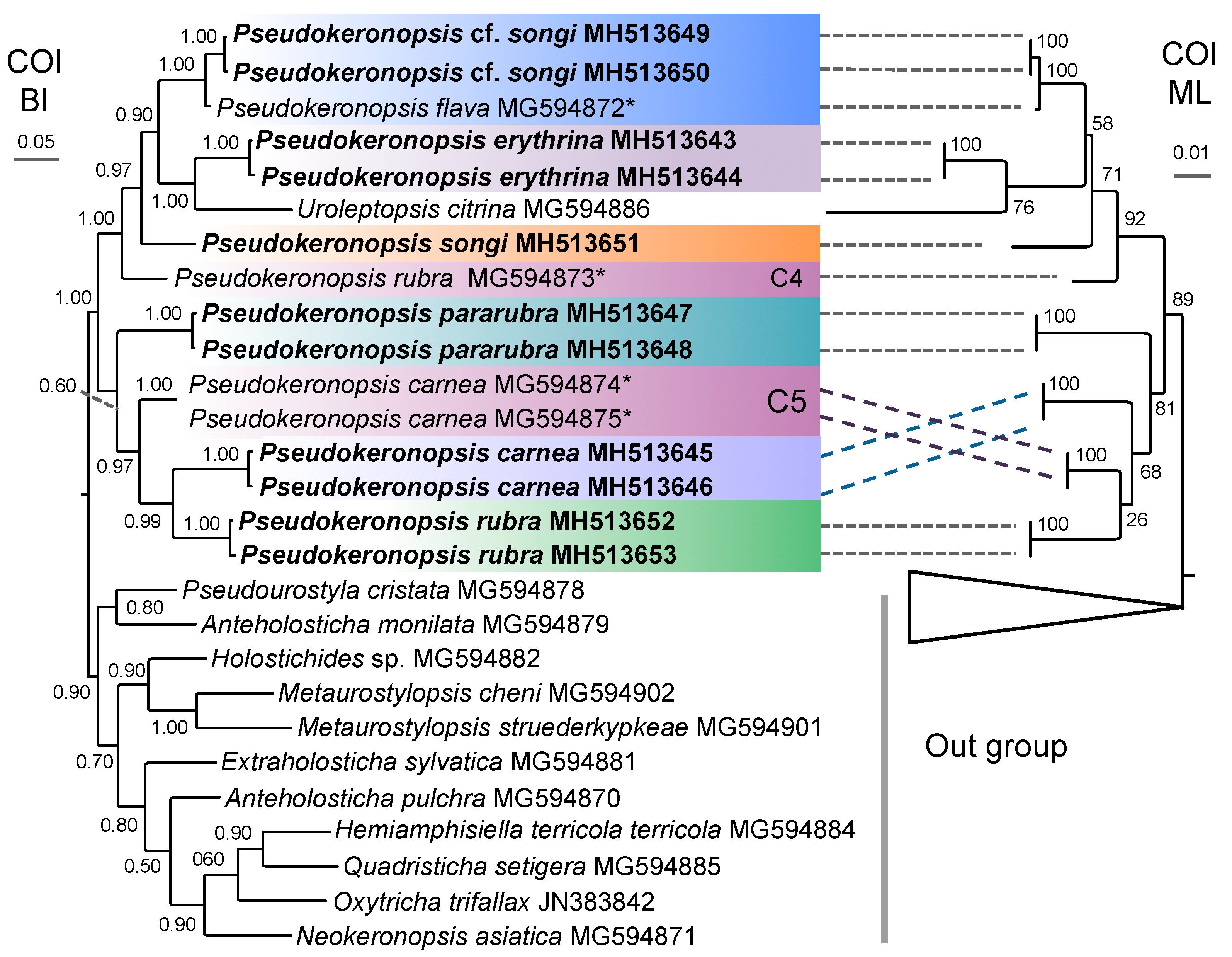
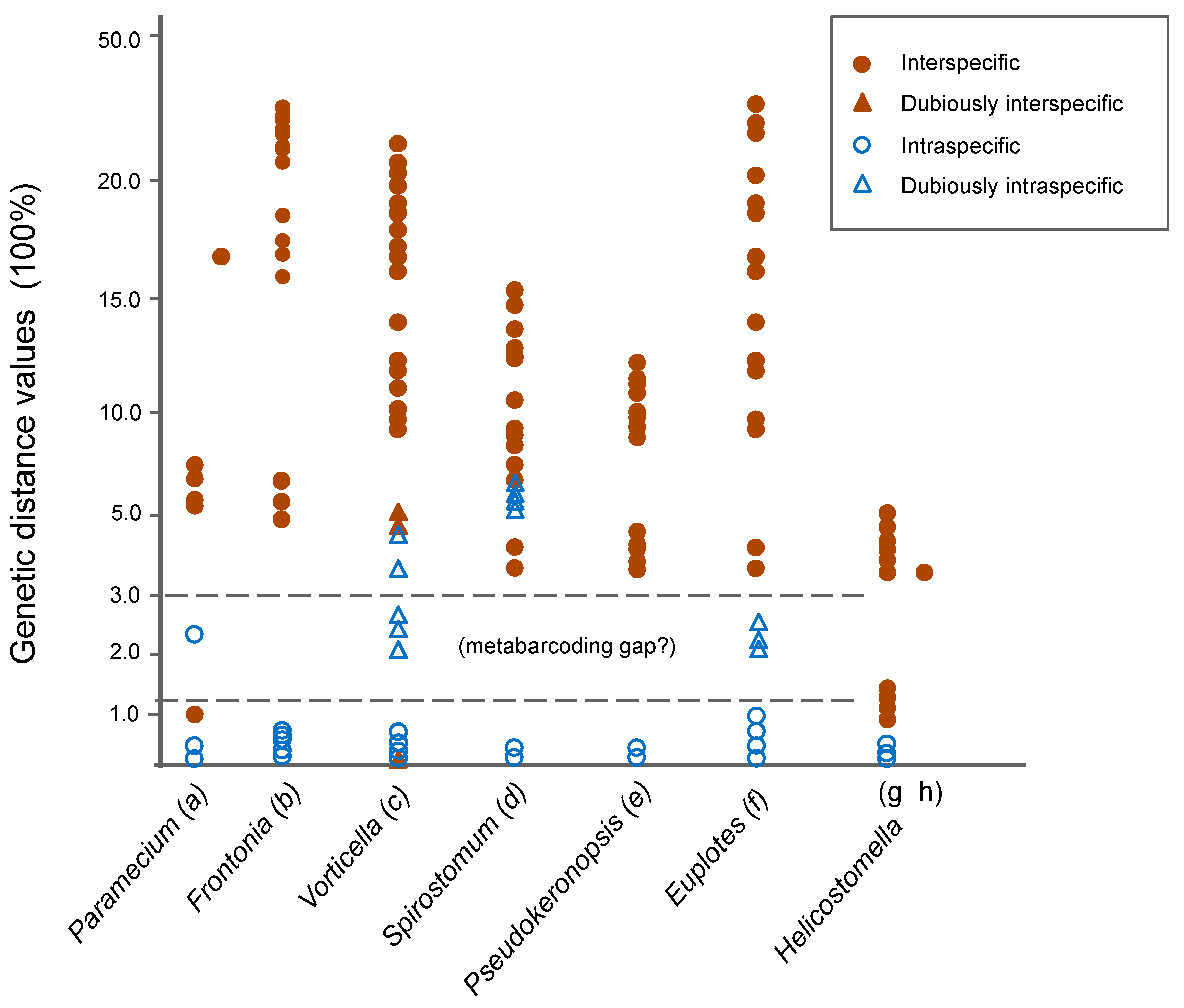
3.1. Molecular Sequences, Genetic Distance, and Species Delimitation Analyses
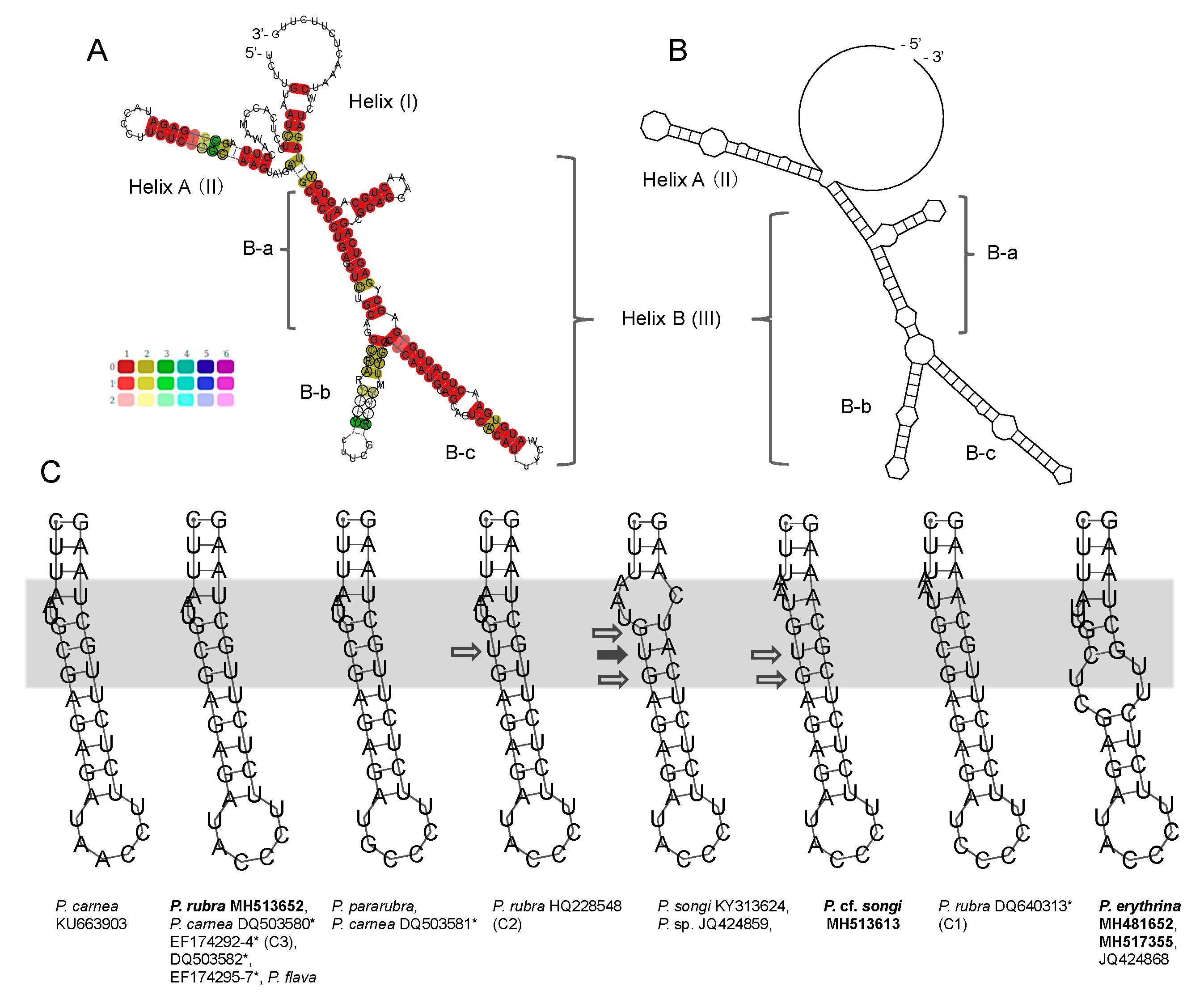
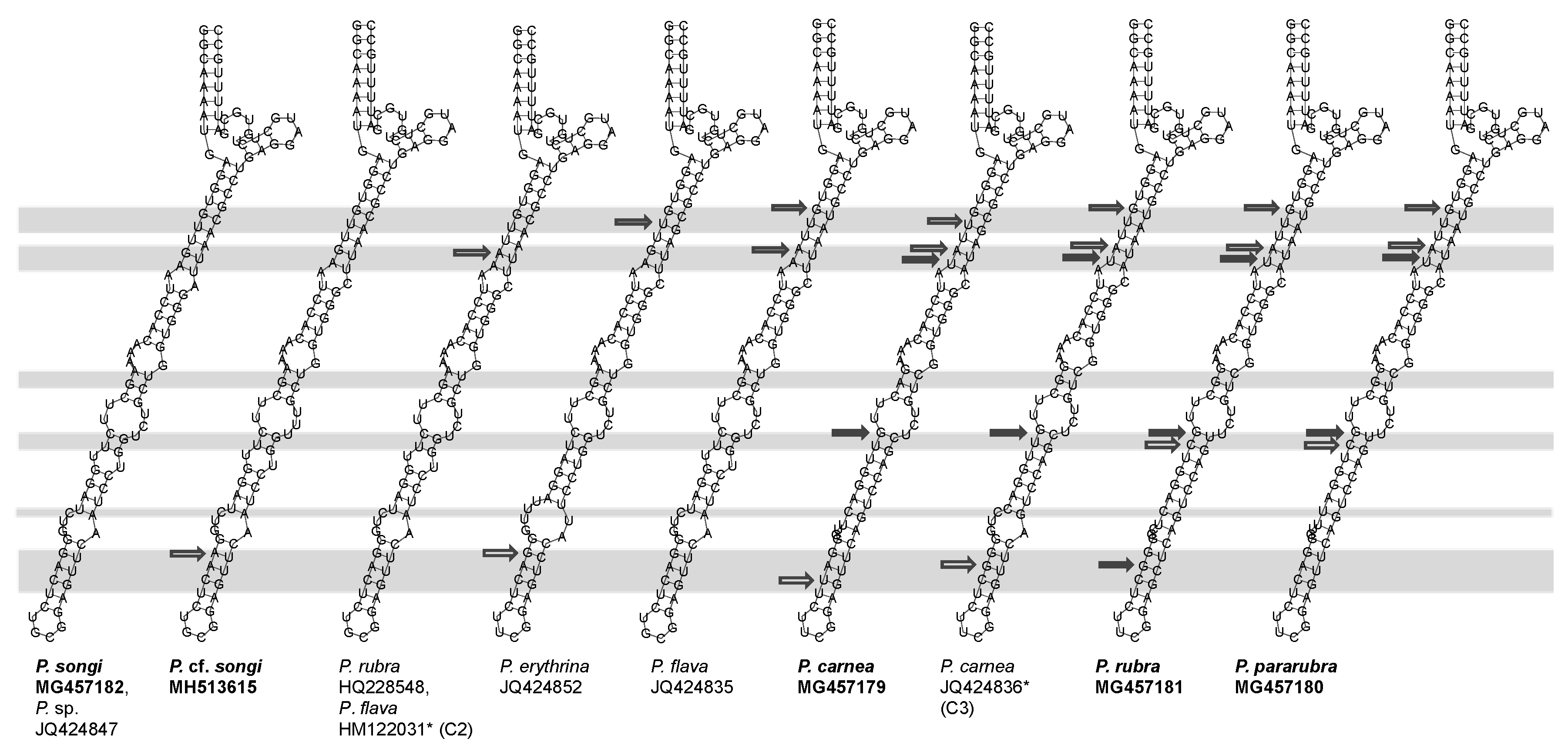
3.2. ITS2 and LSU-D2 Secondary Structures
3.3. Molecular Phylogenetic Analyses
4. Discussion
4.1. Suitable Barcodes for Species Delimitation of Pseudokeronopsis
4.2. Appropriate Metabarcoding Markers for Ciliate Diversity Estimation
4.3. The Species Delimitation Efficiency of CBCs in Putative Secondary Structures
4.4. Phylogenetic Relationships within Pseudokeronopsis
4.5. Confirmation and Revision of Pseudokeronopsis Sequences in GenBank
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- April, J.; Mayden, R.L.; Hanner, R.H.; Bernatchez, L. Genetic calibration of species diversity among North America’s freshwater fishes. Proc. Natl. Acad. Sci. USA 2011, 108, 10602–10607. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; de Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. 2003, 270, S96–S99. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, J.; Audic, S.; Adl, S.; Bass, D.; Belbahri, L.; Berney, C.; Bowser, S.S.; Cepicka, I.; Decelle, J.; Dunthorn, M.; et al. CBOL Protist Working Group: Barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012, 10, e1001419. [Google Scholar] [CrossRef]
- Stoeck, T.; Przybos, E.; Dunthorn, M. The D1-D2 region of the large subunit ribosomal DNA as barcode for ciliates. Mol. Ecol. Resour. 2014, 14, 458–468. [Google Scholar] [CrossRef]
- Elwood, H.J.; Olsen, G.J.; Sogin, M.L. The small–subunit ribosomal RNA gene sequences from the hypotrichous ciliates Oxytricha nova and Stylonychia pustulata. Mol. Biol. Evol. 1985, 2, 399–410. [Google Scholar] [PubMed]
- Lynn, H.D. The Ciliated Protozoa: Characterization, Classification and Guide to the Literature; Springer: Dordrecht, The Netherlands, 2008. [Google Scholar]
- Dunthorn, M.; Klier, J.; Bunge, J.; Stoeck, T. Comparing the hyper-variable V4 and V9 regions of the small subunit rDNA for assessment of ciliate environmental diversity. J. Eukaryot. Microbiol. 2012, 59, 185–187. [Google Scholar] [CrossRef]
- Santoferrara, L.F.; McManus, G.B.; Alder, V.A. Utility of genetic markers and morphology for species discrimination within the order Tintinnida (Ciliophora, Spirotrichea). Protist 2013, 164, 24–36. [Google Scholar] [CrossRef]
- Zhao, F.; Filker, S.; Stoeck, T.; Xu, K. Ciliate diversity and distribution patterns in the sediments of a seamount and adjacent abyssal plains in the tropical Western Pacific Ocean. BMC Microbiol. 2017, 17, 192. [Google Scholar] [CrossRef] [PubMed]
- Lara, E.; Acosta-Mercado, D. A molecular perspective on ciliates as soil bioindicators. Eur. J. Soil Bol. 2012, 49, 107–111. [Google Scholar] [CrossRef]
- Li, J.; Zhan, Z.; Xu, K. Systematics and molecular phylogeny of the ciliate genus Pseudokeronopsis (Ciliophora, Hypotrichia). J. Eukaryot. Microbiol. 2017, 64, 850–872. [Google Scholar] [CrossRef] [PubMed]
- Strüder-Kypke, M.C.; Lynn, D.H. Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst. Biodivers. 2010, 8, 131–148. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Gentekaki, E.; Zhan, A.; Al-Farraj, S.A.; Song, W. Utility of combining morphological characters, nuclear and mitochondrial genes: An attempt to resolve the conflicts of species identification for ciliated protists. Mol. Phylogenet. Evol. 2016, 94, 718–729. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.; Warren, A.; Song, W. Species delimitation for the molecular taxonomy and ecology of a widely distributed microbial eukaryotes genus Euplotes (Alveolata, Ciliophora). Proc. Biol. Sci. 2018, 285, 20172159. [Google Scholar] [CrossRef] [PubMed]
- Berger, H. Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monogr. Biol. 2006, 85, 731–980. [Google Scholar]
- Borror, A.C.; Wicklow, B.J. The suborder Urostylina, Jankowski (Ciliophora, Hypotrichida): Morphology, systematics and identification of species. Acta Protozool. 1983, 22, 97–126. [Google Scholar]
- Chen, X.; Clamp, J.C.; Song, W. Phylogeny and systematic revision of the family Pseudokeronopsidae (Protista, Ciliophora, Hypotricha), with description of a new estuarine species of Pseudokeronopsis. Zool. Scr. 2011, 40, 659–671. [Google Scholar] [CrossRef]
- Song, W.; Warren, A.; Roberts, D.; Wilbert, N.; Li, L.; Sun, P.; Hu, X.; Ma, H. Comparison and redefinition of four marine, colored Pseudokeronopsis spp. (Ciliophora, Hypotrichida), with emphasis on their living morphology. Acta Protozool. 2006, 45, 271–287. [Google Scholar]
- Yi, Z.; Chen, Z.; Warren, A.; Roberts, D.M.; Al-Rasheid, K.A.S.; Miao, M.; Gao, S.; Shao, C.; Song, W. Molecular phylogeny of Pseudokeronopsis (Protozoa, Ciliophora, Urostylida), with reconsideration of three closely related species at inter- and intra-specific levels inferred from the small subunit ribosomal RNA gene and the ITS1-5.8S-ITS2 region sequences. J. Zool. 2008, 275, 268–275. [Google Scholar] [CrossRef]
- Anesi, A.; Buonanno, F.; Di Giuseppe, G.; Ortenzi, C.; Guella, G. Metabolites from the euryhaline ciliate Pseudokeronopsis erythrina. Eur. J. Org. Chem. 2016, 7, 1330–1336. [Google Scholar] [CrossRef]
- Buonanno, F.; Anesi, A.; Giuseppe, G.D.; Guella, G.; Ortenzi, C. Chemical Defense by Erythrolactones in the Euryhaline Ciliated Protist, Pseudokeronopsis erythrina. Zool. Sci. 2017, 34, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Huang, L.; Yang, R.; Lin, X.; Song, W. Actin evolution in ciliates (Protist, Alveolata) is characterized by high diversity and three duplication events. Mol. Biol. Evol. 2016, 96, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, Z.; Song, W.; Berger, H. Three-gene based phylogeny of the Urostyloidea (Protista, Ciliophora, Hypotricha), with notes on classification of some core taxa. Mol. Phylogenet. Evol. 2014, 70, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Barth, D.; Krenek, S.; Fokin, S.I.; Berendonk, T.U. Intraspecific genetic variation in Paramecium revealed by mitochondrial cytochrome c oxidase I sequences. J. Eukaryot. Microbiol. 2006, 53, 20–25. [Google Scholar] [CrossRef]
- Greczek-Stachura, M.; Potekhin, A.; Przyboś, E.; Rautian, M.; Skoblo, I.; Tarcz, S. Identification of Paramecium bursaria syngens through molecular markers–comparative analysis of three loci in the nuclear and mitochondrial DNA. Protist 2012, 163, 671–685. [Google Scholar] [CrossRef]
- Park, M.H.; Jung, J.H.; Jo, E.; Park, K.M.; Baek, Y.S.; Kim, S.J.; Min, G.S. Utility of mitochondrial COI sequences for species discrimination of Spirotrichea ciliates (Protozoa, Ciliophora). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2019, 30, 148–155. [Google Scholar]
- Tarcz, S.; Przyboś, E.; Surmacz, M. An assessment of haplotype variation in ribosomal and mitochondrial DNA fragments suggests incomplete lineage sorting in some species of the Paramecium aurelia complex (Ciliophora, Protozoa). Mol. Phylogenet. Evol. 2013, 67, 255–265. [Google Scholar] [CrossRef]
- Tarcz, S.; Rautian, M.; Potekhin, A.; Sawka, N.; Beliavskava, A.; Kiselev, I.; Nekrasova, I.; Przyboś, E. Paramecium putrinum (Ciliophora, Protozoa): The first insight into the variation of two DNA fragments–molecular support for the existence of cryptic species. Mol. Phylogenet. Evol. 2014, 73, 140–145. [Google Scholar] [CrossRef]
- Shazib, S.U.A.; Vďačný, P.; Kim, J.H.; Jang, S.W.; Shin, M.K. Molecular phylogeny and species delimitation within the ciliate genus Spirostomum (Ciliophora, Postciliodesmatophora, Heterotrichea), using the internal transcribed spacer region. Mol. Biol. Evol. 2016, 102, 128–144. [Google Scholar] [CrossRef]
- Sun, P.; Clamp, J.C.; Xu, D. Analysis of the secondary structure of ITS transcripts in peritrich ciliates (Ciliophora, Oligohymenophorea): Implications for structural evolution and phylogenetic reconstruction. Mol. Phylogenet. Evol. 2010, 56, 242–251. [Google Scholar] [CrossRef]
- Sun, P.; Clamp, J.C.; Xu, D.; Huang, B.; Shin, M.K.; Turner, F. An ITS–based phylogenetic framework for the genus Vorticella: Finding the molecular and morphological gaps in a taxonomically difficult group. Proc. Biol. Sci. 2013, 280, 20131177. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Sun, P.; Shin, M.K.; Kim, Y.O. Species boundaries in tintinnid ciliates: A case study—Morphometric variability, molecular characterization, and temporal distribution of Helicostomella species (Ciliophora, Tintinnina). J. Eukaryot. Microbiol. 2012, 59, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Chantangsi, C.; Lynn, D.H.; Brandl, M.T.; Cole, J.C.; Hetrick, N.; Ikonomi, P. Barcoding ciliates: A comprehensive study of 75 isolates of the genus Tetrahymena. Int. J. Syst. Evol. Microbiol. 2007, 57, 2412–2425. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.; Jung, J.; Min, G. Redescription of two marine ciliates (Ciliophora: Urostylida: Pseudstokesokeronopsidae), Pseudokeronopsis carnea and Uroleptopsis citrina, from Korea. Korean J. Syst. Zool. 2011, 27, 220–227. [Google Scholar] [CrossRef]
- Li, Y.; Niu, Y.; Liu, L. Phylogenetic studies of four species of ciliate inferred from 16S-like small subunit rRNA gene sequences. J. For. Res. 2008, 19, 119–124. [Google Scholar] [CrossRef]
- Smith, D.; Leary, P.; Craggs, J.; Bythell, J.; Sweet, M. Healthy and White Syndrome-Affected Echinopora lamellosa in Aquaria and Experimental Treatment with the Antibiotic Ampicillin. PLoS ONE 2015, 10, e0121780. [Google Scholar] [CrossRef]
- Foissner, W. An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol. 2014, 64, 271–292. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Goggin, C.L.; Murphy, N.E. Conservation of sequence in the internal transcribed spacers and 5.8S ribosomal RNA among geographically separated isolates of parasitic scuticociliates (Ciliophora, Orchitophryidae). Dis. Aquat. Org. 2000, 40, 79–83. [Google Scholar] [CrossRef]
- Gong, J.; Kim, S.J.; Kim, S.Y.; Min, G.S.; Roberts, D.M.; Warren, A.; Choi, J.K. Taxonomic redescriptions of two ciliates, Protogastrostyla pulchra ng, n. comb. and Hemigastrostyla enigmatica (Ciliophora: Spirotrichea, Stichotrichia), with phylogenetic analyses based on 18S and 28S rRNA gene sequences. J. Eukaryot. Microbiol. 2007, 54, 468–478. [Google Scholar]
- Katoh, K.; Standley, D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Coleman, A.W. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 2005, 52, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Wuyts, J.; De Rijk, P.; Van de Peer, Y.; Winkelmans, T.; De Wachter, R. The European large subunit ribosomal RNA database. Nucleic Acids Res. 2001, 29, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Wuyts, J.; Van de Peer, Y.; Winkelmans, T.; De Wachter, R. The European database on small subunit ribosomal RNA. Nucleic Acids Res. 2002, 30, 183–185. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or Bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA 2012, 5, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Wolf, M.; Friedrich, J.; Dandekar, T.; Müller, T. CBCAnalyzer: Inferring phylogenies based on compensatory base changes in RNA secondary structures. Silico Biol. 2005, 5, 291–294. [Google Scholar]
- Yi, Z.; Song, W. Evolution of the order Urostylida (Protozoa, Ciliophora): New hypotheses based on multi-gene information and identification of localized incongruence. PLoS ONE 2011, 6, e17471. [Google Scholar] [CrossRef]
- Song, W.; Sun, P.; Ji, D. Redefinition of the yellow hypotrichous ciliate, Pseudokeronopsis flava (Hypotrichida: Ciliophora). J. Mar. Biol. Assoc. U. K. 2004, 84, 1137–1142. [Google Scholar] [CrossRef]
- Jung, S.J.; Bae, M.J.; Oh, M.J.; Lee, J. Sequence conservation in the internal transcribed spacers and 5.8S ribosomal RNA of parasitic scuticociliates Miamiensis avidus (Ciliophora, Scuticociliatia). Parasitol. Int. 2011, 60, 216–219. [Google Scholar] [CrossRef]
- Santoferrara, L.F.; Tian, M.; Alder, V.A.; McManus, G.B. Discrimination of closely related species in tintinnid ciliates: New insights on crypticity and polymorphism in the genus Helicostomella. Protist 2015, 166, 78–92. [Google Scholar] [CrossRef]
- Gentekaki, E.; Lynn, D.H. High-level genetic diversity but no population structure inferred from nuclear and mitochondrial markers of the peritrichous ciliate Carchesium polypinum in the Grand River Basin (North America). Appl. Environ. Microbiol. 2009, 75, 3187–3195. [Google Scholar] [CrossRef]
- Kher, C.P.; Doerder, F.P.; Cooper, J.; Ikonomi, P.; Achilles-Day, U.; Küpper, F.C.; Lynn, D.H. Barcoding Tetrahymena: Discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox-1) barcode. Protist 2011, 162, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.H.; Strüder-Kypke, M. Species of Tetrahymena identical by small subunit rRNA gene sequences are discriminated by mitochondrial cytochrome c oxidase I gene sequences. J. Eukaryot. Microbiol. 2006, 53, 385–387. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Breiner, H.W.; Filker, S.; Ostermaier, V.; Kammerlander, B.; Sonntag, B. A morphogenetic survey on ciliate plankton from a mountain lake pinpoints the necessity of lineage-specific barcode markers in microbial ecology. Environ. Microbiol. 2014, 16, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Tarcz, S. Intraspecific differentiation of Paramecium novaurelia strains (Ciliophora, Protozoa) inferred from phylogenetic analysis of ribosomal and mitochondrial DNA variation. Eur. J. Protistol. 2013, 49, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Tarcz, S.; Potekhin, A.; Rautian, M.; Przyboś, E. Variation in ribosomal and mitochondrial DNA sequences demonstrates the existence of intraspecific groups in Paramecium multimicronucleatum (Ciliophora, Oligohymenophorea). Mol. Phylogenet. Evol. 2012, 63, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Heeger, F.; Bourne, E.C.; Baschien, C.; Yurkov, A.; Bunk, B.; Spröer, C.; Overmann, J.; Mazzoni, C.J.; Monaghan, M.T. Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments. Mol. Ecol. Resour. 2018, 18, 1500–1514. [Google Scholar] [CrossRef]
- Wagner, J.; Coupland, P.; Browne, H.P.; Lawley, T.D.; Francis, S.C.; Parkhill, J. Evaluation of PacBio sequencing for full-length bacterial 16S rRNA gene classification. BMC Microbiol. 2016, 16, 274. [Google Scholar] [CrossRef]
- Coleman, A.W. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef]
- Müller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef]
- Ruhl, M.W.; Wolf, M.; Jenkins, T.M. Compensatory base changes illuminate morphologically difficult taxonomy. Mol. Phylogenet. Evol. 2010, 54, 664–669. [Google Scholar] [CrossRef]
- Sorhannus, U.; Ortiz, J.D.; Wolf, M.; Fox, M.G. Microevolution and speciation in Thalassiosira weissflogii (Bacillariophyta). Protist 2010, 161, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Müller, T. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences—A proof of concept. PLoS ONE 2013, 8, e6672. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yi, Z.; Al-Farraj, S.A.; Song, W. Phylogenetic positions and taxonomic assignments of the systematically controversial genera, Spirotrachelostyla, Uroleptopsis and Tunicothrix (Protozoa, Ciliophora, Stichotrichia) based on small subunit rRNA gene sequences. Syst. Biol. 2010, 8, 409–416. [Google Scholar]



| Organisms | Sampling Location | References | Accession Numbers | |||
|---|---|---|---|---|---|---|
| SSU rDNA | ITS1-5.8S-ITS2 | LSU rDNA | COI | |||
| Pseudokeronopsis rubra | Yellow Sea, Qingdao, China | Present study | MG457184 | MG457183 | MG457181 | MH513652, MH513653 |
| Pseudokeronopsis pararubra | Yellow Sea, Qingdao, China | Present study | KU663902 | KU663904 | MG457180 | MH513647, MH513648 |
| Pseudokeronopsis carnea | Yellow Sea, Qingdao, China | Present study | KU663901 | KU663903 | MG457179 | MH513645, MH513646 |
| P. carnea * (C5) | Yellow Sea, Incheon harbor, Korea | [27,35] | JN714476 | - | - | MG594874 |
| P. carnea * (C5) | Yellow Sea, Gwangyang, Korea | [27] | MG603616 | - | - | MG594875 |
| P. carnea * | Yellow Sea, Qingdao, China | [23] | KT984168 | - | - | - |
| P. carnea pop-I * (C3) | Yellow Sea, Qingdao, China | [20] | AY881633 | DQ503580, EF174292-4 | JQ424836 | - |
| P. carnea pop-II * | Yellow Sea, Qingdao, China | [20] | - | DQ503581 | - | - |
| P. carnea pop-III * | Yellow Sea, Qingdao, China | [20] | - | DQ503582, EF174295-7 | - | - |
| Pseudokeronopsis erythrina | Estuary, Yancheng, China | Present study | MH478194 | MH481652, MH517355 | MH478198, MH478200 | MH513643, MH513644 |
| P. erythrina | Estuary of Pearl River, Guangzhou, China | [18,24] | FJ775723 | JQ424868 | JQ424852 | - |
| P. erythrina | Estuary of Pearl River, Guangzhou, China | [23] | KT984173 | - | - | - |
| P. erythrina | Freshwater River Yamuna, Delhi, India | Kaur et al., unpublished | MG994990 | - | - | - |
| P. erythrina | Lake Trasimeno, Perugia, Italy | [22] | KX459375 | - | - | - |
| Pseudokeronopsis cf. songi | Yellow Sea, Qingdao, China | Present study | MH513618, MH513619 | MH513613, MH513614 | MH513615, MH513616 | MH513649, MH513650 |
| Pseudokeronopsis songi | Yellow Sea, Qingdao, China | Present study | KY313623 | KY313624 | MG457182 | MH513651 |
| P. rubra * (C4) | Sea of Japan, Hwajinpo, Korea | [27] | MG603620 | - | MG594873 | |
| P. rubra * (C1) | Yellow Sea, Qingdao, China | [20] | DQ640314 | DQ640313 | - | - |
| P. rubra * | - | Khan and Shin, unpublished | HM140387 | - | - | - |
| P. rubra * | - | [36] | EF535729 | - | - | - |
| P. rubra * | Yellow Sea, Qingdao, China | [27] | KT984169 | - | - | - |
| P. rubra * | South China Sea, Guangdong, China | [27] | KT984170 | - | - | - |
| P. rubra * | Daya Bay, Guangdong, China | [27] | KT984171 | - | - | - |
| P. rubra * | Mangrove wetland, Shenzhen, China | [27] | KT984172 | - | - | - |
| P. rubra * (C2) | Yellow Sea, Incheon, Korea | Jung et al., unpublished | HQ228548 | HQ228548 | HQ228548 | - |
| Pseudokeronopsis flava | Sea of Japan, Pohang-is, Korea | [27] | MG603616 | - | - | MG594872 |
| P. flava | South China Sea, Zhanjiang, China | [20] | AY881634 | DQ503579 | JQ424835 | - |
| P. flava | Yellow Sea, Qingdao, China | [20] | DQ227798 | - | - | - |
| P. flava | Mangrove wetland, Shenzhen, China | [27] | KT984174 | - | - | - |
| P. flava | Yellow Sea, Qingdao, China | [27] | KT984175 | - | - | - |
| P. flava | - | Khan and Shin, unpublished | HM140386 | - | HM122031 | - |
| Pseudokeronopsis sp. | Yellow Sea, Qingdao, China | [24] | JQ424830 | JQ424859 | JQ424847 | - |
| Pseudokeronopsis sp. | Horniman Museum and Gardens Aquarium, London, England | [37] | KP793002 | - | - | - |
| Sequences | Reference | Morphological Data * | Molecular Analyses | Emendation |
|---|---|---|---|---|
| P. carnea DQ503581 | [20] | Similar to P. pararubra and P. rubra | No difference from P. pararubra in ITS1-5.8S-ITS2. | P. pararubra |
| P. carnea DQ503582, EF174295-7 | [20] | Similar to P. pararubra and P. rubra | No difference from P. rubra in ITS2 sequence. | P. rubra |
| P. carnea DQ503580, EF174292-4, JQ424836, AY881633 (C3) | [20] | Similar to P. pararubra and P. rubra | The ITS1-5.8S-ITS2 genetic distances between C3 and congeners were in the range of 3.23–10.74%, similar to those among congeners (3.68–13.01%), indicating a cryptic species. | Cryptic species related to P. pararubra |
| P. rubra DQ640313, DQ640314 (C1) | [20] | Similar to P. songi | The ITS1-5.8S-ITS2 genetic distances between C1 and congeners were in the range of 3.46–11.93%, indicating a cryptic species. | Cryptic species related to P. songi |
| P. rubra MG603620, MG594873 (C4) | [27] | Not available | The genetic distances between C4 and congeners were in the range of 13.55–21.54%, indicating an uncertain or cryptic species. | Misidentified, uncertain species |
| P. flava MG594872, MG603616 | [27] | Not available | The COI genetic distance between P. flava MG594872 and P. cf. songi was 2.58%, whereas those among congeners ranged from 13.25% to 21.28%, indicating it conspecific with P. cf. songi. | P. cf. songi |
| Pseudokeronopsis sp. JQ424859, JQ424830 | [24] | Not available | The ITS1-5.8S-ITS2 genetic distance between JQ424859 and P. songi was 0.21% (vs. conspecific distances 0–0.42%), indicating that the former belonged to P. songi. | P. songi |
| P. carnea JN714476, MG603616, MG594874, MG594875 (C5) | [27] | Not available | The COI genetic distance between P. carnea MG594874 and MG594875 was zero, whereas those between them and congeners were much higher than conspecific ones (13.55–20.85% vs. 0–2.58%), suggesting they represented an uncertain species. | Misidentified, uncertain species |
| P. rubra HQ228548 (C2), P. flava HM122031 | Jung et al. (unpublished), Khan & Shin (unpublished) | Not available | The ITS1-5.8S-ITS2 genetic distance between C2 and congeners were in the range of 3.67–9.73%, suggesting it represented an uncertain species. P. flava HM122031 and C2 were conspecific, possessing the identical LSU-D2 sequence. | Misidentified, uncertain species |
| P. carnea KT002202-07, P. flava KT002234-44, KR263877-79, KR263883-89, P. rubra HM140387, EF535729, KT984169-72, KT002196-201, KT002222-33, KT002245-48, KR263874-76, KR263880-82, KR263888-91 | [23,36] | Not available | Only SSU rDNA and Actin I sequences were available for these populations, showing six, one, and three nucleotide differences of SSU-V4 region from the valid P. carnea, P. flava, and P. rubra, respectively. | Dubious |
| P. carnea GQ258110 | [57] | Not available | Only alpha-tubulin gene sequence was available. | Dubious |
| P. erythrina KX459375, MG994990 | [22], Kaur et al. unpublished | Not available | Only SSU rDNA sequences were available, showing 1 and 23 nucleotide differences of SSU rDNA from the valid P. erythrina, respectively. | Dubious |
| Species/Populations | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P. pararubra KU663904, P. carnea DQ503581 * | 0 | |||||||||
| 2 | P. carnea DQ503580, EF174292-7 * (C3) | 0 | 0 | ||||||||
| 3 | P. carnea KU663903 | 1 | 1 | - | |||||||
| 4 | P. songi KY313624, Pseudokeronopsis sp. JQ424859 | 2 | 2 | 2 | 0 | ||||||
| 5 | P. rubra DQ640313 * (C1) | 1 | 1 | 1 | 1 | - | |||||
| 6 | P. cf. songi MH513613, MH513614 | 1 | 1 | 1 | 0 | 0 | 0 | ||||
| 7 | P. rubra HQ228548 * (C2) | 1 | 1 | 1 | 0 | 0 | 0 | - | |||
| 8 | P. flava DQ503579 | 2 | 1 | 1 | 2 | 1 | 1 | 0 | - | ||
| 9 | P. erythrinaMH481652, MH517355, JQ424868 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 10 | P. rubraMG457183, P. carnea DQ503582 * | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 |
| Species/Populations | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | P. songi, Pseudokeronopsis sp. JQ424847 | 0 | ||||||||
| 2 | P. cf. songiMH513615, MH513616 | 0 | 0 | |||||||
| 3 | P. flava JQ424835 | 1 | 1 | - | ||||||
| 4 | P. erythrinaMH478198, MH478200, JQ424852 | 2 | 3 | 0 | 0 | |||||
| 5 | P. carneaMG457179 | 7 | 8 | 7 | 9 | - | ||||
| 6 | P. pararubraMG457180 | 5 | 6 | 5 | 8 | 1 | - | |||
| 7 | P. rubraMG457181 | 7 | 8 | 6 | 8 | 2 | 1 | - | ||
| 8 | P. carnea JQ424836 * (C3) | 5 | 6 | 6 | 7 | 1 | 1 | 1 | - | |
| 9 | P. rubra HQ228548 *, P. flava HM122031 (C2) | 0 | 0 | 0 | 1 | 7 | 5 | 7 | 5 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, Z.; Li, J.; Xu, K. Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora). Microorganisms 2019, 7, 493. https://doi.org/10.3390/microorganisms7110493
Zhan Z, Li J, Xu K. Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora). Microorganisms. 2019; 7(11):493. https://doi.org/10.3390/microorganisms7110493
Chicago/Turabian StyleZhan, Zifeng, Ju Li, and Kuidong Xu. 2019. "Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora)" Microorganisms 7, no. 11: 493. https://doi.org/10.3390/microorganisms7110493
APA StyleZhan, Z., Li, J., & Xu, K. (2019). Ciliate Environmental Diversity Can Be Underestimated by the V4 Region of SSU rDNA: Insights from Species Delimitation and Multilocus Phylogeny of Pseudokeronopsis (Protist, Ciliophora). Microorganisms, 7(11), 493. https://doi.org/10.3390/microorganisms7110493





