Genome Analysis of Vallitalea guaymasensis Strain L81 Isolated from a Deep-Sea Hydrothermal Vent System
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Reclassification of Abyssivirga alkaniphila L81T
3.2. Emended Description of Vallitalea guaymasensis
3.3. Emended Description of Vallitalea Gen
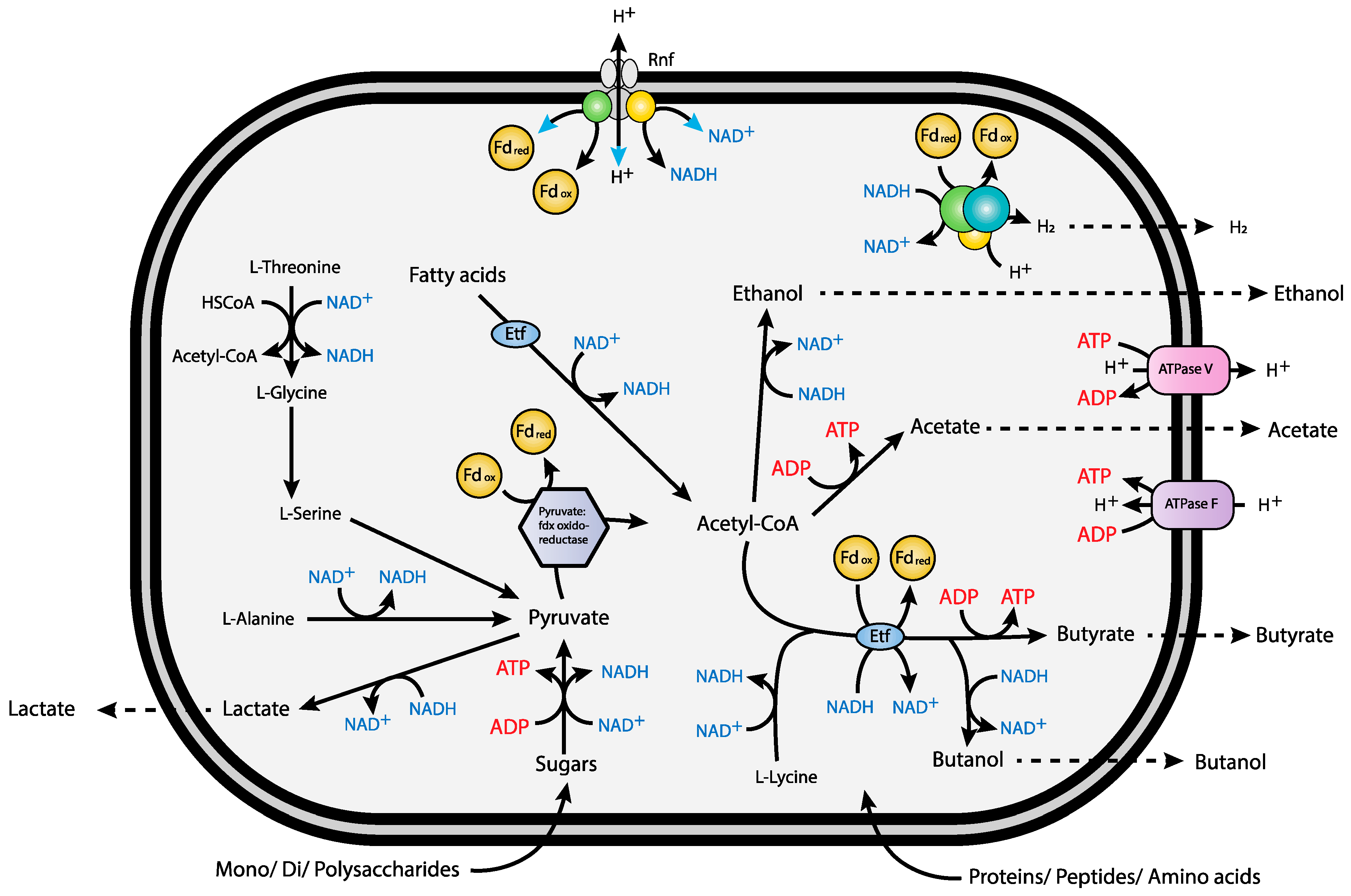
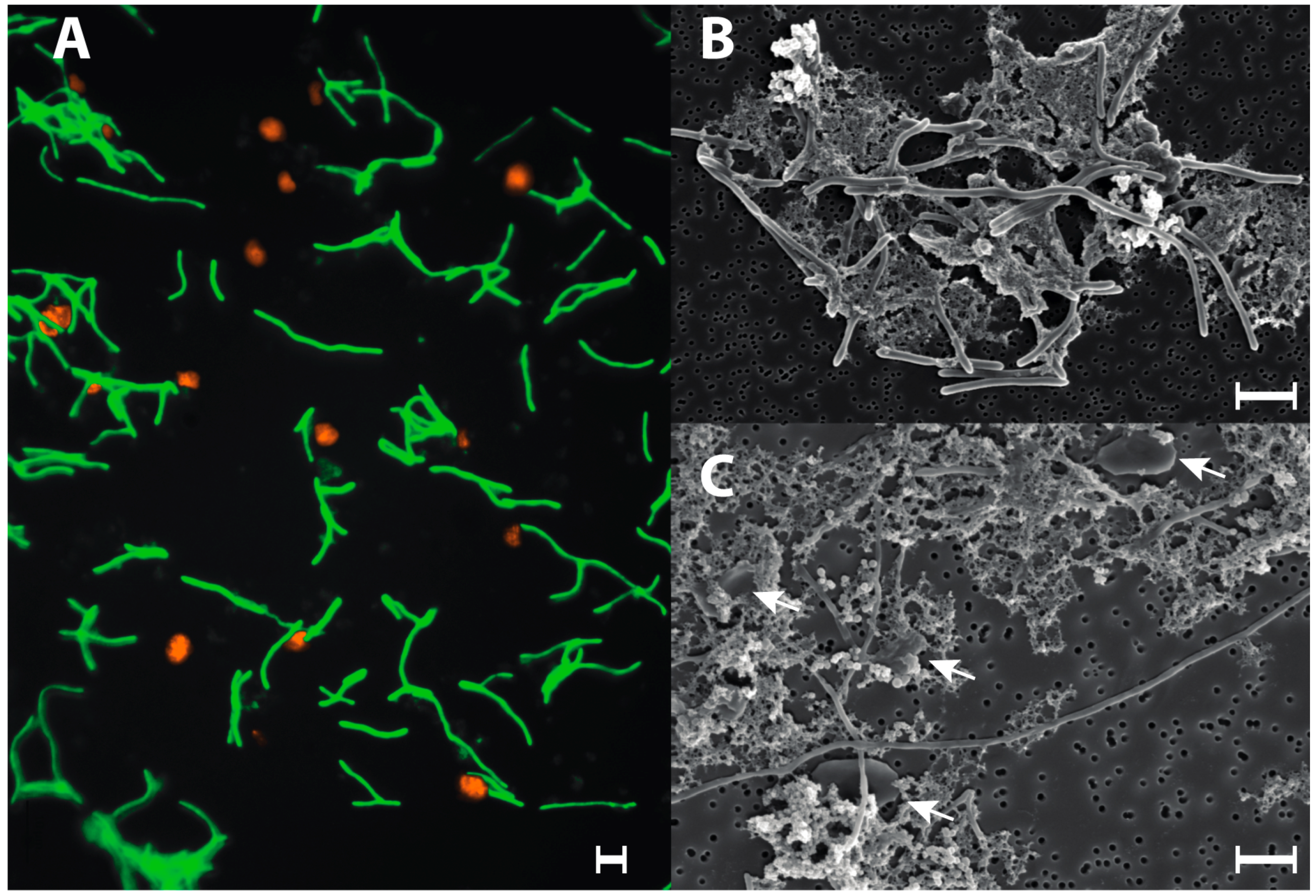
3.4. Syntrophic Growth
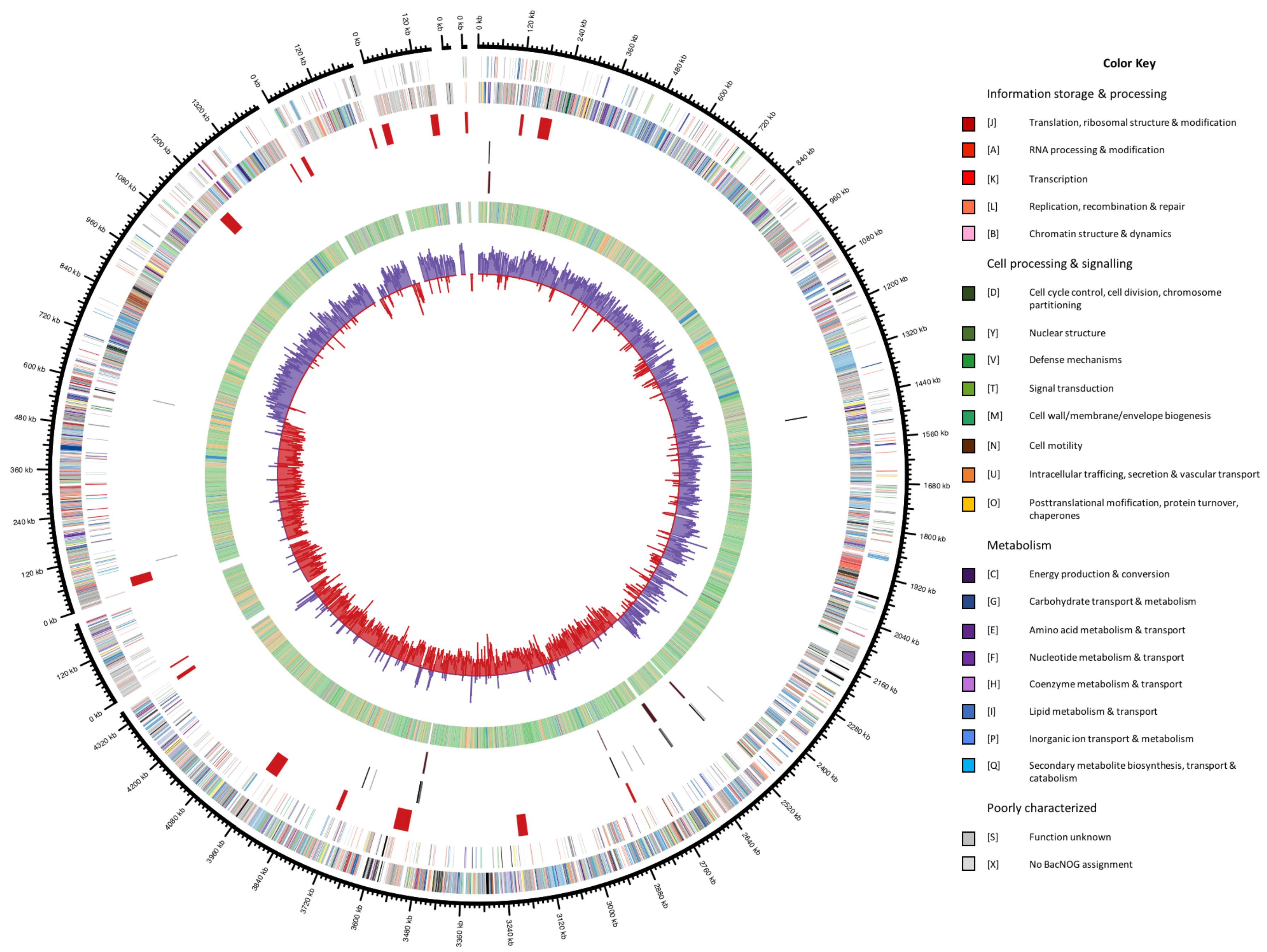
3.5. General Genomic Features
3.6. Carbohydrate Metabolism and Transport
3.7. Protein Metabolism and Transport
3.8. Alkane Activation and Degradation
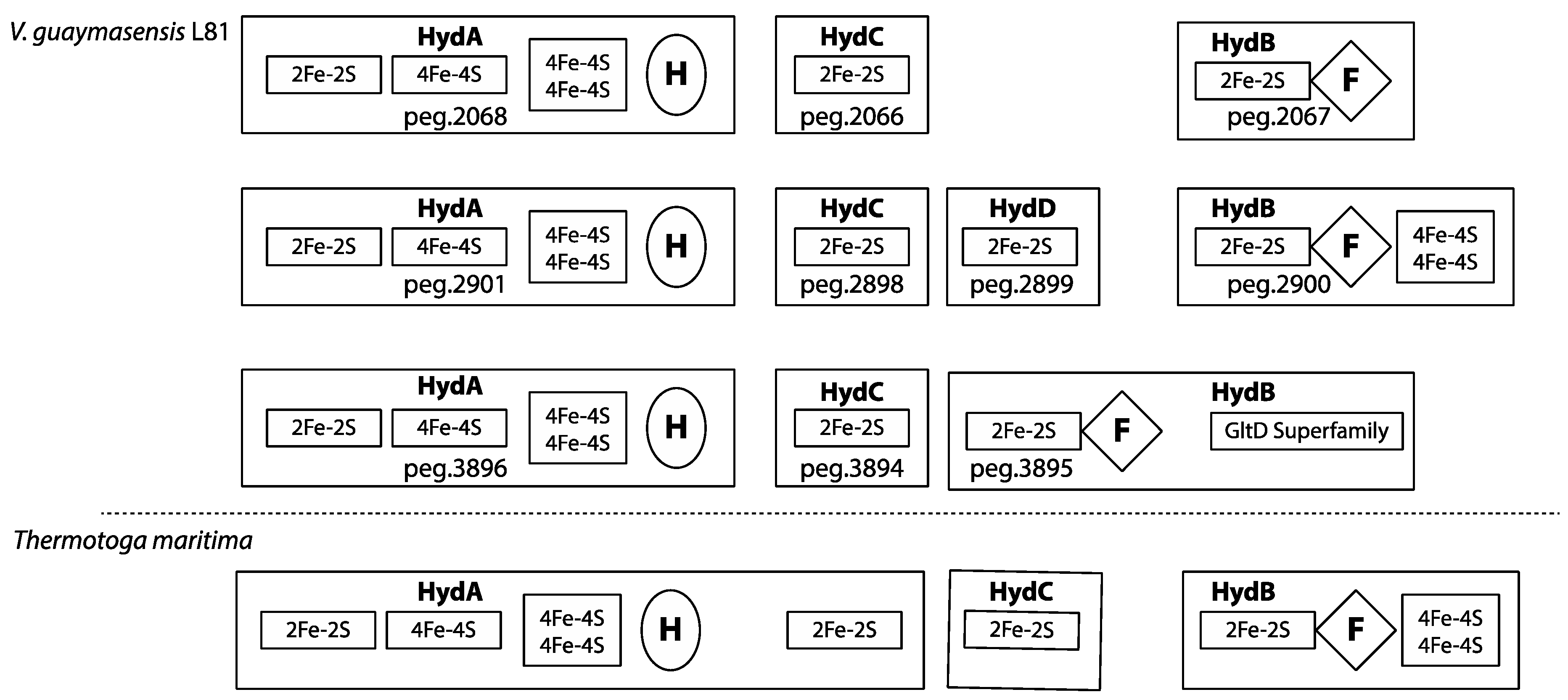
3.9. Redox-Components
3.10. Polyketid Synthesis
3.11. Intracellular Compartments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rogers, K.L.; Amend, J.P. Energetics of potential heterotrophic metabolisms in the marine hydrothermal system of Vulcano Island, Italy. Geochim. Cosmochim. Acta 2006, 70, 6180–6200. [Google Scholar] [CrossRef]
- Slobodkina, G.B.; Kolganova, T.V.; Tourova, T.P.; Kostrikina, N.A.; Jeanthon, C.; Bonch-Osmolovskaya, E.A.; Slobodkin, A.I. Clostridium tepidiprofundi sp. nov.; a moderately thermophilic bacterium from a deep-sea hydrothermal vent. Int. J. Syst. Evolut. Microbiol. 2008, 58 Pt 4, 852–855. [Google Scholar] [CrossRef] [PubMed]
- Stokke, R.; Dahle, H.; Roalkvam, I.; Wissuwa, J.; Daae, F.L.; Tooming-Klunderud, A.; Thorseth, I.H.; Pedersen, R.B.; Steen, I.H. Functional interactions among filamentous Epsilonproteobacteria and Bacteroidetes in a deep-sea hydrothermal vent biofilm. Environ. Microbiol. 2015, 17, 4063–4077. [Google Scholar] [CrossRef] [PubMed]
- Schouw, A.; Eide, T.L.; Stokke, R.; Pedersen, R.B.; Steen, I.H.; Bødtker, G. Abyssivirga alkaniphila gen. nov.; sp. nov.; an alkane-degrading, anaerobic bacterium from a deep-sea hydrothermal vent system, and emended descriptions of Natranaerovirga pectinivora and Natranaerovirga hydrolytica. Int. J. Syst. Evolut. Microbiol. 2016, 66, 1724–1734. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.B.; Rapp, H.T.; Thorseth, I.H.; Lilley, M.D.; Barriga, F.J.; Baumberger, T.; Flesland, K.; Fonseca, R.; Früh-Green, G.L.; Jorgensen, S.L. Discovery of a black smoker vent field and vent fauna at the Arctic Mid-Ocean Ridge. Nat. Commun. 2010, 1, 126. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.B.; Thorseth, I.H.; Nygård, T.; Lilley, M.D.; Kelley, D.S. Diversity of hydrothermal systems on slow spreading ocean ridges. Geophys. Monogr. Ser. 2010, 188. [Google Scholar] [CrossRef]
- Baumberger, T.; Früh-Green, G.L.; Thorseth, I.H.; Lilley, M.D.; Hamelin, C.; Bernasconi, S.M.; Okland, I.E.; Pedersen, R.B. Fluid composition of the sediment-influenced Loki’s Castle vent field at the ultra-slow spreading Arctic Mid-Ocean Ridge. Geochim. Cosmochim. Acta 2016, 187, 156–178. [Google Scholar] [CrossRef]
- Eickmann, B.; Thorseth, I.H.; Peters, M.; Strauss, H.; Bröcker, M.; Pedersen, R.B. Barite in hydrothermal environments as a recorder of subseafloor processes: A multiple-isotope study from the Loki’s Castle vent field. Geobiology 2014. [Google Scholar] [CrossRef] [PubMed]
- Dahle, H.; Roalkvam, I.; Thorseth, I.H.; Pedersen, R.B.; Steen, I.H. The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 2013, 5, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.L.; Roalkvam, I.; Steen, I.H.; Dahle, H. Lutibacter profundi sp. nov.; isolated from a deep-sea hydrothermal system on the Arctic Mid-Ocean Ridge and emended description of the genus Lutibacter. Int. J. Syst. Evolut. Microbiol. 2016, 66, 2671–2677. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, J.; Bauer, S.L.; Steen, I.H.; Stokke, R. Complete genome sequence of Lutibacter profundi LP1T isolated from an arctic deep-sea hydrothermal vent system. Stand. Genom. Sci. 2017, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Postec, A.; Olivier, B.; Fardeau, M.-L. Objection to the proposition of the novel genus Abyssivirga. Int. J. Syst. Evolut. Microbiol. 2016, 67, 174. [Google Scholar] [CrossRef]
- Ben Aissa, F.; Postec, A.; Erauso, G.; Payri, C.; Pelletier, B.; Hamdi, M.; Ollivier, B.; Fardeau, M.-L. Vallitalea pronyensis sp. nov.; isolated from a marine alkaline hydrothermal chimney. Int. J. Syst. Evolut. Microbiol. 2014, 64 Pt 4, 1160–1165. [Google Scholar] [CrossRef] [PubMed]
- Lakhal, R.; Pradel, N.; Postec, A.; Hamdi, M.; Ollivier, B.; Godfroy, A.; Fardeau, M.-L. Vallitalea guaymasensis gen. nov.; sp. nov.; isolated from marine sediment. Int. J. Syst. Evolut. Microbiol. 2013, 63, 3019–3023. [Google Scholar] [CrossRef] [PubMed]
- Teske, A.; Callaghan, A.V.; LaRowe, D.E. Biosphere frontiers of subsurface life in the sedimented hydrothermal system of Guaymas Basin. Front. Microbiol. 2014, 5, 362. [Google Scholar] [CrossRef] [PubMed]
- Sander, R. Compilation of Henry’s law constants (version 4.0) for water as solvent. Atmos. Chem. Phys. 2015, 15, 4399–4981. [Google Scholar] [CrossRef]
- Glöckner, F.; Amann, R.; Alfreider, A.; Pernthaler, J.; Psenner, R.; Trebesius, K.; Schleifer, K.-H. An in-situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 1996, 19, 403–406. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [PubMed]
- Loy, A.; Lehner, A.; Lee, N.; Adamczyk, J.; Meier, H.; Ernst, J.; Schleifer, K.-H.H.; Wagner, M. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 2002, 68, 5064–5081. [Google Scholar] [CrossRef] [PubMed]
- Cashion, P.; Holder-Franklin, M.; McCully, J.; Franklin, M. A rapid method for the base ratio determination of bacterial DNA. Anal. Biochem. 1977, 81, 461–466. [Google Scholar] [CrossRef]
- De Ley, J.; Cattoir, H.; Reynaerts, A. The quantitative measurement of DNA hybridization from renaturation rates. Eur. J. Biochem. 1970, 12, 133–142. [Google Scholar] [CrossRef]
- Huss, V.A.R.; Festl, H.; Schleifer, K.H. Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst. Appl. Microbiol. 1983, 4, 184–192. [Google Scholar] [CrossRef]
- Ryu, E. On the Gram-differentiation of bacteria by the simplest method. J. Jpn. Soc. Vet. Sci. 1938, 15, 205–207. [Google Scholar] [CrossRef]
- Ryu, E. A simple method of staining bacterial flagella. Kitasato Arch. Exp. Med. 1937, 14, 218–219. [Google Scholar]
- Heimbrook, M.E.; Wang, W.L.; Campbell, G. Staining bacterial flagella easily. J. Clin. Microbiol. 1989, 27, 2612–2615. [Google Scholar] [PubMed]
- Roalkvam, I.; Drønen, K.; Stokke, R.; Daae, F.-L.; Dahle, H.; Steen, I.H. Physiological and genomic characterization of Arcobacter anaerophilus IR-1 reveals new metabolic features in Epsilonproteobacteria. Front. Microbiol. 2015, 6, 987. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.A.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.A.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-M.A.; Markowitz, V.M.; Chu, K.; Palaniappan, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Andersen, E.; Huntemann, M.; et al. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017, 45, D507–D516. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.-M.A.; Chu, K.; Szeto, E.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Pagani, I.; Tringe, S.; et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014, 42, D568–D573. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, V.M.; Chen, I.-M.A.; Palaniappan, K.; Chu, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Woyke, T.; Huntemann, M.; et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014, 42, D560–D567. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.; Bruccoleri, R.; Lee, S.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, D.; Pedersen, C.; Greening, C. HydDB: A web tool for hydrogenase classification and analysis. Sci. Rep. 2016, 6, 34212. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.; Murray, R.; Stackebrandt, E.S. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Koonin, E.V.; Wolf, Y.I. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008, 36, 6688–6719. [Google Scholar] [CrossRef] [PubMed]
- Land, M.; Hauser, L.; Jun, S.-R.; Nookaew, I.; Leuze, M.R.; Ahn, T.-H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 2002, 10, 393–395. [Google Scholar] [CrossRef]
- Daubin, V.; Lerat, E.; Perrière, G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.; Somerville, G.; Sonenshein, A. Regulating the intersection of metabolism and pathogenesis in gram-positive bacteria. In Metabolism and Bacterial Pathogenesis, 1st ed.; Department of Health and Human Services: Washington, DC, USA, 2015; Volume 1, pp. 129–165. [Google Scholar]
- Mead, G. The amino acid-fermenting clostridia. J. Gen. Microbiol. 1971, 67, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Schink, B.; Stams, A.J. Syntrophism among prokaryotes. In The Procaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: Berlin, Germany, 2006; Volume 2, pp. 309–335. ISBN 978-0-387-30740-4. [Google Scholar]
- Buckel, W. Unusual enzymes involved in five pathways of glutamate fermentation. Appl. Microbiol. Biotechnol. 2001, 57, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Bes, M.; Merrouch, M.; Joseph, M.; Quéméneur, M.; Payri, C.; Pelletier, B.; Ollivier, B.; Fardeau, M.-L.; Erauso, G.; Postec, A. Acetoanaerobium pronyense sp. nov.; an anaerobic alkaliphilic bacterium isolated from a carbonate chimney of the Prony Hydrothermal Field (New Caledonia). Int. J. Syst. Evolut. Microbiol. 2015, 65, 2574–2580. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Ritari, J.; Boeren, S.; de Waard, P.; Plugge, C.M.; de Vos, W.M. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 2015, 6, 10062. [Google Scholar] [CrossRef] [PubMed]
- Barker, H. Explorations of bacterial metabolism. Annu. Rev. Biochem. 1978, 47, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Callaghan, A.V.; Morris, B.E.L.; Pereira, I.A.C.; McInerney, M.J.; Austin, R.N.; Groves, J.T.; Kukor, J.J.; Suflita, J.M.; Young, L.Y.; Zylstra, G.J.; et al. The genome sequence of Desulfatibacillum alkenivorans AK-01: A blueprint for anaerobic alkane oxidation. Environ. Microbiol. 2012, 14. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, H.; Rabus, R.; Fischer, T.; Armstroff, A.; Behrends, A.; Widdel, F. Anaerobic degradation of n-hexane in a denitrifying bacterium: Further degradation of the initial intermediate (1-methylpentyl) succinate via C-skeleton rearrangement. Arch. Microbiol. 2002, 177, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Schut, G.J.; Adams, M.W. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: A new perspective on anaerobic hydrogen production. J. Bacteriol. 2009, 191, 4451–4457. [Google Scholar] [CrossRef] [PubMed]
- Mulder, D.W.; Shepard, E.M.; Meuser, J.E.; Joshi, N.; King, P.W.; Posewitz, M.C.; Broderick, J.B.; Peters, J.W. Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure 2011. [Google Scholar] [CrossRef] [PubMed]
- Vignais, P.; Billoud, B.; Meyer, J. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 2001, 25, 455–501. [Google Scholar] [CrossRef] [PubMed]
- Greening, C.; Biswas, A.; Carere, C.R.; Jackson, C.J.; Taylor, M.C.; Stott, M.B.; Cook, G.M.; Morales, S.E. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016, 10, 761–777. [Google Scholar] [CrossRef] [PubMed]
- Vignais, P.; Billoud, B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem. Rev. 2007, 107, 4206–4272. [Google Scholar] [CrossRef] [PubMed]
- Calusinska, C.; Happe, T.; Joris, B.; Wilmotte, A. The surprising diversity of clostridial hydrogenases: A comparative genomic perspective. Microbiology 2010, 156, 1575–1588. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.; Hogsett, D.; Lynd, L. Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J. Bacteriol. 2009, 191, 6457–6464. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Kahnt, J.; Kwon, I.; Mackie, R.; Thauer, R. Hydrogen formation and its regulation in Ruminococcus albus: Involvement of an electron-bifurcating [FeFe]-hydrogenase, of a non-electron-bifurcating [FeFe]-Hydrogenase, and of a Putative Hydrogen-Sensing [FeFe]-Hydrogenase. J. Bacteriol. 2014, 196, 3840–3852. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.; Biswas, A.; Morales, S.; Greening, C.; Gaskins, R. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes 2016, 7, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Soboh, B.; Linder, D.; Hedderich, R. A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology 2004, 150, 2451–2463. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.P.; Kahnt, J.; Buckel, W. Reduction of ferredoxin or oxygen by flavin-based electron bifurcation in Megasphaera elsdenii. FEBS J. 2015, 282, 3149–3160. [Google Scholar] [CrossRef] [PubMed]
- Costas, A.M.; Poudel, S.; Miller, A.-F.; Schut, G.J.; Ledbetter, R.N.; Fixen, K.R.; Seefeldt, L.C.; Adams, M.W.W.; Harwood, C.S.; Boyd, E.S.; et al. Defining electron bifurcation in the electron-transferring flavoprotein family. J. Bacteriol. 2017, 199, e00440-17. [Google Scholar] [CrossRef] [PubMed]
- Perzov, N.; Padler-Karavani, V.; Nelson, H.; Nelson, N. Features of V-ATPases that distinguish them from F-ATPases. FEBS Lett. 2001, 504, 223–228. [Google Scholar] [CrossRef]
- Visser, M.; Worm, P.; Muyzer, G.; Pereira, I.; Schaap, P.J.; Plugge, C.M.; Kuever, J.; Parshina, S.N.; Nazina, T.N.; Ivanova, A.E.; et al. Genome analysis of Desulfotomaculum kuznetsovii strain 17T reveals a physiological similarity with Pelotomaculum thermopropionicum strain SIT. Stand. Genom. Sci. 2013, 8, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Kevany, B.M.; Rasko, D.A.; Thomas, M.G. Characterization of the complete Zwittermicin A biosynthesis gene cluster from Bacillus cereus. Appl. Environ. Microbiol. 2009, 75, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, E.; Müller, S.; Hertlein, G.; Heid, N.; Süssmuth, R.D.; Genersch, E. Biological effects of paenilamicin, a secondary metabolite antibiotic produced by the honey bee pathogenic bacterium Paenibacillus larvae. MicrobiologyOpen 2014, 3, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Homburg, S.; Oswald, E.; Hacker, J.; Dobrindt, U. Expression analysis of the colibactin gene cluster coding for a novel polyketide in Escherichia coli. FEMS Microbiol. Lett. 2007, 275, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Jahns, C.; Hoffmann, T.; Müller, S.; Gerth, K.; Washausen, P.; Höfle, G.; Reichenbach, H.; Kalesse, M.; Müller, R. Pellasoren: Structure elucidation, biosynthesis, and total synthesis of a cytotoxic secondary metabolite from Sorangium cellulosum. Angew. Chem. 2012, 51, 5239–5243. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Borchert, S.; Conrad, B.; Feesche, J.; Hofemeister, B.; Hofemeister, J.; Entian, K.-D. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J. Bacteriol. 2002, 184, 1703–1711. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W.; Jaskolla, T.W.; Bochmann, S.; Kötter, P.; Wichelhaus, T.; Karas, M.; Stein, T.; Entian, K.-D. Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl. Environ. Microbiol. 2011, 77, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Kaletta, C.; Schnell, N.; Entian, K.-D. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 1992, 58, 132–142. [Google Scholar] [PubMed]
- Rince, A.; Dufour, A.; Le Pogam, S.; Thuault, D.; Bourgeois, C.M.; Le Pennec, J.P. Cloning, expression, and nucleotide sequence of genes involved in production of lactococcin DR, a bacteriocin from Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 1994, 60, 1652–1657. [Google Scholar] [PubMed]
- Rincé, A.; Dufour, A.; Uguen, P.; Le Pennec, J.P.; Haras, D. Characterization of the lacticin 481 operon: The Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 1997, 63, 4252–4260. [Google Scholar] [PubMed]
- Kampa, A.; Gagunashvili, A.N.; Gulder, T.A.M.; Morinaka, B.I.; Daolio, C.; Godejohann, M.; Miao, V.P.W.; Piel, J.; Andrésson, O.S. Metagenomic natural product discovery in lichen provides evidence for a family of biosynthetic pathways in diverse symbioses. Proc. Natl. Acad. Sci. USA 2013, 110, E3129–E3137. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, D.; Haines, A.S.; Song, Z.; Murphy, A.C.; Hothersall, J.; Stephens, E.R.; Gurney, R.; Cox, R.J.; Crosby, J.; Willis, C.L.; et al. A natural plasmid uniquely encodes two biosynthetic pathways creating a potent anti-MRSA antibiotic. PLoS ONE 2011, 6, e18031. [Google Scholar] [CrossRef] [PubMed]
- Mattheus, W.; Gao, L.-J.J.; Herdewijn, P.; Landuyt, B.; Verhaegen, I.; Masschelein, I.; Volckaert, G.; Lavigne, R. Isolation and purification of a new kalimantacin/batumin-related polyketide antibiotic and elucidation of its biosynthesis gene cluster. Chem. Biol. 2010, 17, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Lincke, T.; Behnken, S.; Hertweck, C. Induced biosynthesis of cryptic polyketide metabolites in a Burkholderia thailandensis quorum sensing mutant. J. Am. Chem. Soc. 2010, 132, 13966–13968. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Lincke, T.; Hertweck, C. Assembly and absolute configuration of short-lived polyketides from Burkholderia thailandensis. Angew. Chem. 2012, 51, 5470–5474. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Ishida, K.; Jenke-Kodama, H.; Dittmann, E.; Gurgui, C.; Hochmuth, T.; Taudien, S.; Platzer, M.; Hertweck, C.; Piel, J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 2008, 26, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and Functional Characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef] [PubMed]
- Moldenhauer, J.; Chen, X.-H.; Borriss, R.; Piel, J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT Family. Angew. Chem. 2007, 46, 8195–8197. [Google Scholar] [CrossRef] [PubMed]
- Dehn, R.; Katsuyama, Y.; Weber, A.; Gerth, K.; Jansen, R.; Steinmetz, H.; Höfle, G.; Müller, R.; Kirschning, A. Molecular basis of elansolid biosynthesis: Evidence for an unprecedented quinone methide initiated intramolecular Diels–Alder cycloaddition/macrolactonization. Angew. Chem. 2011, 50, 3882–3887. [Google Scholar] [CrossRef] [PubMed]
- Moebius, N.; Ross, C.; Scherlach, K.; Rohm, B.; Roth, M.; Hertweck, C. Biosynthesis of the respiratory toxin bongkrekic acid in the pathogenic bacterium Burkholderia gladioli. Chem. Biol. 2012, 19, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, T.; Egami, Y.; Nakashima, Y.; Wakimoto, Y.; Mori, T.; Awakawa, T.; Ito, T.; Kenmoku, H.; Asakawa, Y.; Piel, J.; et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat. Chem. Biol. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Hamer, H.A.; Sirasani, G.; Balskus, E.P. Cylindrocyclophane biosynthesis involves functionalization of an unactivated carbon center. J. Am. Chem. Soc. 2012, 134, 18518–18521. [Google Scholar] [CrossRef] [PubMed]
- Preisitsch, M.; Heiden, S.E.; Beerbaum, M.; Niedermeyer, T.H.J.; Schneefeld, M.; Herrmann, J.; Kumpfmüller, J.; Thürmer, A.; Neidhardt, I.; Wiesner, C.; et al. Effects of halide ions on the carbamidocyclophane biosynthesis in Nostoc sp. CAVN2. Mar. Drugs 2016, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Pag, U.; Sahl, H.-G.G. Multiple activities in lantibiotics—Models for the design of novel antibiotics? Curr. Pharm. Des. 2002, 8, 815–833. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Chaudhry, V.; Mittal, G.; Liao, L.M.; Matos, C.O.; Khatri, N.; Franco, O.L.; Patil, P.B.; Korpole, S. Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. Strain A3. Antimicrob. Agents Chemother. 2016, 60, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Heather, Z.; Holden, M.T.; Steward, K.F.; Parkhill, J.; Song, L.; Challis, G.L.; Robinson, C.; Davis-Poynter, N.; Waller, A.S. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol. Microbiol. 2008, 70, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- Axen, S.D.; Erbilgin, O.; Kerfeld, C.A. A Taxonomy of Bacterial Microcompartment Loci Constructed by a Novel Scoring Method. PLoS Comput. Biol. 2014, 10, e1003898. [Google Scholar] [CrossRef] [PubMed]
- Buchrieser, C.; Rusniok, C.; Kunst, F.; Cossart, P.; Glaser, P. The Listeria Consortium. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 2003, 35, 207–213. [Google Scholar] [CrossRef]
- Bobik, T.A.; Havemann, G.D.; Busch, R.J.; Williams, D.S.; Aldrich, H.C. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J. Bacteriol. 1999, 181, 5967–5975. [Google Scholar] [PubMed]
- Bobik, T.; Lehman, B.; Yeates, T. Bacterial microcompartments: Widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol. Microbiol. 2015, 98, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Kerfeld, C.; Heinhorst, S.; Cannon, G. Bacterial microcompartments. Annu. Rev. Microbiol. 2010, 64, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Rondon, M.R.; Kazmierczak, R.; Escalante-Semerena, J.C. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J. Bacteriol. 1995, 177, 5434–5439. [Google Scholar] [CrossRef] [PubMed]
- Stojiljkovic, I.; Bäumler, A.J.; Heffron, F. Ethanolamine utilization in Salmonella typhimurium: Nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 1995, 177, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, C.; Sinha, S.; Chun, S.; Yeates, T.O.; Bobik, T.A. Diverse Bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 2014, 78, 438–468. [Google Scholar] [CrossRef] [PubMed]
- Petit, E.; LaTouf, G.W.; Coppi, M.V.; Warnick, T.A.; Currie, D.; Romashko, I.; Deshpande, S.; Haas, K.; Alvelo-Maurosa, J.G.; Wardman, C.; et al. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS ONE 2013, 8, e54337. [Google Scholar] [CrossRef] [PubMed]
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. MMBR 1997, 61, 262–280. [Google Scholar] [PubMed]
- Buckel, W.; Thauer, R.K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta BBA Bioenerg. 2013, 1827, 94–113. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.P.; Klomann, K.; Seubert, A.; Buckel, W. Reduction of flavodoxin by electron bifurcation and sodium ion-dependent reoxidation by NAD+ catalyzed by ferredoxin-NAD+ reductase (Rnf). J. Biol. Chem. 2016, 291, 11993–12002. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, N.P.; Mowafy, A.M.; Demmer, J.K.; Upadhyay, V.; Koelzer, S.; Jayamani, E.; Kahnt, J.; Hornung, M.; Demmer, U.; Ermler, U.; et al. Studies on the mechanism of electron bifurcation catalyzed by electron transferring flavoprotein (Etf) and butyryl-CoA dehydrogenase (Bcd) of Acidaminococcus fermentans. J. Biol. Chem. 2014, 289, 5145–5157. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, G.; Jayamani, E.; Mai, G.; Buckel, W. Energy Conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 2008, 190, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Biegel, E.; Schmidt, S.; Gonzalez, J.M.M.; Müller, V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell. Mol. Life Sci. CMLS 2011, 68, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, P.-L.; Zhang, T.; Dar, S.A.; Leang, C.; Lovley, D.B. The Rnf complex of Clostridium ljungdahlii is a proton-translocating Ferredoxin: NAD+ oxidoreductase essential for autotrophic growth. MBio 2013, 4, e00406-12. [Google Scholar] [CrossRef]
- Dolfing, J.; Larter, S.R.; Head, I.M. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2008, 2, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.M.; Head, I.M.; Gray, N.D.; Adams, J.J.; Rowan, A.K.; Aitken, C.M.; Bennett, B.; Huang, H.; Brown, A.; Bowler, B.F.; et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 2008, 451, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Richnow, H.H.; Rosselló-Mora, R.; Michaelis, W.; Widdel, F. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature 1999, 401, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Jarling, R.; Kühner, S.; Janke, E.B.; Gruner, A.; Drozdowska, M.; Golding, B.T.; Rabus, R.; Wilkes, H. Versatile transformations of hydrocarbons in anaerobic bacteria: Substrate ranges and regio- and stereo-chemistry of activation reactions. Front. Microbiol. 2015, 6, 880. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, H.; Buckel, W.; Golding, B.T.; Rabus, R. Metabolism of hydrocarbons in n-Alkane-utilizing anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 2016, 26, 138–151. [Google Scholar] [CrossRef] [PubMed]
- De Bok, F.; Plugge, C.; Stams, A.J. Interspecies electron transfer in methanogenic propionate degrading consortia. Water Res. 2004, 38, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Kosaka, T.; Hori, K.; Hotta, Y.; Watanabe, K. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl. Environ. Microbiol. 2005, 71, 7838–7845. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Kosaka, T.; Hotta, Y.; Watanabe, K. Simulating the contribution of coaggregation to interspecies hydrogen fluxes in syntrophic methanogenic consortia. Appl. Environ. Microbiol. 2006, 72, 5093–5096. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Hashimoto, K.; Watanabe, K. Microbial interspecies electron transfer via electric currents through conductive minerals. Proc. Natl. Acad. Sci. USA 2012, 109, 10042–10046. [Google Scholar] [CrossRef] [PubMed]
- Kouzuma, A.; Kato, S.; Watanabe, K. Microbial interspecies interactions: Recent findings in syntrophic consortia. Front. Microbiol. 2015, 6, 477. [Google Scholar] [CrossRef] [PubMed]
- Stams, A.J. Metabolic interactions between anaerobic bacteria in methanogenic environments. Antonie van Leeuwenhoek 1994, 66, 271–294. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Watanabe, K. Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ. 2010, 25, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, T.; Kato, S.; Ishii, S.; Watanabe, K. Flagellum mediates symbiosis. Science 2009, 323, 1574. [Google Scholar] [CrossRef] [PubMed]
| Category | RAST | IMG-ER |
|---|---|---|
| Genome size | 6419149 bp | 6419149 bp |
| Contigs | 7 | 7 |
| GC content | 31.2% | 31.2% |
| Coding sequences | 5628 | 5651 |
| RNA genes | 67 (1.2%) | 121 (2.1%) |
| rRNA genes | 11 | 17 |
| 5S rRNA | 6 | |
| 16S rRNA | 6 | |
| 23S rRNA | 5 | |
| tRNA genes | 56 | 56 |
| Other RNA genes | 48 | |
| Genes with function prediction | 3820 (67.87%) | 4043 (70.05%) |
| Genes without function prediction | 1808 (32.13%) | 1608 (27.86%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schouw, A.; Vulcano, F.; Roalkvam, I.; Hocking, W.P.; Reeves, E.; Stokke, R.; Bødtker, G.; Steen, I.H. Genome Analysis of Vallitalea guaymasensis Strain L81 Isolated from a Deep-Sea Hydrothermal Vent System. Microorganisms 2018, 6, 63. https://doi.org/10.3390/microorganisms6030063
Schouw A, Vulcano F, Roalkvam I, Hocking WP, Reeves E, Stokke R, Bødtker G, Steen IH. Genome Analysis of Vallitalea guaymasensis Strain L81 Isolated from a Deep-Sea Hydrothermal Vent System. Microorganisms. 2018; 6(3):63. https://doi.org/10.3390/microorganisms6030063
Chicago/Turabian StyleSchouw, Anders, Francesca Vulcano, Irene Roalkvam, William Peter Hocking, Eoghan Reeves, Runar Stokke, Gunhild Bødtker, and Ida Helene Steen. 2018. "Genome Analysis of Vallitalea guaymasensis Strain L81 Isolated from a Deep-Sea Hydrothermal Vent System" Microorganisms 6, no. 3: 63. https://doi.org/10.3390/microorganisms6030063
APA StyleSchouw, A., Vulcano, F., Roalkvam, I., Hocking, W. P., Reeves, E., Stokke, R., Bødtker, G., & Steen, I. H. (2018). Genome Analysis of Vallitalea guaymasensis Strain L81 Isolated from a Deep-Sea Hydrothermal Vent System. Microorganisms, 6(3), 63. https://doi.org/10.3390/microorganisms6030063





