Multi-Regional Study on the Microbial Community Structure, Core Microbiome and Functional Characteristics in Deep Fracture Waters
Abstract
1. Introduction
2. Materials and Methods
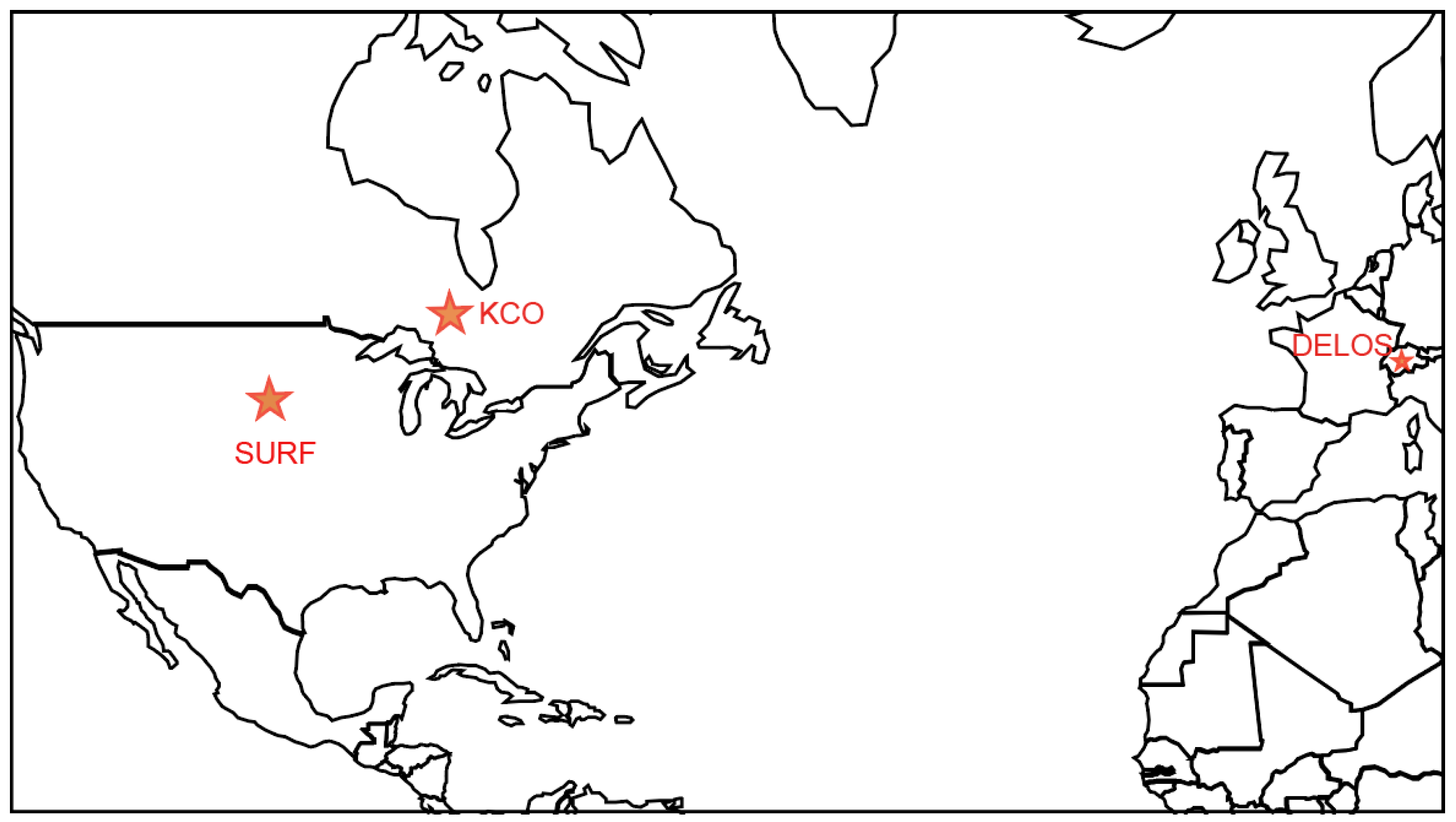
2.1. Study Area
2.2. Data Acquisition
2.3. High-Throughput Sequencing Data Processing
2.4. Water Chemistry Data Processing
2.5. Diversity and Statistical Analyses
2.6. Functional Prediction Analyses
3. Results
3.1. The Hydrochemical Properties of Deep Fracture Waters Are Spatially Heterogenous Across Study Regions
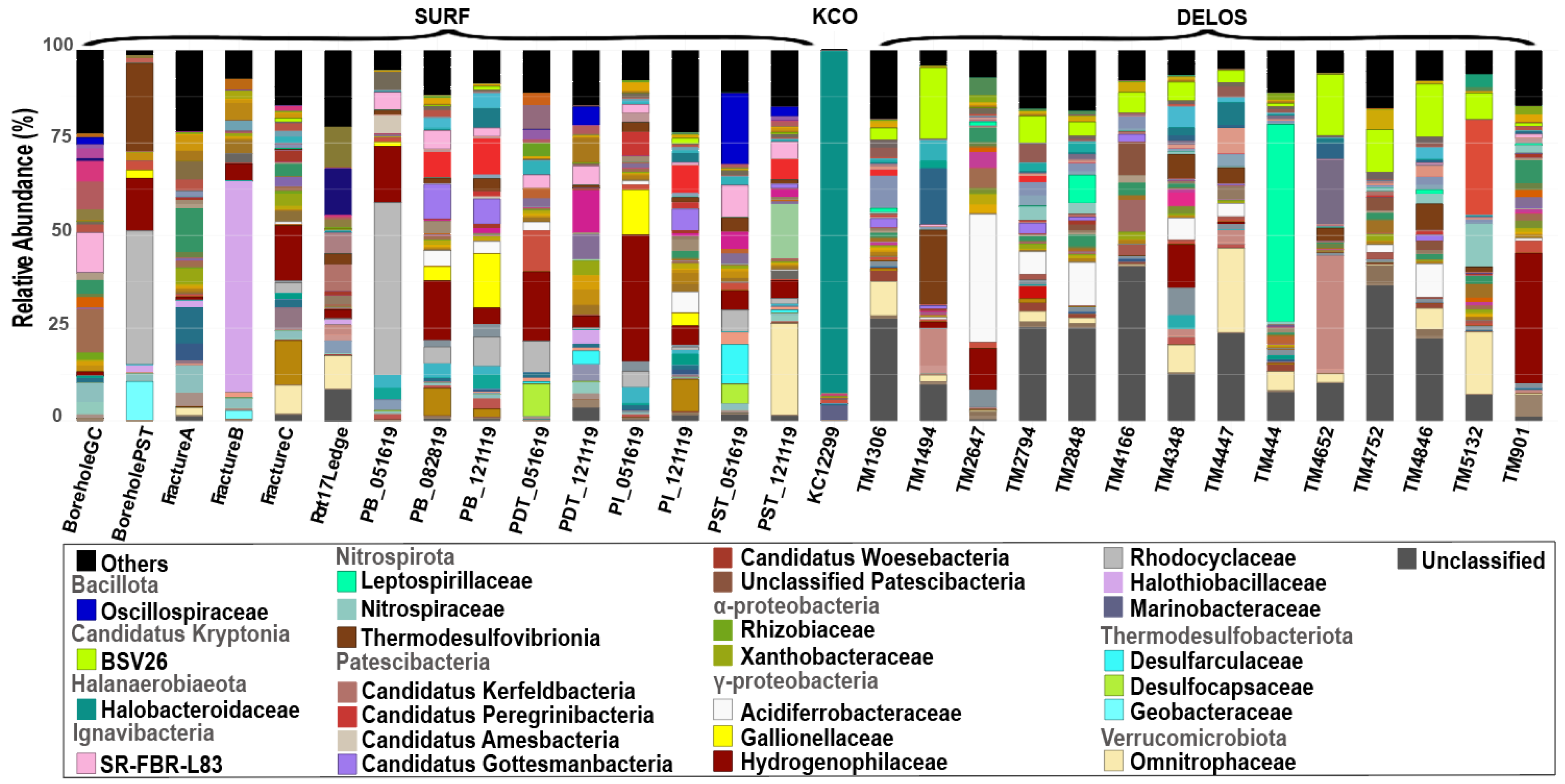
3.2. Different Microbial Community Compositions Within-Site and Across-Sites
3.3. Alpha Diversity Analysis
3.4. Samples from Different Regions Form Distinct Clusters
3.5. Limited Shared Microbial Taxa Among Three Regions
3.6. FAPROTAX Functional Prediction Reveals Metabolic Niche Partitioning of Groundwater Microbiota in C–N–S–Fe Cycling Across the Three Study Regions
4. Discussion
4.1. Environmental Filtration Exerts a Driving Influence on the Microbial Community Structure Within Deep Groundwater Environments
4.2. Significant Differences in the Microbial Community Composition Across the Three Aquifers Revealed by Bray–Curtis and Weighted Unifrac Distances
4.3. Core Microbiome Is Present in Deep Groundwater Environments
4.4. The Functional Characteristics of Deep Fractures Are Collectively Shaped by Both Core Microbial Genera and Dominant Genera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magnabosco, C.; Lin, L.H.; Dong, H.; Bomberg, M.; Ghiorse, W.; Stan-Lotter, H.; Pedersen, K.; Kieft, T.L.; van Heerden, E.; Onstott, T.C. The biomass and biodiversity of the continental subsurface. Nat. Geosci. 2018, 11, 707–717. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, M.E.; Simo, J.A.; Freiberg, P.G. Stratigraphic and geochemical controls on naturally occurring arsenic in groundwater, eastern Wisconsin, USA. Hydrogeol. J. 2000, 8, 161–176. [Google Scholar] [CrossRef]
- Phelps, T.J.; Raione, E.G.; White, D.C.; Fliermans, C.B. Microbial activities in deep subsurface environments. Geomicrobiol. J. 1989, 7, 79–91. [Google Scholar] [CrossRef]
- Reith, F. Life in the deep subsurface. Geology 2011, 39, 287–288. [Google Scholar] [CrossRef]
- Morais, S.; Vidal, E.; Cario, A.; Marre, S.; Ranchou-Peyruse, A. Microfluidics for studying the deep underground biosphere: From applications to fundamentals. FEMS Microbiol. Ecol. 2024, 100, fiae151. [Google Scholar] [CrossRef]
- Neu, A.T.; Allen, E.E.; Roy, K. Defining and quantifying the core microbiome: Challenges and prospects. Proc. Natl. Acad. Sci. USA 2021, 118, e2104429118. [Google Scholar] [CrossRef]
- Lowe, B.A.; Marsh, T.L.; Isaacs-Cosgrove, N.; Kirkwood, R.N.; Kiupel, M.; Mulks, M.H. Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiol. 2012, 12, 20. [Google Scholar] [CrossRef]
- Hernandez-Agreda, A.; Gates, R.D.; Ainsworth, T.D. Defining the Core Microbiome in Corals’ Microbial Soup. Trends Microbiol. 2017, 25, 125–140. [Google Scholar] [CrossRef]
- Forster, R.J.; Mohan, A.M.; Bibby, K.J.; Lipus, D.; Hammack, R.W.; Gregory, K.B. The Functional Potential of Microbial Communities in Hydraulic Fracturing Source Water and Produced Water from Natural Gas Extraction Characterized by Metagenomic Sequencing. PLoS ONE 2014, 9, e107682. [Google Scholar] [CrossRef]
- Konno, U.; Kouduka, M.; Komatsu, D.D.; Ishii, K.; Fukuda, A.; Tsunogai, U.; Ito, K.; Suzuki, Y. Novel Microbial Populations in Deep Granitic Groundwater from Grimsel Test Site, Switzerland. Microb. Ecol. 2013, 65, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Hallbeck, L.; Pedersen, K. Characterization of microbial processes in deep aquifers of the Fennoscandian Shield. Appl. Geochem. 2008, 23, 1796–1819. [Google Scholar] [CrossRef]
- Ino, K.; Konno, U.; Kouduka, M.; Hirota, A.; Togo, Y.S.; Fukuda, A.; Komatsu, D.; Tsunogai, U.; Tanabe, A.S.; Yamamoto, S.; et al. Deep microbial life in high-quality granitic groundwater from geochemically and geographically distinct underground boreholes. Environ. Microbiol. Rep. 2016, 8, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.; Liu, S.V.; Li, G.; Huang, H.; Phelps, T.J.; Zhou, J. Isolation and Characterization of Metal-ReducingThermoanaerobacter Strains from Deep Subsurface Environments of the Piceance Basin, Colorado. Appl. Environ. Microbiol. 2002, 68, 6013–6020. [Google Scholar] [CrossRef]
- Payler, S.J.; Biddle, J.F.; Sherwood Lollar, B.; Fox-Powell, M.G.; Edwards, T.; Ngwenya, B.T.; Paling, S.M.; Cockell, C.S. An Ionic Limit to Life in the Deep Subsurface. Front. Microbiol. 2019, 10, 426. [Google Scholar] [CrossRef]
- Shu, W.-S.; Huang, L.-N. Microbial diversity in extreme environments. Nat. Rev. Microbiol. 2021, 20, 219–235. [Google Scholar] [CrossRef]
- Gittins, D.A.; Bhatnagar, S.; Hubert, C.R.J.; Goordial, J. Environmental Selection and Biogeography Shape the Microbiome of Subsurface Petroleum Reservoirs. mSystems 2023, 8, e0088422. [Google Scholar] [CrossRef]
- Liu, W.; Liu, L.; Yang, X.; Deng, M.; Wang, Z.; Wang, P.; Yang, S.; Li, P.; Peng, Z.; Yang, L.; et al. Long-term nitrogen input alters plant and soil bacterial, but not fungal beta diversity in a semiarid grassland. Glob. Change Biol. 2021, 27, 3939–3950. [Google Scholar] [CrossRef]
- Acciardo, A.S.; Arnet, M.; Gholizadeh Doonechaly, N.; Ceccato, A.; Rodriguez, P.; Tran, H.N.H.; Wenning, Q.; Zimmerman, E.; Hertrich, M.; Brixel, B.; et al. Spatial and temporal groundwater biogeochemical variability help inform subsurface connectivity within a high-altitude Alpine catchment (Riale di Ronco, Switzerland). Front. Microbiol. 2025, 16, 1522714. [Google Scholar] [CrossRef]
- Rast, M.; Galli, A.; Ruh, J.B.; Guillong, M.; Madonna, C. Geology along the Bedretto tunnel: Kinematic and geochronological constraints on the evolution of the Gotthard Massif (Central Alps). Swiss J. Geosci. 2022, 115, 8. [Google Scholar] [CrossRef]
- Ma, X.; Hertrich, M.; Amann, F.; Bröker, K.; Gholizadeh Doonechaly, N.; Gischig, V.; Hochreutener, R.; Kästli, P.; Krietsch, H.; Marti, M.; et al. Multi-disciplinary characterizations of the BedrettoLab—A new underground geoscience research facility. Solid Earth 2022, 13, 301–322. [Google Scholar] [CrossRef]
- Hafner, S. Petrographie des Südwestlichen Gotthardmassivs Zwischen St.Gotthardpass und Nufenenpass. Ph.D. Thesis, ETH Zurich, Zurich, Switzerland, 1958. [Google Scholar]
- Jamieson, J.W.; Wing, B.A.; Farquhar, J.; Hannington, M.D. Neoarchaean seawater sulphate concentrations from sulphur isotopes in massive sulphide ore. Nat. Geosci. 2012, 6, 61–64. [Google Scholar] [CrossRef]
- Kneafsey, T.J.; Blankenship, D.A.; Dobson, P.F.; Morris, J.; White, M.D.; Fu, P.; Schwering, P.C. The EGS Collab Project: Learnings from Experiment 1; Stanford University: Stanford, CA, USA, 2020. [Google Scholar]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2015, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Dekas, A.E.; Hawkins, A.J.; Parada, A.E.; Gorbatenko, O.; Li, K.; Horne, R.N. Microbial Community Composition in Deep-Subsurface Reservoir Fluids Reveals Natural Interwell Connectivity. Water Resour. Res. 2020, 56, e2019WR025916. [Google Scholar] [CrossRef]
- Zhang, Y.; Horne, R.N.; Hawkins, A.J.; Primo, J.C.; Gorbatenko, O.; Dekas, A.E. Geological activity shapes the microbiome in deep-subsurface aquifers by advection. Proc. Natl. Acad. Sci. USA 2022, 119, e2113985119. [Google Scholar] [CrossRef]
- Zhang, Y.; Dekas, A.E.; Hawkins, A.J.; Primo, J.C.; Gorbatenko, O.; Huang, T.; Pang, Z.; Horne, R.N. Transportability of exogenous microbial community correlates with interwell connectivity in deep aquifers. Water Res. 2025, 285, 124008. [Google Scholar] [CrossRef]
- Ford, S.E.; Slater, G.F.; Engel, K.; Warr, O.; Lollar, G.S.; Brady, A.; Neufeld, J.D.; Lollar, B.S. Deep terrestrial indigenous microbial community dominated by Candidatus Frackibacter. Commun. Earth Environ. 2024, 5, 795. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Watson, M.; McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinform. J. 2013, 7, 1–8. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Dobson, P.F.; Kneafsey, T.J.; Nakagawa, S.; Sonnenthal, E.L.; Voltolini, M.; Smith, J.T.; Borglin, S.E. Fracture Sustainability in Enhanced Geothermal Systems: Experimental and Modeling Constraints. J. Energy Resour. Technol. 2021, 143, 100901. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Charrad, M.; Ghazzali, N.; Boiteau, V.; Niknafs, A. NbClust: AnRPackage for Determining the Relevant Number of Clusters in a Data Set. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]
- Gentle, J.E.; Kaufman, L.; Rousseuw, P.J. Finding Groups in Data: An Introduction to Cluster Analysis. Biometrics 1991, 47, 788. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Shi, C.; Liu, L.; Han, J.; Yang, Q.; Wang, Y.; Li, X.; Fu, W.; Gao, H.; Huang, H.; et al. Majorbio Cloud 2024: Update single-cell and multiomics workflows. iMeta 2024, 3, e217. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Peng, C.; Liu, Y.; Qin, Y.; Sun, D.; Jia, J.; Xie, Z.; Gong, B. Dissolved Oxygen Decline in Northern Beibu Gulf Summer Bottom Waters: Reserve Management Insights from Microbiome Analysis. Microorganisms 2025, 13, 1945. [Google Scholar] [CrossRef]
- Abdi, H. Z-scores. Encycl. Meas. Stat. 2007, 3, 1055–1058. [Google Scholar]
- Li, L.; Wing, B.A.; Bui, T.H.; McDermott, J.M.; Slater, G.F.; Wei, S.; Lacrampe-Couloume, G.; Lollar, B.S. Sulfur mass-independent fractionation in subsurface fracture waters indicates a long-standing sulfur cycle in Precambrian rocks. Nat. Commun. 2016, 7, 13252. [Google Scholar] [CrossRef]
- Warr, O.; Young, E.D.; Giunta, T.; Kohl, I.E.; Ash, J.L.; Sherwood Lollar, B. High-resolution, long-term isotopic and isotopologue variation identifies the sources and sinks of methane in a deep subsurface carbon cycle. Geochim. Cosmochim. Acta 2021, 294, 315–334. [Google Scholar] [CrossRef]
- Lollar, B.S.; Lacrampe-Couloume, G.; Voglesonger, K.; Onstott, T.C.; Pratt, L.M.; Slater, G.F. Isotopic signatures of CH4 and higher hydrocarbon gases from Precambrian Shield sites: A model for abiogenic polymerization of hydrocarbons. Geochim. Cosmochim. Acta 2008, 72, 4778–4795. [Google Scholar] [CrossRef]
- Chen, P.; Mei, T.; He, X.; Lin, Y.; He, Z.; Kong, X. Impacts of Lead and Nanoplastic Co-Exposure on Decomposition, Microbial Diversity, and Community Assembly Mechanisms in Karst Riverine Miscanthus Litter. Microorganisms 2025, 13, 2172. [Google Scholar] [CrossRef]
- Uhl, B.; Schall, P.; Bässler, C. Achieving structural heterogeneity and high multi-taxon biodiversity in managed forest ecosystems: A European review. Biodivers. Conserv. 2024, 34, 3327–3358. [Google Scholar] [CrossRef]
- Cassol, I.; Ibañez, M.; Bustamante, J.P. Key features and guidelines for the application of microbial alpha diversity metrics. Sci. Rep. 2025, 15, 622. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Peng, C.; Cao, H.; Song, J.; Gong, B.; Li, L.; Wang, L.; He, Y.; Liang, M.; Lin, J.; et al. Microbial functional assemblages predicted by the FAPROTAX analysis are impacted by physicochemical properties, but C, N and S cycling genes are not in mangrove soil in the Beibu Gulf, China. Ecol. Indic. 2022, 139, 108887. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can We Use Functional Annotation of Prokaryotic Taxa (FAPROTAX) to Assign the Ecological Functions of Soil Bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Chandler, L.; Harford, A.J.; Hose, G.C.; Humphrey, C.L.; Chariton, A.; Greenfield, P.; Davis, J. Saline mine-water alters the structure and function of prokaryote communities in shallow groundwater below a tropical stream. Environ. Pollut. 2021, 284, 117318. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, H.; Chen, H.; Xiong, H.; Tong, L.; Guo, G. Is redox zonation an appropriate method for determining the stage of natural remediation in deep contaminated groundwater? Sci. Total Environ. 2024, 928, 172224. [Google Scholar] [CrossRef]
- Xu, F.; Li, P. Biogeochemical mechanisms of iron (Fe) and manganese (Mn) in groundwater and soil profiles in the Zhongning section of the Weining Plain (northwest China). Sci. Total Environ. 2024, 939, 173506. [Google Scholar] [CrossRef]
- Ma, J.; Liu, H.; Zhang, C.; Ding, K.; Chen, R.; Liu, S. Joint response of chemistry and functional microbial community to oxygenation of the reductive confined aquifer. Sci. Total Environ. 2020, 720, 137587. [Google Scholar] [CrossRef]
- Durand, S.; Guillier, M. Transcriptional and Post-transcriptional Control of the Nitrate Respiration in Bacteria. Front. Mol. Biosci. 2021, 8, 667758. [Google Scholar] [CrossRef] [PubMed]
- Caddey, S.W.; Bachman, R.L.; Campbell, T.J.; Reid, R.R.; Otto, R.P. The Homestake Gold Mine, an Early Proterozoic Iron-Formation-Hosted Gold Deposit, Lawrence County, South Dakota; US Government Printing Office: Reston, VA, USA, 1991. [CrossRef]
- Brisson, V.; Schmidt, J.; Northen, T.R.; Vogel, J.P.; Gaudin, A. A New Method to Correct for Habitat Filtering in Microbial Correlation Networks. Front. Microbiol. 2019, 10, 585. [Google Scholar] [CrossRef] [PubMed]
- Horner-Devine, M.C.; Carney, K.M.; Bohannan, B.J.M. An ecological perspective on bacterial biodiversity. Proc. R. Soc. London. Ser. B Biol. Sci. 2004, 271, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Fujii, M.; Ibrahim, M.G.; Elreedy, A. On the potential of halophiles enriched from hypersaline sediments for biohydrogen production from saline wastewater. J. Clean. Prod. 2022, 341, 130901. [Google Scholar] [CrossRef]
- Wang, S.; Hu, Y.; Fan, T.; Fang, W.; Liu, X.; Xu, L.; Li, B.; Wei, X. Microbial Community Structure and Co-Occurrence Patterns in Closed and Open Subsidence Lake Ecosystems. Water 2023, 15, 1829. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Teng, D.; Yang, X.; Zhang, Y.; Li, Y. Metagenomic Insights into Microbial Community Structure, Function, and Salt Adaptation in Saline Soils of Arid Land, China. Microorganisms 2022, 10, 2183. [Google Scholar] [CrossRef]
- Nixon, S.L.; Daly, R.A.; Borton, M.A.; Solden, L.M.; Welch, S.A.; Cole, D.R.; Mouser, P.J.; Wilkins, M.J.; Wrighton, K.C.; Suen, G. Genome-Resolved Metagenomics Extends the Environmental Distribution of the Verrucomicrobia Phylum to the Deep Terrestrial Subsurface. mSphere 2019, 4, e00613-19. [Google Scholar] [CrossRef]
- Kothari, A.; Roux, S.; Zhang, H.; Prieto, A.; Soneja, D.; Chandonia, J.-M.; Spencer, S.; Wu, X.; Altenburg, S.; Fields, M.W.; et al. Ecogenomics of Groundwater Phages Suggests Niche Differentiation Linked to Specific Environmental Tolerance. mSystems 2021, 6, e0053721. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef]
- Ricotta, C.; Podani, J. On some properties of the Bray-Curtis dissimilarity and their ecological meaning. Ecol. Complex. 2017, 31, 201–205. [Google Scholar] [CrossRef]
- Boeraş, I.; Burcea, A.; Coman, C.; Bănăduc, D.; Curtean-Bănăduc, A. Bacterial Microbiomes in the Sediments of Lotic Systems Ecologic Drivers and Role: A Case Study from the Mureş River, Transylvania, Romania. Water 2021, 13, 3518. [Google Scholar] [CrossRef]
- Shah, T.; Liu, Q.; Yin, G.; Shah, Z.; Li, H.; Wang, J.; Wang, B.; Xia, X. Structural and Functional Differences in the Gut and Lung Microbiota of Pregnant Pomona Leaf-Nosed Bats. Microorganisms 2025, 13, 1887. [Google Scholar] [CrossRef] [PubMed]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K.; Tringe, S.G. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, T.D.; Krause, L.; Bridge, T.; Torda, G.; Raina, J.-B.; Zakrzewski, M.; Gates, R.D.; Padilla-Gamiño, J.L.; Spalding, H.L.; Smith, C.; et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015, 9, 2261–2274. [Google Scholar] [CrossRef]
- Pinar-Méndez, A.; Wangensteen, O.S.; Præbel, K.; Galofré, B.; Méndez, J.; Blanch, A.R.; García-Aljaro, C. Monitoring Bacterial Community Dynamics in a Drinking Water Treatment Plant: An Integrative Approach Using Metabarcoding and Microbial Indicators in Large Water Volumes. Water 2022, 14, 1435. [Google Scholar] [CrossRef]
- Suzuki, T.; Okamura, Y.; Calugay, R.J.; Takeyama, H.; Matsunaga, T. Global Gene Expression Analysis of Iron-Inducible Genes in Magnetospirillum magneticum AMB-1. J. Bacteriol. 2006, 188, 2275–2279. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Li, X.; Huang, T.; Li, Y.; Pang, Z.; Zhang, Y. Multi-Regional Study on the Microbial Community Structure, Core Microbiome and Functional Characteristics in Deep Fracture Waters. Microorganisms 2026, 14, 45. https://doi.org/10.3390/microorganisms14010045
Li X, Huang T, Li Y, Pang Z, Zhang Y. Multi-Regional Study on the Microbial Community Structure, Core Microbiome and Functional Characteristics in Deep Fracture Waters. Microorganisms. 2026; 14(1):45. https://doi.org/10.3390/microorganisms14010045
Chicago/Turabian StyleLi, Xiaoxuan, Tianming Huang, Yiman Li, Zhonghe Pang, and Yuran Zhang. 2026. "Multi-Regional Study on the Microbial Community Structure, Core Microbiome and Functional Characteristics in Deep Fracture Waters" Microorganisms 14, no. 1: 45. https://doi.org/10.3390/microorganisms14010045
APA StyleLi, X., Huang, T., Li, Y., Pang, Z., & Zhang, Y. (2026). Multi-Regional Study on the Microbial Community Structure, Core Microbiome and Functional Characteristics in Deep Fracture Waters. Microorganisms, 14(1), 45. https://doi.org/10.3390/microorganisms14010045







