Role of SCFAs for Fimbrillin-Dependent Biofilm Formation of Actinomyces oris
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Human Saliva Collection
2.3. Biofilm Formation Assay in 96-Well and 6-Well Plates
2.4. Flow Cell Biofilm Formation Assay
2.5. Statistical Analysis
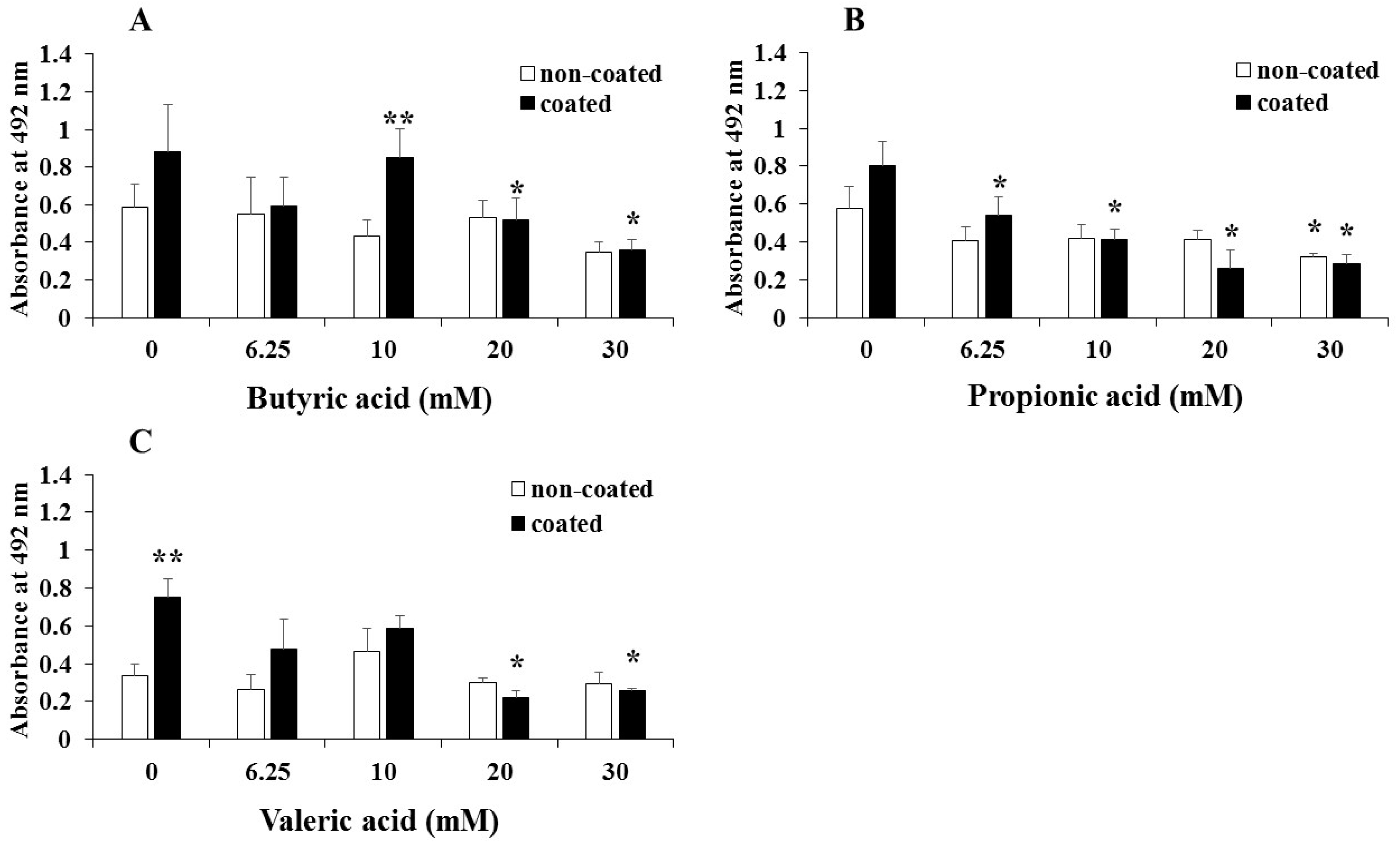
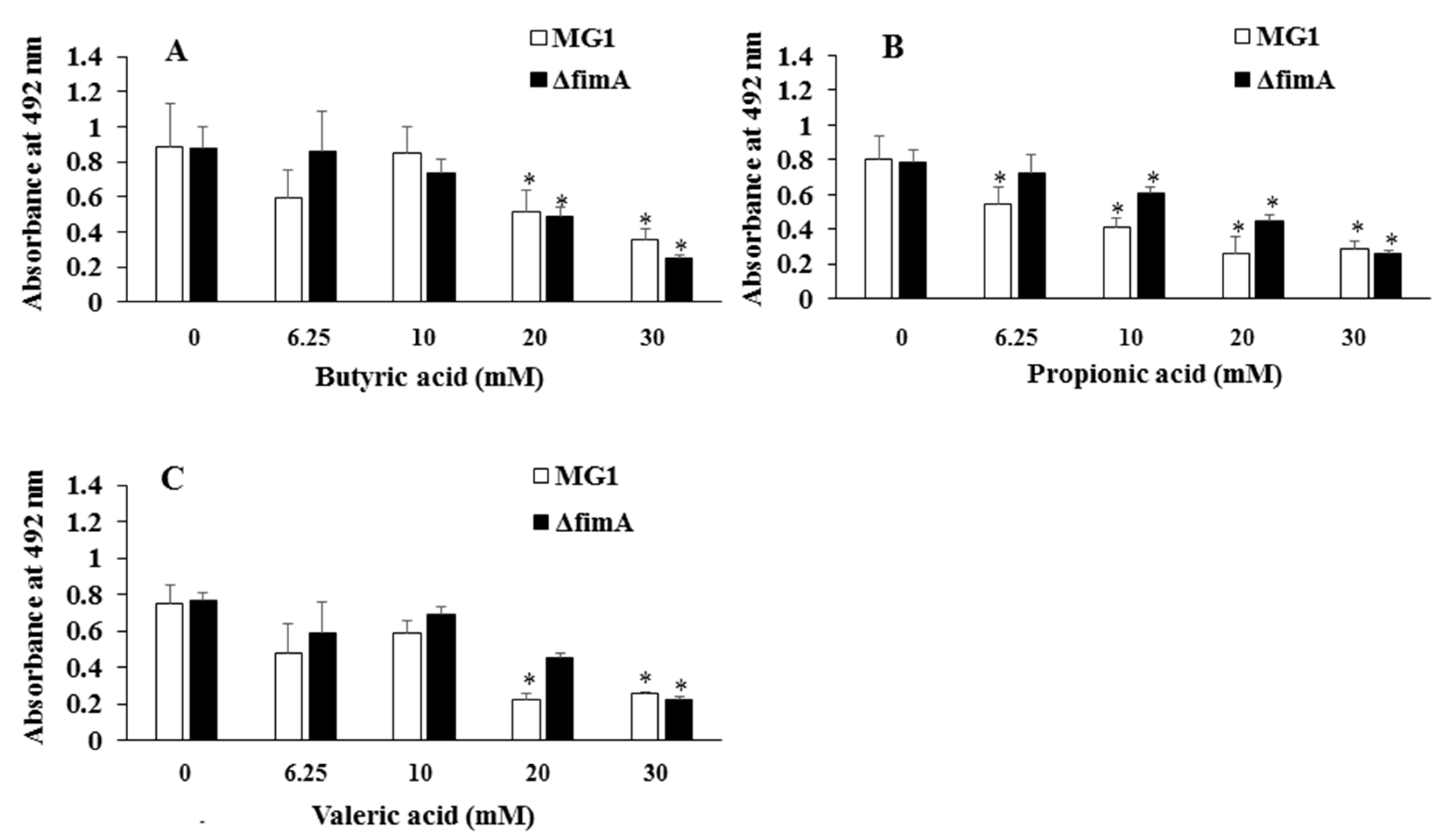
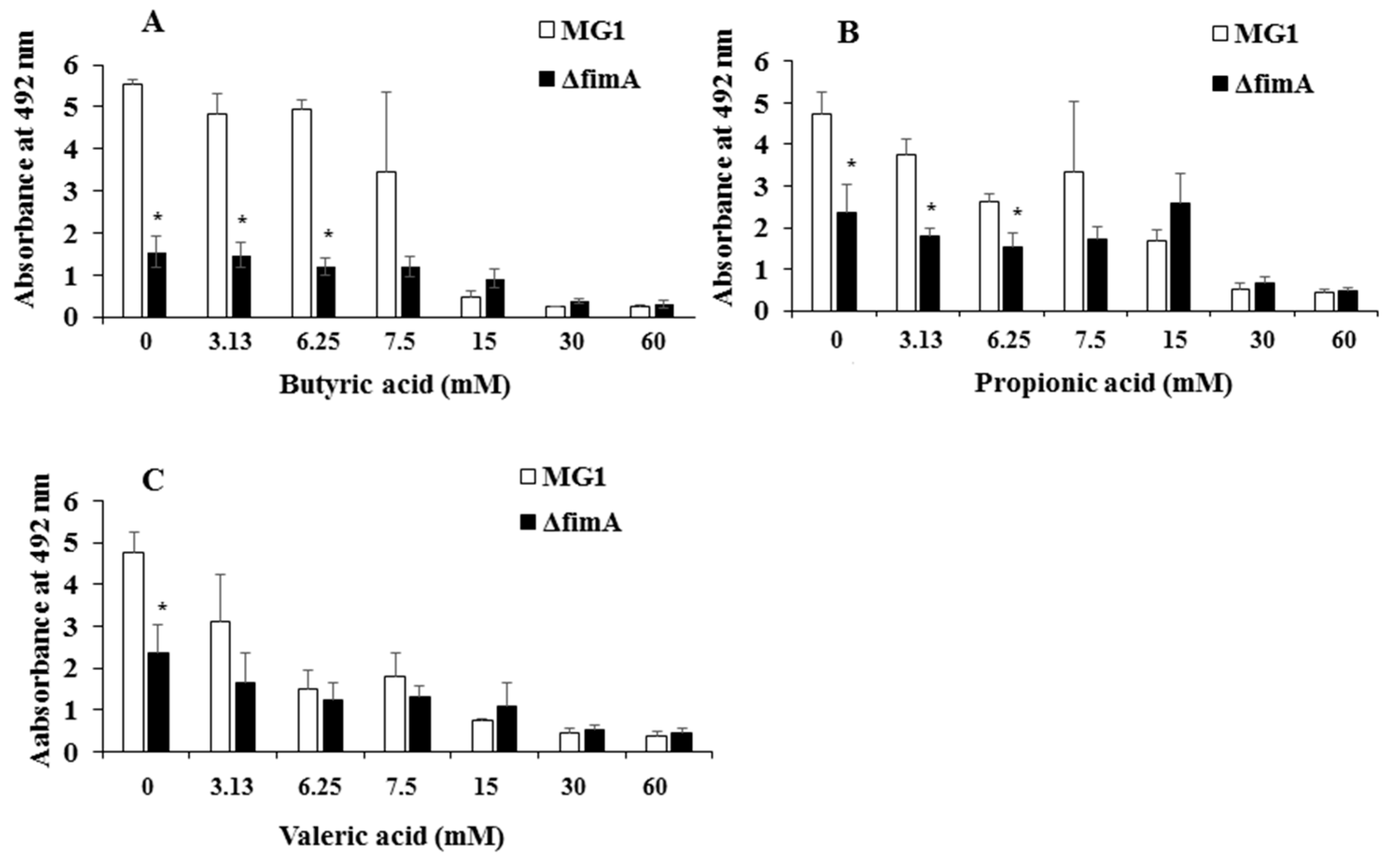
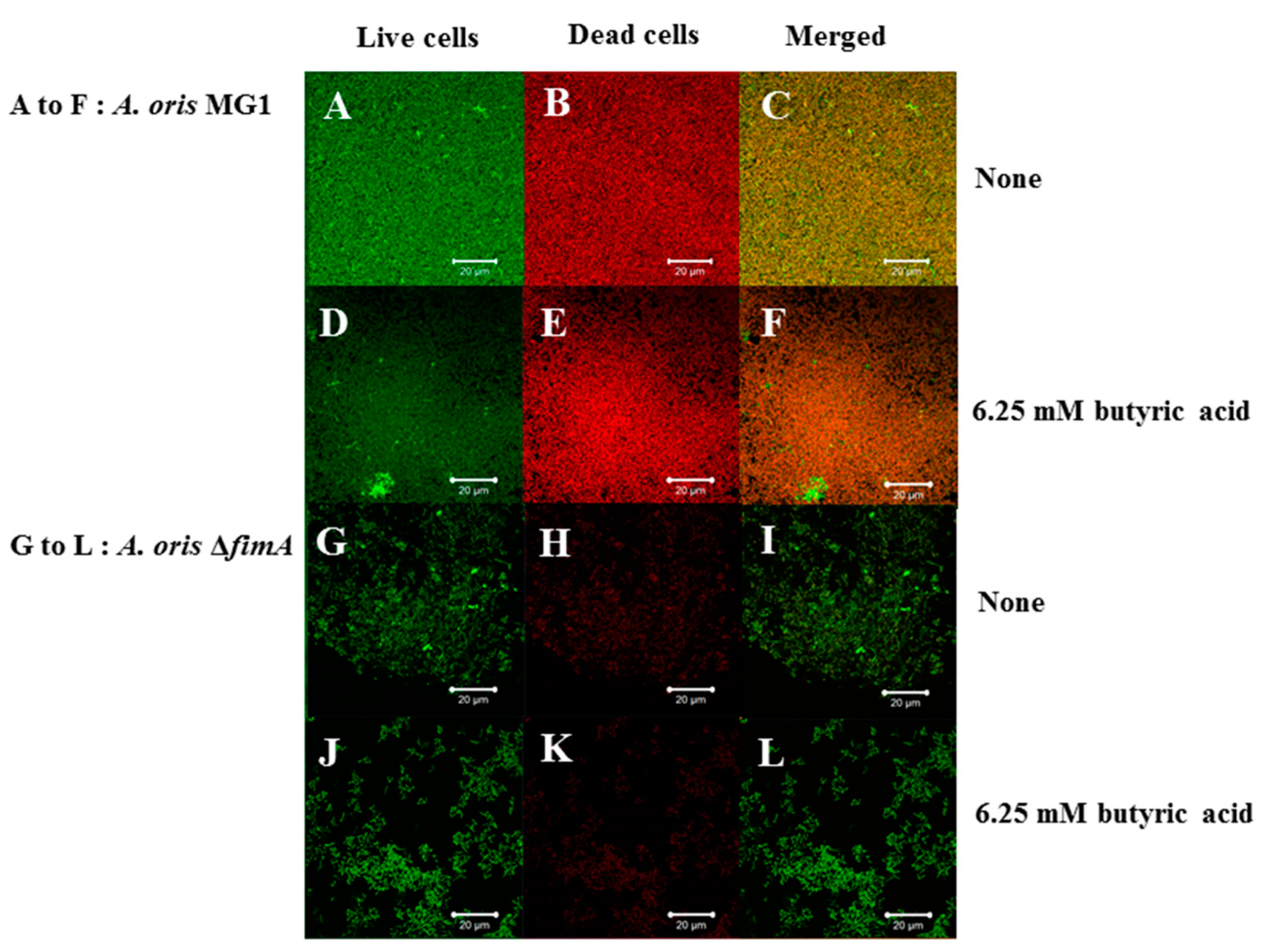
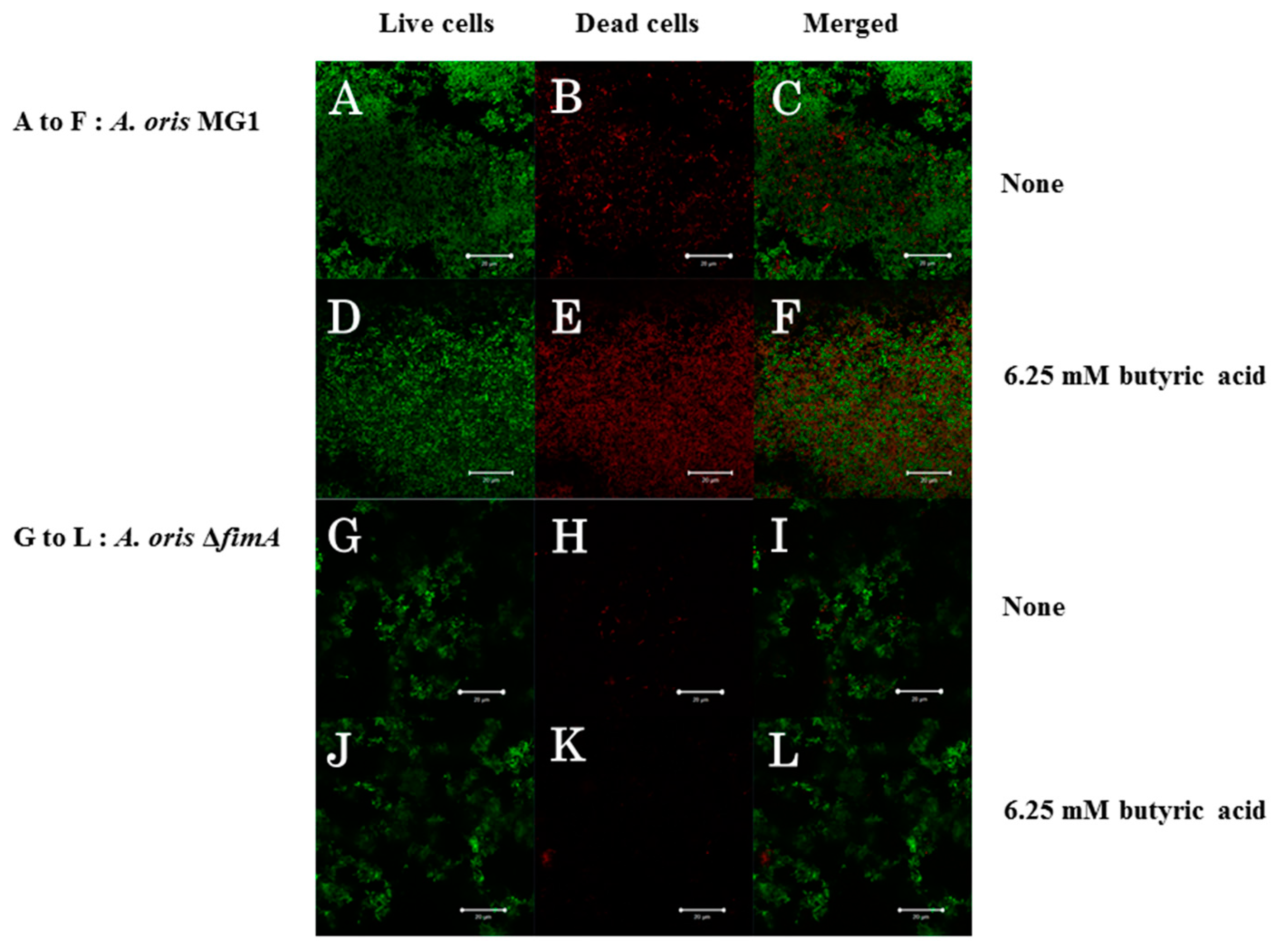
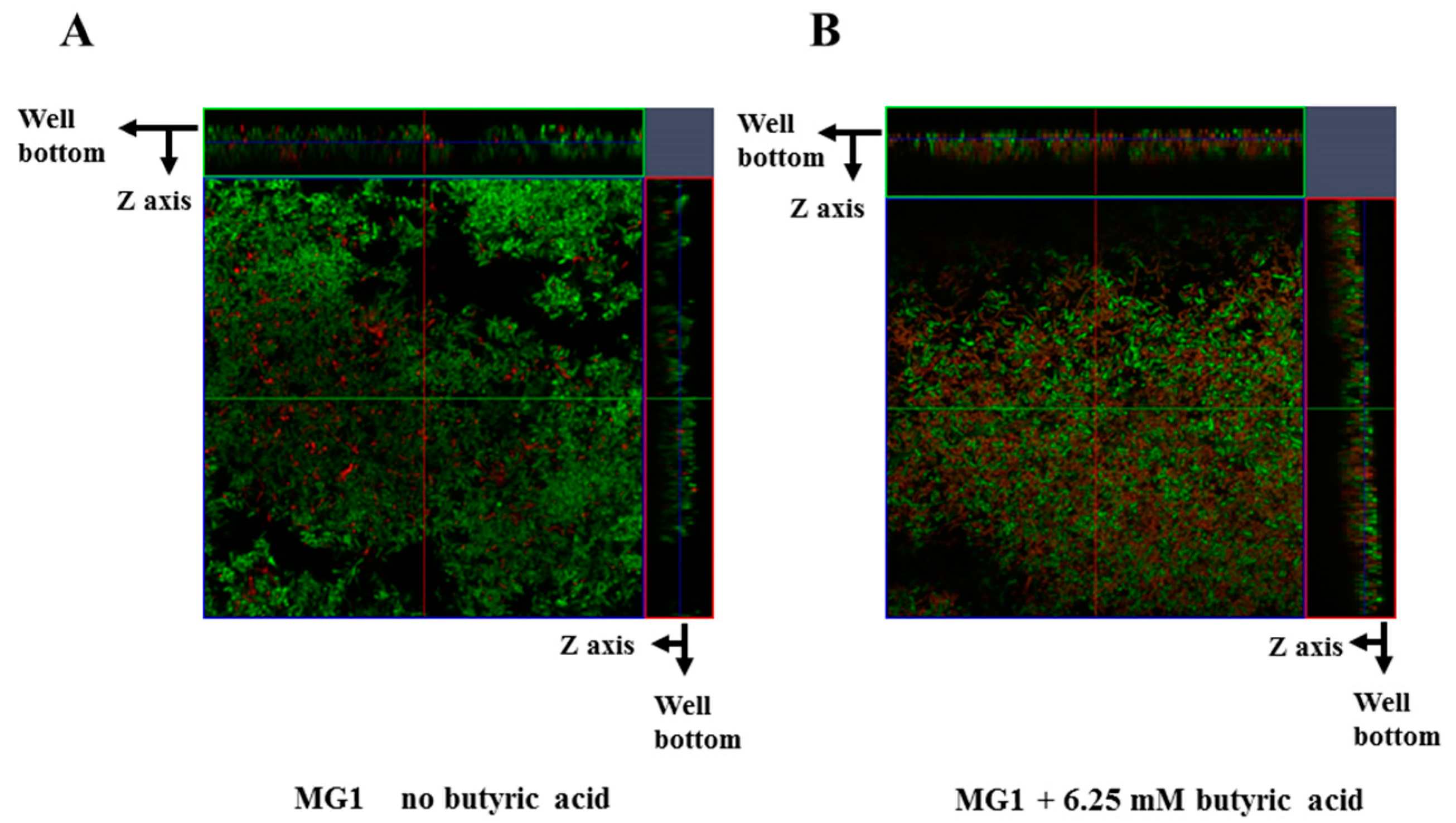
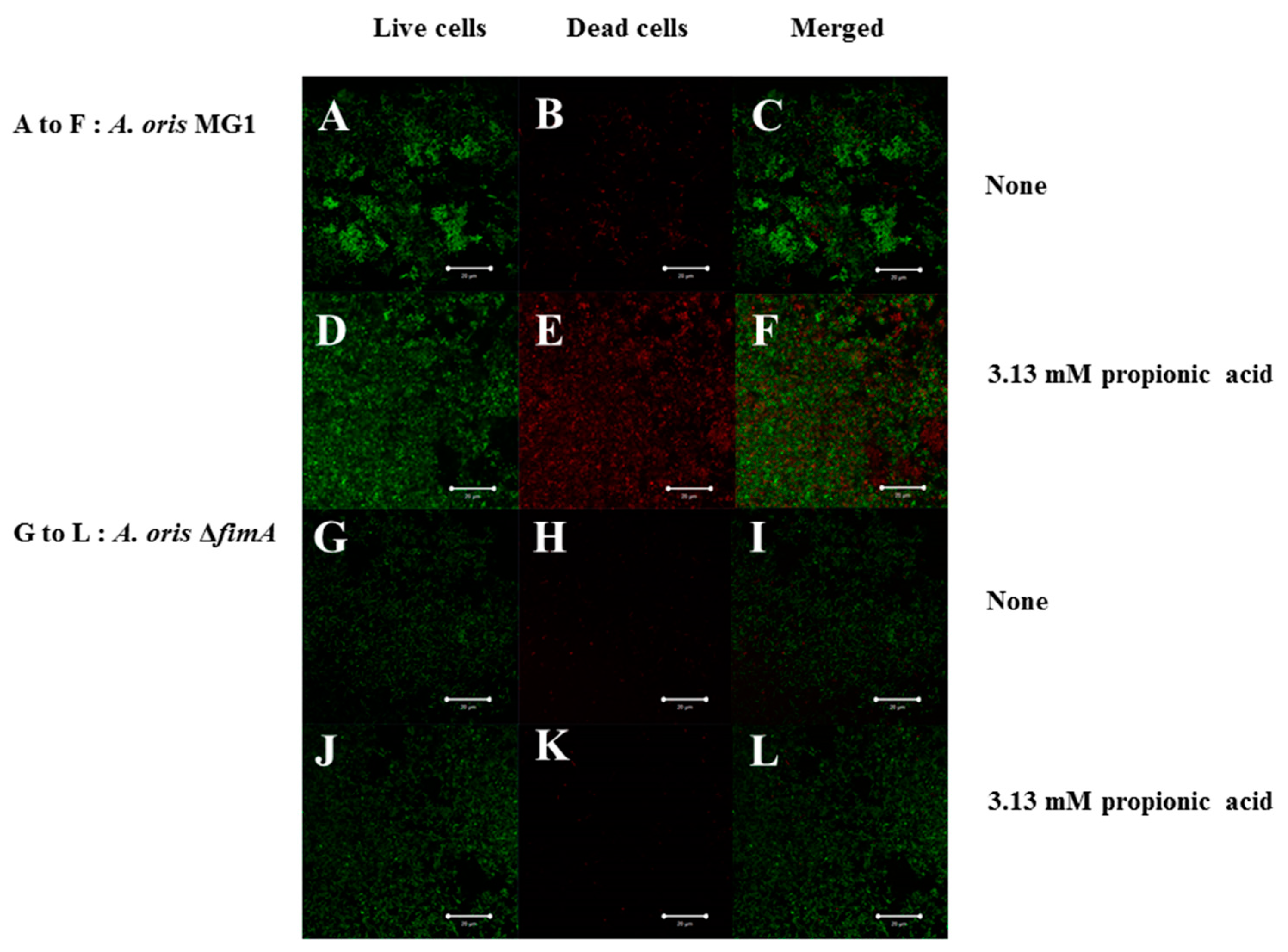
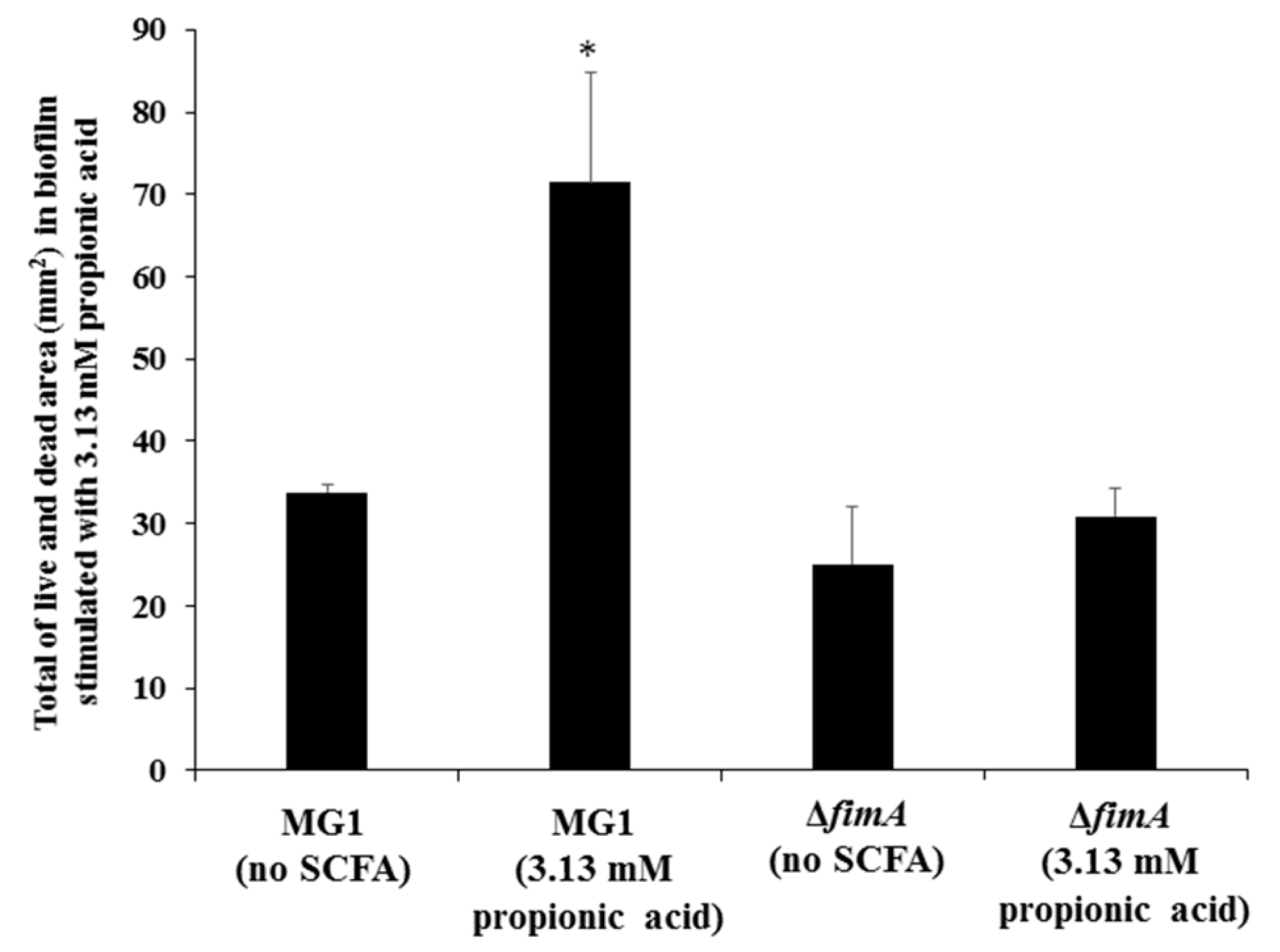
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Periasamy, S.; Jakubovics, N.S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 2010, 8, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Nyvad, B.; Kilian, M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 1987, 95, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.; Wunder, A.; Auschill, T.M.; Follo, M.; Braun, G.; Hellwig, E.; Arweiler, N.B. The in vivo dynamics of Streptococcus spp., Actinomyces naeslundii, Fusobacterium nucleatum and Veillonella spp. in dental plaque biofilm as analysed by five-colour multiplex fluorescence in situ hybridization. J. Med. Microbiol. 2007, 56, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Do, T.; Henssge, U.; Gilbert, S.C.; Clark, D.; Beighton, D. Evidence for recombination between a sialidase (nanH) of Actinomyces naeslundii and Actinomyces oris, previously named ‘Actinomyces naeslundii genospecies 1 and 2’. FEMS Microbiol. Lett. 2008, 288, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Henssge, U.; Do, T.; Gilbert, S.C.; Cox, S.; Clark, D.; Wickstom, C.; Ligtenberg, A.J.M.; Radford, D.R.; Beighton, D. Application of MLST and pilus gene sequence comparisons to investigate the population structures of Actinomyces naeslundii and Actinomyces oris. PLoS ONE 2009, 6, 21430–21441. [Google Scholar] [CrossRef] [PubMed]
- Paddick, J.S.; Brailsford, S.R.; Kidd, E.A.; Gilbert, S.C.; Clark, D.T.; Alam, S.; Killick, Z.J.; Beighton, D. Effect of the environment on genotypic diversity of Actinomyces naeslundii and Streptococcus oralis in the oral biofilm. Appl. Environ. Microbiol. 2003, 69, 6475–6480. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.K.; Ragsdale, P.A. Synthesis and function of Actinomyces naeslundii T14V Type 1 fimbriae require the expression of additional fimbria-associated genes. Infect. Immun. 1997, 65, 2629–2639. [Google Scholar] [PubMed]
- Arai, T.; Ochiai, K.; Senpuku, H. Actinomyces naeslundii GroEL-dependent initial attachment and biofilm formation in a flow cell system. J. Microbiol. Methods 2015, 109, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Burne, R.A. Oral streptococci ... products of their environment. J. Dent. Res. 1998, 77, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Desaymard, C.; Ivanyi, L. Comparison of in vitro immunogenicity, tolerogenicity and mitogenicity of dinitrophenyl-levan conjugates with varying epitope density. Immunology 1976, 30, 647–653. [Google Scholar] [PubMed]
- Shilo, M.; Wolman, B. Activities of bacterial levans and of lipopolysaccharides in the processes of inflammation and infection. Br. J. Exp. Pathol. 1958, 39, 652–660. [Google Scholar] [PubMed]
- Mishra, A.; Das, A.; Cisar, J.O.; Ton-That, H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J. Bacteriol. 2007, 189, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Mishra, A.; Yang, J.; Cisar, J.O.; Das, A.; Ton-That, H. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J. Bacteriol. 2011, 193, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Wu, C.; Yang, J.; Cisar, J.O.; Das, A.; Ton-That, H. The type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol. Microbiol. 2010, 77, 841–854. [Google Scholar] [PubMed]
- Cisar, J.O.; Curl, S.H.; Kolenbrander, P.E.; Vatter, A.E. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosus T14V. Infect. Immun. 1983, 40, 759–765. [Google Scholar] [PubMed]
- McIntire, F.C.; Crosby, L.K.; Vatter, A.E.; Cisar, J.O.; McNeil, M.R.; Bush, C.A.; Tjoa, S.S.; Fennessey, P.V. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 1988, 170, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Niederman, R.; Buyle-Bodin, Y.; Lu, B.Y.; Robinson, P.; Naleway, C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J. Dent. Res. 1997, 76, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Bendyk, A.; Marino, V.; Zilm, P.S.; Howe, P.; Bafold, P.M. Effect of dietary omega-3 polyunsaturated fatty acids on experimental periodontitis in the mouse. J. Periodontal. Res. 2009, 44, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Geum-Sun, A. Correlation of microorganisms and carboxylic acid in oral cavity. Int. J. Clin. Prev. Dent. 2015, 11, 165–170. [Google Scholar] [CrossRef]
- Margolis, H.C.; Duckworth, J.H.; Moreno, E.C. Composition and buffer capacity of pooled starved plaque fluid from caries-free and caries-susceptible individuals. J. Dent. Res. 1988, 67, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Yoneda, S.; Kawarai, T.; Narisawa, N.; Tuna, E.B.; Sato, N.; Tsugane, T.; Saeki, Y.; Ochiai, K.; Senpuku, H. Effects of short-chain fatty acids on Actinomyces naeslundii biofilm formation. Mol. Oral Microbiol. 2013, 28, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Motegi, M.; Takagi, Y.; Yonezawa, H.; Hanada, N.; Terajima, J.; Watanabe, H.; Senpuku, H. Assessment of genes associated with Streptococcus mutans biofilm morphology. Appl. Environ. Microbiol. 2016, 72, 6277–6287. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, R.J.; Hay, D.I.; Cisar, J.O.; Clark, W.B. Adsorbed salivary proline-rich protein 1 and statherin: Receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect. Immun. 1988, 56, 2990–2993. [Google Scholar] [PubMed]
- Li, T.; Khah, M.K.; Slavnic, S.; Johansson, I.; Strömberg, N. Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect. Immun. 2001, 69, 7224–7233. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, S.; Sandberg, A.L.; Cisar, J.O. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral Actinomyces and Streptococci. J. Dent. Res. 2004, 83, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.J., Jr.; Gordon, S.M.; Cisar, J.O.; Kolenbrander, P.E. Coaggregation-mediated interactions of Streptococci and Actinomyces detected in initial human dental plaque. J. Bacteriol. 2003, 185, 3400–3409. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, L.J.; Burne, R.A. Roles of fructosyltransferase and levanase-sucrase of Actinomyces naeslundii in fructan and sucrose metabolism. Infect. Immun. 2001, 69, 5395–5402. [Google Scholar] [CrossRef] [PubMed]
- Thurnheer, T.; Guggenheim, B.; Gmür, R. Characterization of monoclonal antibodies for rapid identification of Actinomyces naeslundii in clinical samples. FEMS Microbiol. Lett. 1997, 150, 255–262. [Google Scholar] [CrossRef]
- Bowden, G.H.; Nolette, N.; Ryding, H.; Cleghorn, B.M. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J. Dent. Res. 1999, 78, 1800–1809. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sharma, P.K.; Busscher, H.J.; van der Mei, H.C.; Krom, B.P. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 2010, 76, 3405–3408. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, K.; Hammarström, K.-J.; Falsen, E.; Dahlen, G.; Gibbons, R.; Hay, D.I.; Strömberg, N. Actinomyces naeslundii genospecies 1 and 2 express different binding specificities to N-acetyl-β-d-galactosamine, whereas Actinomyces odontolyticus expresses a different binding specificity in colonizing the human mouth. Oral Microbiol. Immunol. 1998, 13, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, K.; Holm, C.; Öhman, U.; Strömberg, N. Actinomyces naeslundii displays variant fimP and fimA fimbrial subunit genes corresponding to different types of acidic proline-rich protein and beta-linked galactosamine binding specificity. Infect. Immun. 1998, 66, 4403–4410. [Google Scholar] [PubMed]
- Cisar, J.O.; Sandberg, A.L.; Mergenhagen, S.E. Lectin-dependent attachment of Actinomyces naeslundii to receptors on epithelial cells. Infect. Immun. 1984, 46, 459–464. [Google Scholar]
- Cisar, J.O.; Vatter, A.E.; Clark, W.B.; Curl, S.H.; Hurst-Calderone, S.; Sandberg, A.L. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 1988, 56, 2984–2989. [Google Scholar] [PubMed]
- Whittaker, C.J.; Klier, C.M.; Kolenbrander, P.E. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 1996, 50, 513–552. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, I.; Shimizu, T.; Senpuku, H. Role of SCFAs for Fimbrillin-Dependent Biofilm Formation of Actinomyces oris. Microorganisms 2018, 6, 114. https://doi.org/10.3390/microorganisms6040114
Suzuki I, Shimizu T, Senpuku H. Role of SCFAs for Fimbrillin-Dependent Biofilm Formation of Actinomyces oris. Microorganisms. 2018; 6(4):114. https://doi.org/10.3390/microorganisms6040114
Chicago/Turabian StyleSuzuki, Itaru, Takehiko Shimizu, and Hidenobu Senpuku. 2018. "Role of SCFAs for Fimbrillin-Dependent Biofilm Formation of Actinomyces oris" Microorganisms 6, no. 4: 114. https://doi.org/10.3390/microorganisms6040114
APA StyleSuzuki, I., Shimizu, T., & Senpuku, H. (2018). Role of SCFAs for Fimbrillin-Dependent Biofilm Formation of Actinomyces oris. Microorganisms, 6(4), 114. https://doi.org/10.3390/microorganisms6040114




