Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Nucleotide Sequences
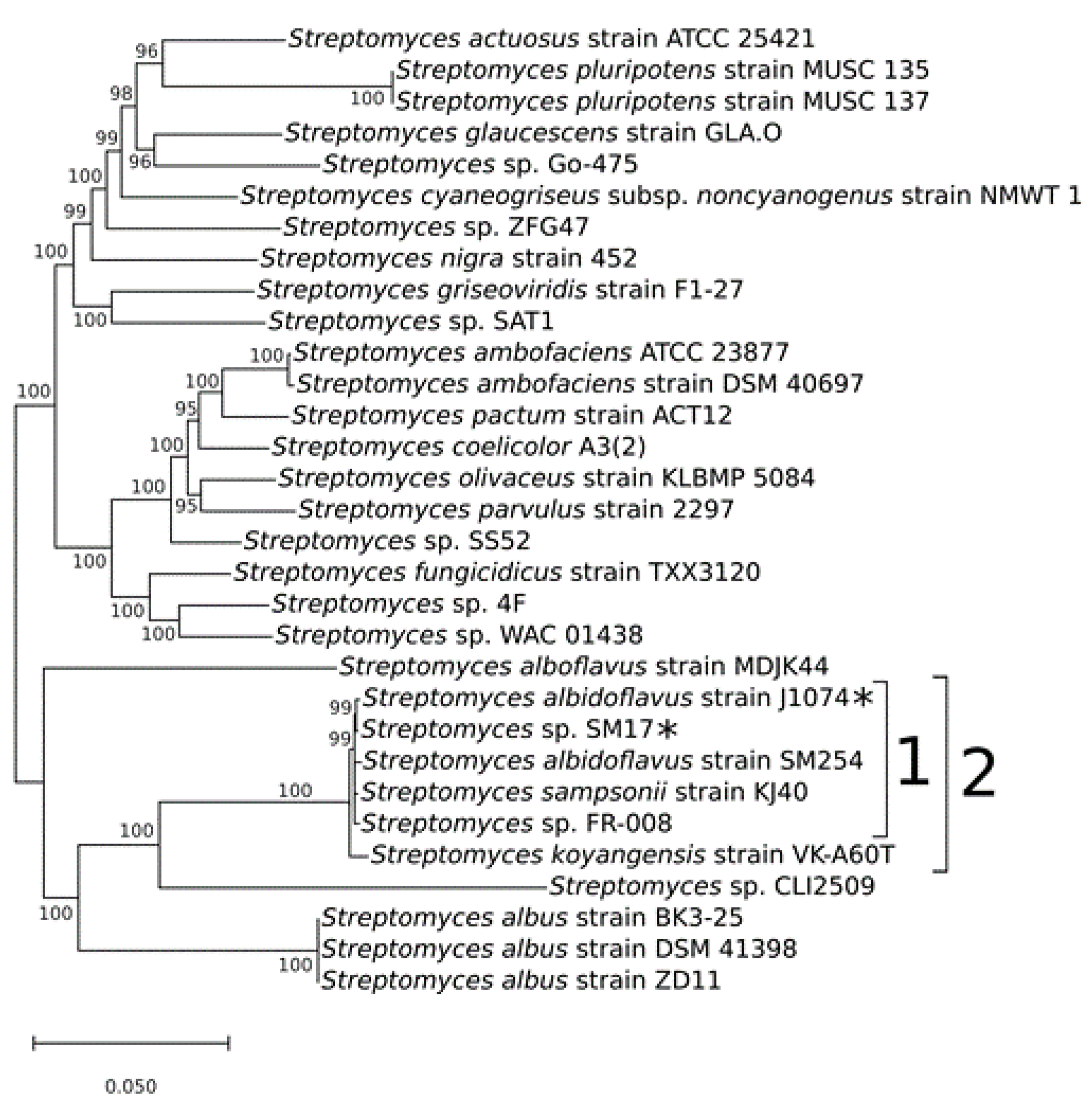
2.2. Phylogenetic Analyses
2.3. Prediction of Secondary Metabolites Biosynthetic Gene Clusters
2.4. Gene Synteny Analysis
2.5. Diagrams and Figures
2.6. Strains Culture, Maintenance, and Secondary Metabolites Production
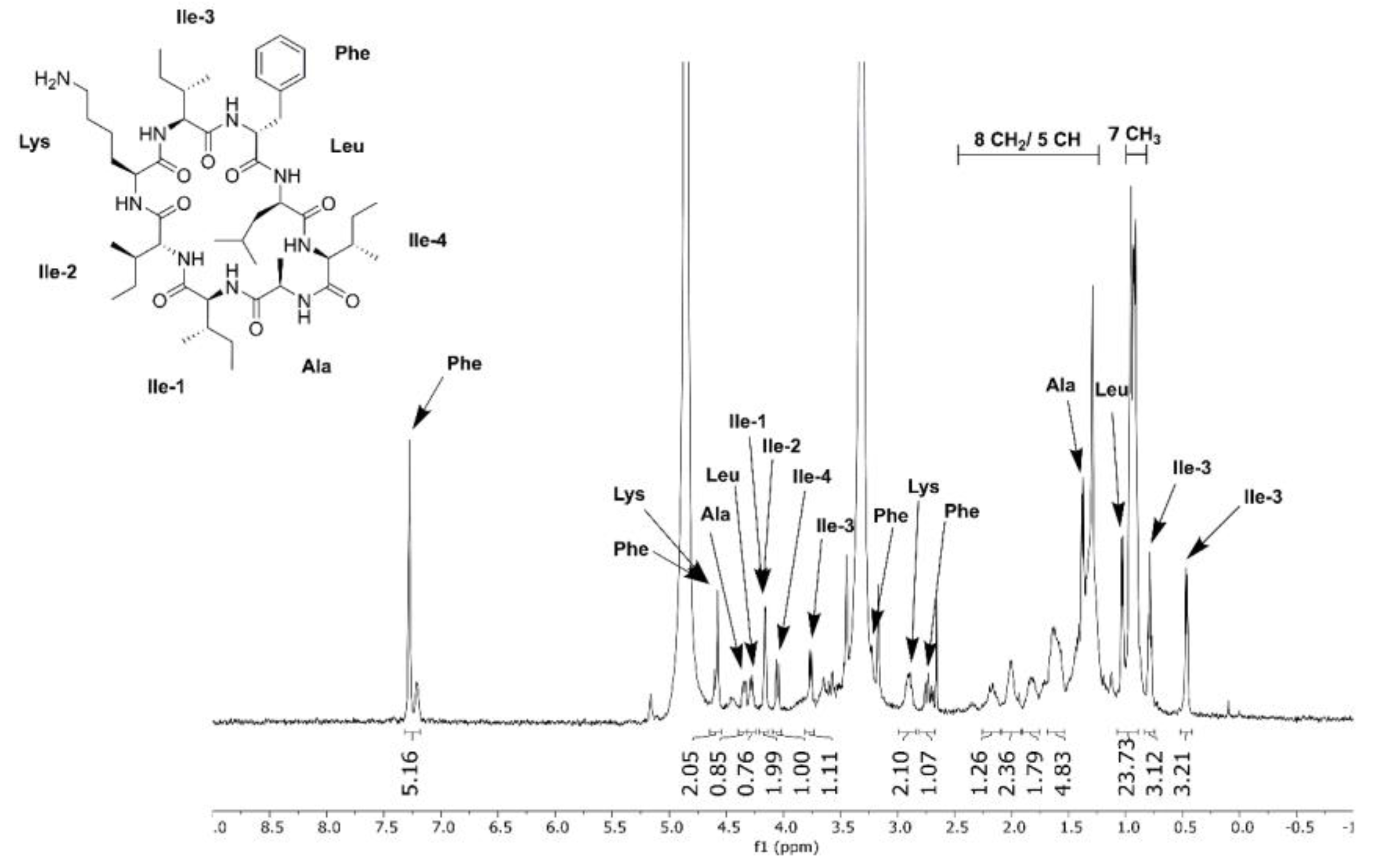
2.7. Metabolic Profiling, Compound Isolation, and Chemical Structure Analysis
3. Results and Discussion
3.1. Multi-locus Sequence Analysis and Taxonomy Assignment of the Streptomyces sp. SM17 Isolate
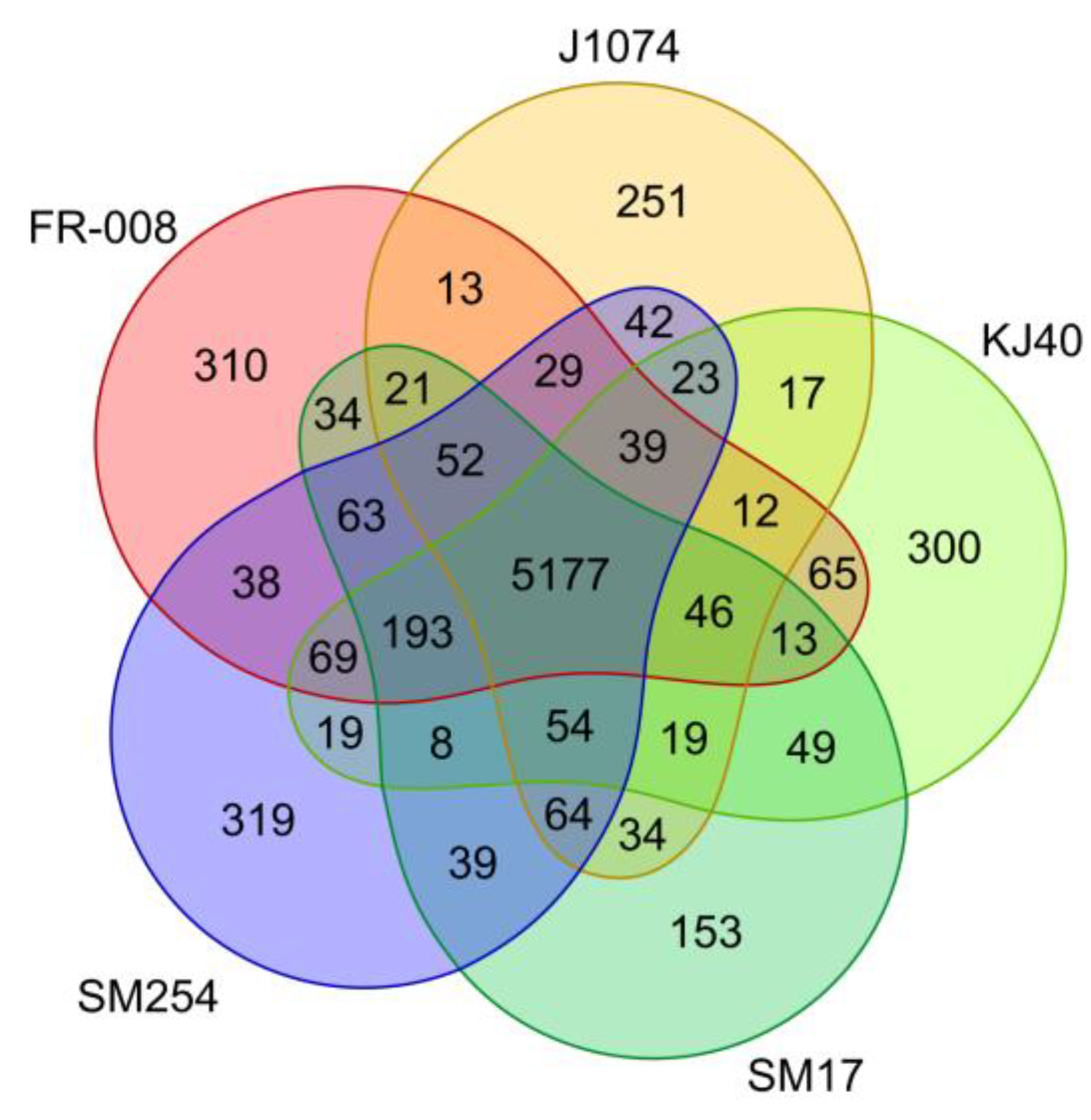
3.2. Analysis of Groups of Orthologous Genes in the Albidoflavus Phylogroup
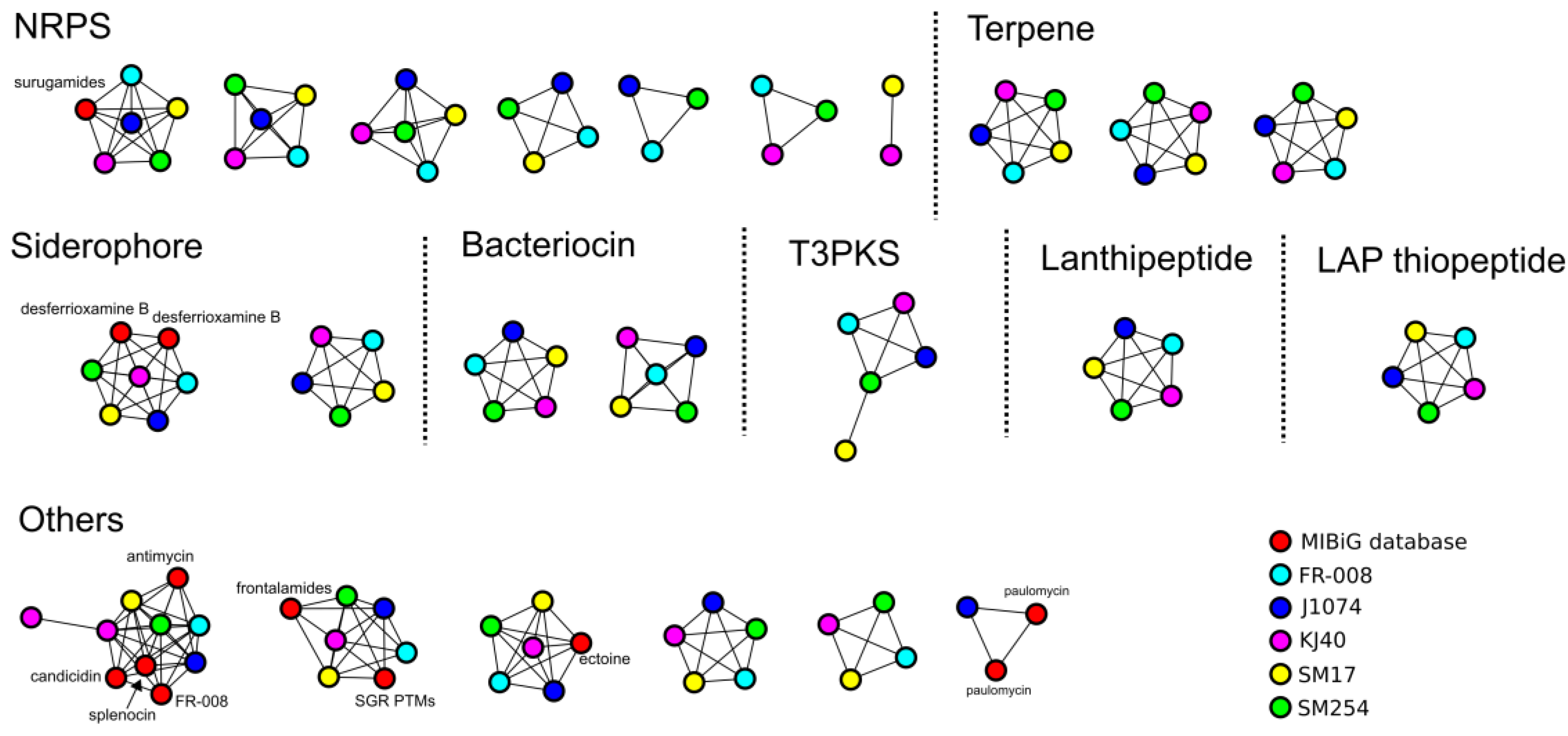
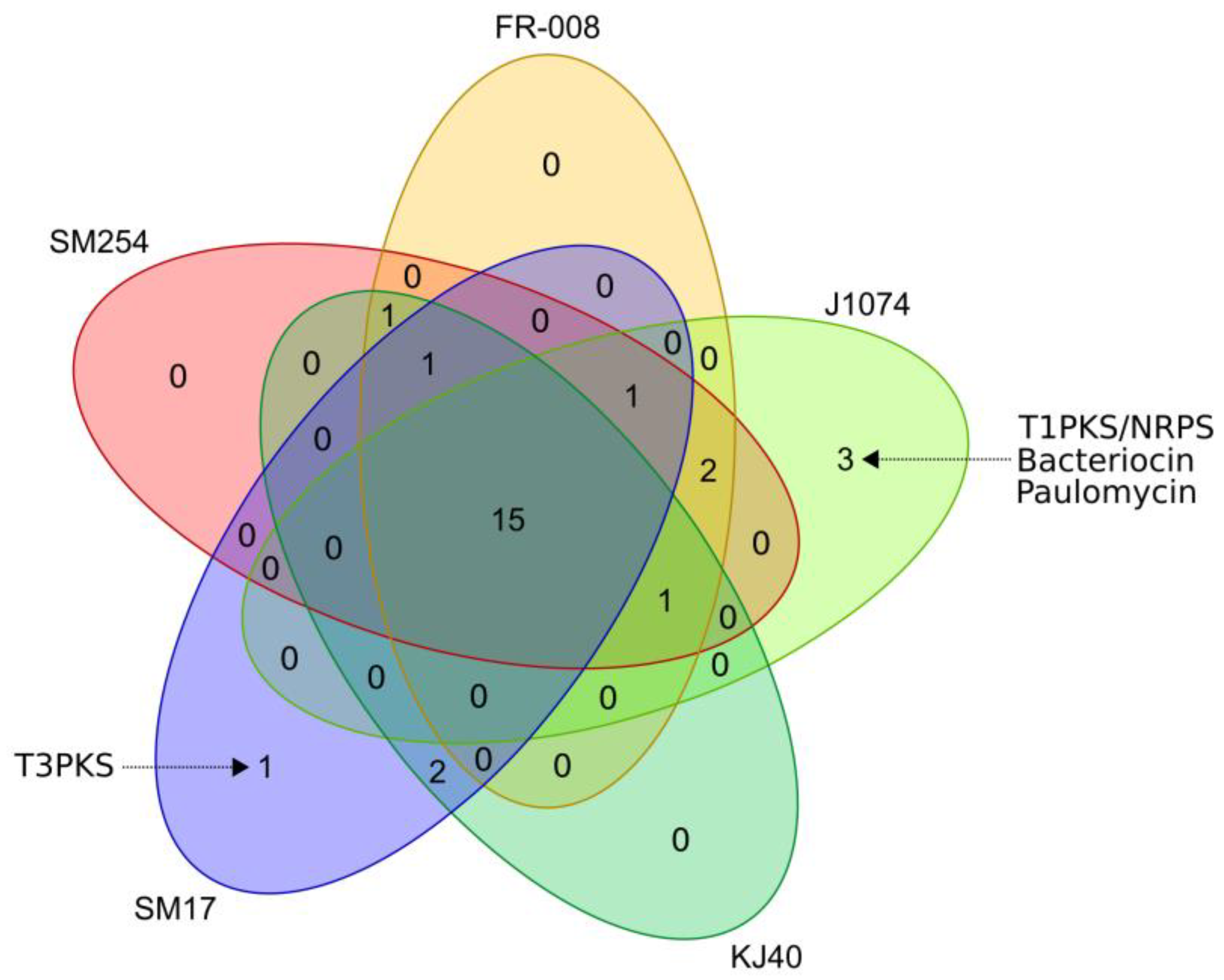
3.3. Prediction of Secondary Metabolites Biosynthetic Gene Clusters in the Albidoflavus Phylogroup
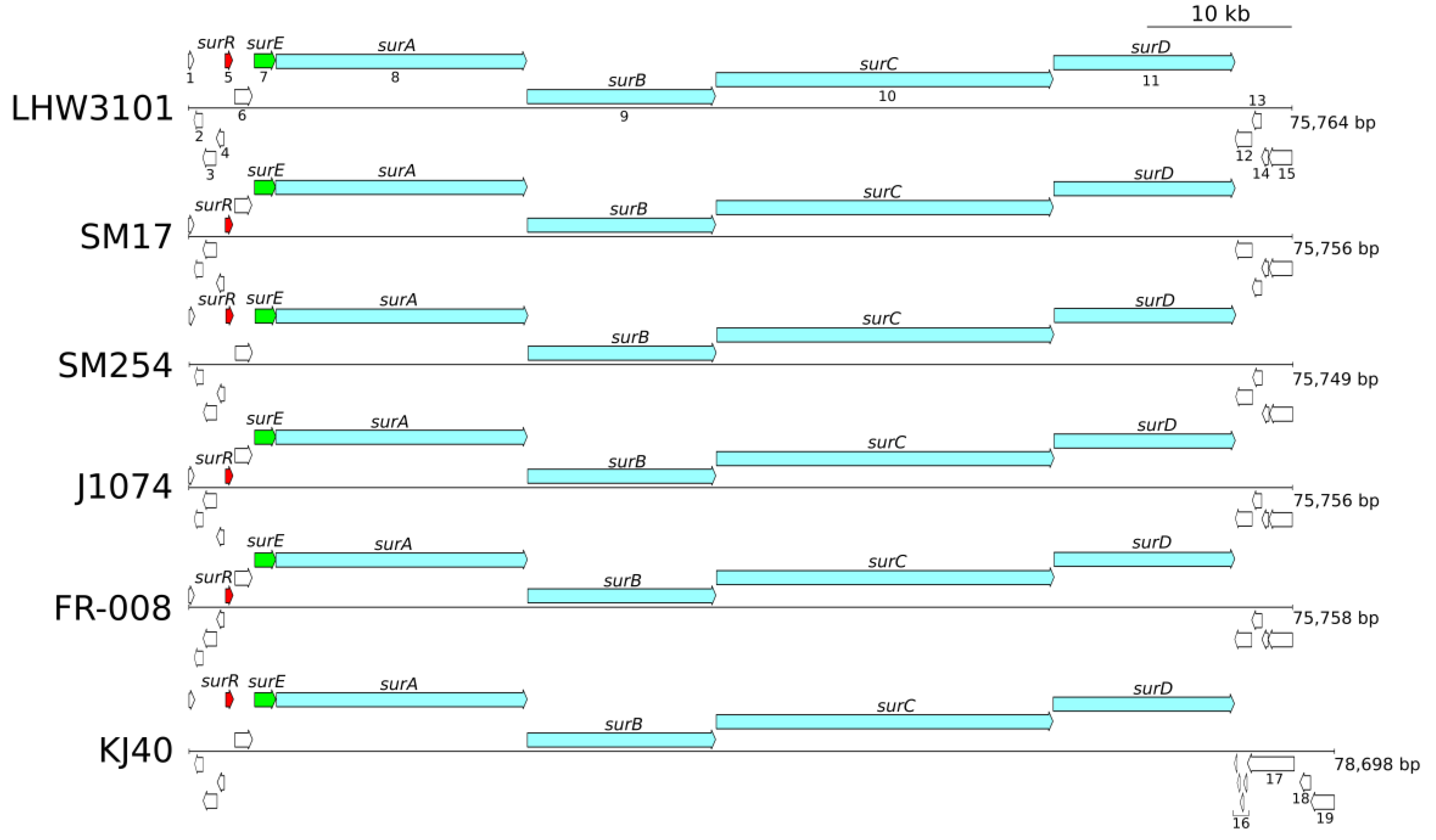
3.4. Phylogeny and Gene Synteny Analysis of Sur BGC Homologs
3.5. Growth, Morphology, Phenotype, and Metabolism Assessment of Streptomyces sp. SM17 in Complex Media
3.6. Differential Production of Surugamide A by Streptomyces sp. SM17 and S. albidoflavus J1074
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hwang, K.-S.; Kim, H.U.; Charusanti, P.; Palsson, B.Ø.; Lee, S.Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014, 32, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Thabit, A.K.; Crandon, J.L.; Nicolau, D.P. Antimicrobial resistance: Impact on clinical and economic outcomes and the need for new antimicrobials. Expert Opin. Pharmacother. 2015, 16, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Demers, D.; Knestrick, M.; Fleeman, R.; Tawfik, R.; Azhari, A.; Souza, A.; Vesely, B.; Netherton, M.; Gupta, R.; Colon, B.; et al. Exploitation of Mangrove Endophytic Fungi for Infectious Disease Drug Discovery. Mar. Drugs 2018, 16, 376. [Google Scholar] [CrossRef] [PubMed]
- Indraningrat, A.; Smidt, H.; Sipkema, D. Bioprospecting Sponge-Associated Microbes for Antimicrobial Compounds. Mar. Drugs 2016, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Calcabrini, C.; Catanzaro, E.; Bishayee, A.; Turrini, E.; Fimognari, C. Marine Sponge Natural Products with Anticancer Potential: An Updated Review. Mar. Drugs 2017, 15, 310. [Google Scholar] [CrossRef]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a Cytotoxic Polyketide from a Streptomyces Strain Isolated from the Marine Sponge Halichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Kunz, A.; Labes, A.; Wiese, J.; Bruhn, T.; Bringmann, G.; Imhoff, J. Nature’s Lab for Derivatization: New and Revised Structures of a Variety of Streptophenazines Produced by a Sponge-Derived Streptomyces Strain. Mar. Drugs 2014, 12, 1699–1714. [Google Scholar] [CrossRef]
- Sathiyanarayanan, G.; Gandhimathi, R.; Sabarathnam, B.; Seghal Kiran, G.; Selvin, J. Optimization and production of pyrrolidone antimicrobial agent from marine sponge-associated Streptomyces sp. MAPS15. Bioprocess Biosyst. Eng. 2014, 37, 561–573. [Google Scholar] [CrossRef]
- Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A.D.W. Draft Genome Sequence of the Antimycin-Producing Bacterium Streptomyces sp. Strain SM8, Isolated from the Marine Sponge Haliclona simulans. Genome Announc. 2018, 6, e01535-17. [Google Scholar] [CrossRef]
- Viegelmann, C.; Margassery, L.; Kennedy, J.; Zhang, T.; O’Brien, C.; O’Gara, F.; Morrissey, J.; Dobson, A.; Edrada-Ebel, R. Metabolomic Profiling and Genomic Study of a Marine Sponge-Associated Streptomyces sp. Mar. Drugs 2014, 12, 3323–3351. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Crossman, L.; Almeida, E.; Margassery, L.; Kennedy, J.; Dobson, A. Diverse and Abundant Secondary Metabolism Biosynthetic Gene Clusters in the Genomes of Marine Sponge Derived Streptomyces spp. Isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Baker, P.; Piper, C.; Cotter, P.D.; Walsh, M.; Mooij, M.J.; Bourke, M.B.; Rea, M.C.; O’Connor, P.M.; Ross, R.P.; et al. Isolation and Analysis of Bacteria with Antimicrobial Activities from the Marine Sponge Haliclona simulans Collected from Irish Waters. Mar. Biotechnol. 2009, 11, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Ninomiya, A.; Naruse, M.; Sun, Y.; Miyazaki, M.; Nogi, Y.; Okada, S.; Matsunaga, S. Surugamides A–E, Cyclic Octapeptides with Four d-Amino Acid Residues, from a Marine Streptomyces sp.: LC–MS-Aided Inspection of Partial Hydrolysates for the Distinction of d- and l-Amino Acid Residues in the Sequence. J. Org. Chem. 2013, 78, 6746–6750. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kuranaga, T.; Sano, A.; Ninomiya, A.; Takada, K.; Wakimoto, T. The Revised Structure of the Cyclic Octapeptide Surugamide A. Chem. Pharm. Bull. 2019, 67, 476–480. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef] [PubMed]
- Thankachan, D.; Fazal, A.; Francis, D.; Song, L.; Webb, M.E.; Seipke, R.F. A trans-Acting Cyclase Offloading Strategy for Nonribosomal Peptide Synthetases. ACS Chem. Biol. 2019, 14, 845–849. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lin, X.; Xu, C.; Shen, Y.; Wang, S.-P.; Liao, H.; Li, L.; Deng, H.; Lin, H.-W. Investigation of Penicillin Binding Protein (PBP)-like Peptide Cyclase and Hydrolase in Surugamide Non-ribosomal Peptide Biosynthesis. Cell Chem. Biol. 2019, 26, 737–744.e4. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, A.; Katsuyama, Y.; Kuranaga, T.; Miyazaki, M.; Nogi, Y.; Okada, S.; Wakimoto, T.; Ohnishi, Y.; Matsunaga, S.; Takada, K. Biosynthetic Gene Cluster for Surugamide A Encompasses an Unrelated Decapeptide, Surugamide F. ChemBioChem 2016, 17, 1709–1712. [Google Scholar] [CrossRef]
- Kuranaga, T.; Matsuda, K.; Sano, A.; Kobayashi, M.; Ninomiya, A.; Takada, K.; Matsunaga, S.; Wakimoto, T. Total Synthesis of the Nonribosomal Peptide Surugamide B and Identification of a New Offloading Cyclase Family. Angew. Chemie Int. Ed. 2018, 57, 9447–9451. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Kobayashi, M.; Kuranaga, T.; Takada, K.; Ikeda, H.; Matsunaga, S.; Wakimoto, T. SurE is a trans-acting thioesterase cyclizing two distinct non-ribosomal peptides. Org. Biomol. Chem. 2019, 17, 1058–1061. [Google Scholar] [CrossRef] [PubMed]
- Koshla, O.T.; Rokytskyy, I.V.; Ostash, I.S.; Busche, T.; Kalinowski, J.; Mösker, E.; Süssmuth, R.D.; Fedorenko, V.O.; Ostash, B.O. Secondary Metabolome and Transcriptome of Streptomyces albus J1074 in Liquid Medium SG2. Cytol. Genet. 2019, 53, 1–7. [Google Scholar] [CrossRef]
- Chater, K.F.; Wilde, L.C. Restriction of a bacteriophage of Streptomyces albus G involving endonuclease SalI. J. Bacteriol. 1976, 128, 644–650. [Google Scholar] [PubMed]
- Chater, K.F.; Wilde, L.C. Streptomyces albus G Mutants Defective in the SalGI Restriction-Modification System. Microbiology 1980, 116, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yang, C.; Zhang, W.; Zhu, Y.; Ma, L.; Fang, Z.; Zhang, C. Albumycin, a new isoindolequinone from Streptomyces albus J1074 harboring the fluostatin biosynthetic gene cluster. J. Antibiot. (Tokyo) 2019, 72, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Zaburannyi, N.; Rabyk, M.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genomics 2014, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A. Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 2018, 49, 316–324. [Google Scholar] [CrossRef]
- Bilyk, O.; Sekurova, O.N.; Zotchev, S.B.; Luzhetskyy, A. Cloning and Heterologous Expression of the Grecocycline Biosynthetic Gene Cluster. PLoS ONE 2016, 11, e0158682. [Google Scholar] [CrossRef]
- Jiang, G.; Zhang, Y.; Powell, M.M.; Zhang, P.; Zuo, R.; Zhang, Y.; Kallifidas, D.; Tieu, A.M.; Luesch, H.; Loria, R.; et al. High-Yield Production of Herbicidal Thaxtomins and Thaxtomin Analogs in a Nonpathogenic Streptomyces Strain. Appl. Environ. Microbiol. 2018, 84, e00164-18. [Google Scholar] [CrossRef]
- Labeda, D.P.; Doroghazi, J.R.; Ju, K.-S.; Metcalf, W.W. Taxonomic evaluation of Streptomyces albus and related species using multilocus sequence analysis and proposals to emend the description of Streptomyces albus and describe Streptomyces pathocidini sp. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 894–900. [Google Scholar] [CrossRef]
- Labeda, D.P.; Dunlap, C.A.; Rong, X.; Huang, Y.; Doroghazi, J.R.; Ju, K.-S.; Metcalf, W.W. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek 2017, 110, 563–583. [Google Scholar] [CrossRef] [PubMed]
- Almeida, E.L.; Carillo Rincón, A.F.; Jackson, S.A.; Dobson, A.D. Comparative genomics of marine sponge-derived Streptomyces spp. isolates SM17 and SM18 with their closest terrestrial relatives provides novel insights into environmental niche adaptations and secondary metabolite biosynthesis potential. Front. Microbiol. 2019, 10, 1713. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Waddell, P.J.; Steel, M. General Time-Reversible Distances with Unequal Rates across Sites: Mixing Γ and Inverse Gaussian Distributions with Invariant Sites. Mol. Phylogenet. Evol. 1997, 8, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Muñoz, J.C.; Selem-Mojica, N.; Mullowney, M.W.; Kautsar, S.; Tryon, J.H.; Parkinson, E.I.; Santos, E.L.C.D.L.; Yeong, M.; Cruz-Morales, P.; Abubucker, S.; et al. A computational framework for systematic exploration of biosynthetic diversity from large-scale genomic data. bioRxiv 2018, 10, 445270. [Google Scholar]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.-A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Dusa, A. Venn: Draw Venn Diagrams 2018. Available online: https://cran.r-project.org/web/packages/venn/index.html (accessed on 25 September 2019).
- R Core Team R: A Language and Environment for Statistical Computing, version 3.5.3; Vienna, Austria, 2018. Available online: https://www.R-project.org/ (accessed on 25 September 2019).
- RStudio Team. RStudio Team RStudio: Integrated Development Environment for RStudio Inc.; RStudio Inc.: Boston, MA, USA, 2015; Volume 14. [Google Scholar]
- Glaeser, S.P.; Kämpfer, P. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst. Appl. Microbiol. 2015, 38, 237–245. [Google Scholar] [CrossRef]
- Gadagkar, S.R.; Rosenberg, M.S.; Kumar, S. Inferring species phylogenies from multiple genes: Concatenated sequence tree versus consensus gene tree. J. Exp. Zool. Part B Mol. Dev. Evol. 2005, 304B, 64–74. [Google Scholar] [CrossRef]
- Li, S.; Zhang, B.; Zhu, H.; Zhu, T. Cloning and Expression of the Chitinase Encoded by ChiKJ406136 from Streptomyces sampsonii (Millard & Burr) Waksman KJ40 and Its Antifungal Effect. Forests 2018, 9, 699. [Google Scholar]
- Liu, Q.; Xiao, L.; Zhou, Y.; Deng, K.; Tan, G.; Han, Y.; Liu, X.; Deng, Z.; Liu, T. Development of Streptomyces sp. FR-008 as an emerging chassis. Synth. Syst. Biotechnol. 2016, 1, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Badalamenti, J.P.; Erickson, J.D.; Salomon, C.E. Complete Genome Sequence of Streptomyces albus SM254, a Potent Antagonist of Bat White-Nose Syndrome Pathogen Pseudogymnoascus destructans. Genome Announc. 2016, 4, e00290-16. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Doroghazi, J.R.; Cheng, K.; Zhang, L.; Buckley, D.H.; Huang, Y. Classification of Streptomyces phylogroup pratensis (Doroghazi and Buckley, 2010) based on genetic and phenotypic evidence, and proposal of Streptomyces pratensis sp. nov. Syst. Appl. Microbiol. 2013, 36, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Allenby, N. Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 2018, 365, fny240. [Google Scholar] [CrossRef]
- Li, Y.; Pinto-Tomás, A.A.; Rong, X.; Cheng, K.; Liu, M.; Huang, Y. Population Genomics Insights into Adaptive Evolution and Ecological Differentiation in Streptomycetes. Appl. Environ. Microbiol. 2019, 85, e02555-18. [Google Scholar] [CrossRef]
- Hoz, J.F.-D.L.; Méndez, C.; Salas, J.A.; Olano, C. Novel Bioactive Paulomycin Derivatives Produced by Streptomyces albus J1074. Molecules 2017, 22, 1758. [Google Scholar] [CrossRef]
- Chen, S.; Huang, X.; Zhou, X.; Bai, L.; He, J.; Jeong, K.J.; Lee, S.Y.; Deng, Z. Organizational and Mutational Analysis of a Complete FR-008/Candicidin Gene Cluster Encoding a Structurally Related Polyene Complex. Chem. Biol. 2003, 10, 1065–1076. [Google Scholar] [CrossRef]
- Zhao, Z.; Deng, Z.; Pang, X.; Chen, X.-L.; Zhang, P.; Bai, L.; Li, H. Production of the antibiotic FR-008/candicidin in Streptomyces sp. FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 2015, 161, 539–552. [Google Scholar]
- Hamm, P.S.; Caimi, N.A.; Northup, D.E.; Valdez, E.W.; Buecher, D.C.; Dunlap, C.A.; Labeda, D.P.; Lueschow, S.; Porras-Alfaro, A. Western Bats as a Reservoir of Novel Streptomyces Species with Antifungal Activity. Appl. Environ. Microbiol. 2017, 83, e03057-16. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, Z.; Wang, M.; Ai, G.; Chen, Y. Identification and Analysis of the Paulomycin Biosynthetic Gene Cluster and Titer Improvement of the Paulomycins in Streptomyces paulus NRRL 8115. PLoS ONE 2015, 10, e0120542. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Quadri, L.E.; Sello, J.; Keating, T.A.; Weinreb, P.H.; Walsh, C.T. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 1998, 5, 631–645. [Google Scholar] [CrossRef]
- Lautru, S.; Oves-Costales, D.; Pernodet, J.-L.; Challis, G.L. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 2007, 153, 1405–1412. [Google Scholar] [CrossRef][Green Version]
- Glavinas, H.; Krajcsi, P.; Cserepes, J.; Sarkadi, B. The role of ABC transporters in drug resistance, metabolism and toxicity. Curr. Drug Deliv. 2004, 1, 27–42. [Google Scholar] [CrossRef]
- Polgar, O.; Bates, S.E. ABC transporters in the balance: Is there a role in multidrug resistance? Biochem. Soc. Trans. 2005, 33, 241–245. [Google Scholar] [CrossRef]
- Podlesek, Z.; Comino, A.; Herzog-Velikonja, B.; Zgur-Bertok, D.; Komel, R.; Grabnar, M. Bacillus licheniformis bacitracin-resistance ABC transporter: Relationship to mammalian multidrug resistance. Mol. Microbiol. 1995, 16, 969–976. [Google Scholar] [CrossRef]
- Ohki, R.; Tateno, K.; Okada, Y.; Okajima, H.; Asai, K.; Sadaie, Y.; Murata, M.; Aiso, T. A Bacitracin-Resistant Bacillus subtilis Gene Encodes a Homologue of the Membrane-Spanning Subunit of the Bacillus licheniformis ABC Transporter. J. Bacteriol. 2003, 185, 51–59. [Google Scholar] [CrossRef]
- Kumar, S.; He, G.; Kakarla, P.; Shrestha, U.; Ranjana, K.C.; Ranaweera, I.; Willmon, T.M.; Barr, S.R.; Hernandez, A.J.; Varela, M.F. Bacterial Multidrug Efflux Pumps of the Major Facilitator Superfamily as Targets for Modulation. Infect. Disord. Drug Targets 2016, 16, 28–43. [Google Scholar] [CrossRef]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Nodwell, J.R. The TetR Family of Regulators. Microbiol. Mol. Biol. Rev. 2013, 77, 440–475. [Google Scholar] [CrossRef] [PubMed]
- Koskiniemi, S.; Lamoureux, J.G.; Nikolakakis, K.C.; de Roodenbeke, C.T.K.; Kaplan, M.D.; Low, D.A.; Hayes, C.S. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. USA 2013, 110, 7032–7037. [Google Scholar] [CrossRef] [PubMed]
- Manteca, Á.; Yagüe, P. Streptomyces as a Source of Antimicrobials: Novel Approaches to Activate Cryptic Secondary Metabolite Pathways. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Manteca, A.; Alvarez, R.; Salazar, N.; Yague, P.; Sanchez, J. Mycelium Differentiation and Antibiotic Production in Submerged Cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 2008, 74, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Stülke, J.; Hillen, W. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1999, 2, 195–201. [Google Scholar] [CrossRef]
- Kremling, A.; Geiselmann, J.; Ropers, D.; de Jong, H. Understanding carbon catabolite repression in Escherichia coli using quantitative models. Trends Microbiol. 2015, 23, 99–109. [Google Scholar] [CrossRef]
- Deutscher, J. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2008, 11, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R.; Titgemeyer, F. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002, 209, 141–148. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Ruiz-Villafán, B.; Tierrafría, V.H.; Rodríguez-Sanoja, R.; Sánchez, S. Carbon Catabolite Regulation of Secondary Metabolite Formation and Morphological Differentiation in Streptomyces coelicolor. Appl. Biochem. Biotechnol. 2016, 180, 1152–1166. [Google Scholar] [CrossRef]
- Inoue, O.O.; Schmidell Netto, W.; Padilla, G.; Facciotti, M.C.R. Carbon catabolite repression of retamycin production by Streptomyces olindensis ICB20. Braz. J. Microbiol. 2007, 38, 58–61. [Google Scholar] [CrossRef]
- Gallo, M.; Katz, E. Regulation of secondary metabolite biosynthesis: Catabolite repression of phenoxazinone synthase and actinomycin formation by glucose. J. Bacteriol. 1972, 109, 659–667. [Google Scholar] [PubMed]
- Magnus, N.; Weise, T.; Piechulla, B. Carbon Catabolite Repression Regulates the Production of the Unique Volatile Sodorifen of Serratia plymuthica 4Rx13. Front. Microbiol. 2017, 8, 2522. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Hewavitharana, A.; Ng, Y.; Shaw, P.; Fuerst, J.; Hodson, M. LC-MS-Based Metabolomics Study of Marine Bacterial Secondary Metabolite and Antibiotic Production in Salinispora arenicola. Mar. Drugs 2015, 13, 249–266. [Google Scholar] [CrossRef] [PubMed]

| Strain | Media | Percent (w/w) of Extract | Concentration of Surugamide A (mg/L) Corrected in 5 mg/mL of Extract |
|---|---|---|---|
| SM17 | TSB | 2.44% | 122.01 |
| SM17 | SYP-NaCl | 10.60% | 530.15 |
| SM17 | YD | 1.13% | 56.27 |
| J1074 | TSB | 0.27% | 13.32 |
| J1074 | SYP-NaCl | 3.55% | 176.82 |
| J1074 | YD | 0.09% | 4.26 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, E.L.; Kaur, N.; Jennings, L.K.; Carrillo Rincón, A.F.; Jackson, S.A.; Thomas, O.P.; Dobson, A.D.W. Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074. Microorganisms 2019, 7, 394. https://doi.org/10.3390/microorganisms7100394
Almeida EL, Kaur N, Jennings LK, Carrillo Rincón AF, Jackson SA, Thomas OP, Dobson ADW. Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074. Microorganisms. 2019; 7(10):394. https://doi.org/10.3390/microorganisms7100394
Chicago/Turabian StyleAlmeida, Eduardo L., Navdeep Kaur, Laurence K. Jennings, Andrés Felipe Carrillo Rincón, Stephen A. Jackson, Olivier P. Thomas, and Alan D.W. Dobson. 2019. "Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074" Microorganisms 7, no. 10: 394. https://doi.org/10.3390/microorganisms7100394
APA StyleAlmeida, E. L., Kaur, N., Jennings, L. K., Carrillo Rincón, A. F., Jackson, S. A., Thomas, O. P., & Dobson, A. D. W. (2019). Genome Mining Coupled with OSMAC-Based Cultivation Reveal Differential Production of Surugamide A by the Marine Sponge Isolate Streptomyces sp. SM17 When Compared to Its Terrestrial Relative S. albidoflavus J1074. Microorganisms, 7(10), 394. https://doi.org/10.3390/microorganisms7100394







