The CRISPR-Cas9 System in Entamoeba histolytica Trophozoites: ehcp112 Gene Knockout and Effects on Other Genes in the V1 Virulence Locus
Abstract
1. Introduction
2. Materials and Methods
2.1. E. histolytica Cultures
2.2. In Silico sgRNA Design and Production
2.3. In Vitro Transcription to Produce sgRNAs
2.4. PCR Assays
2.5. Production and Purification of Cas9 Endonuclease
2.6. RNP Formation and In Vitro Assays
2.7. Soaking of E. histolytica Trophozoites with RNPs
2.8. Total RNA Extraction and RT-qPCR Assays
2.9. Growth Rate Assays
2.10. Laser Confocal Immunofluorescence
2.11. Transmission Electron Microscopy (TEM)
2.12. Scanning Electron Microscopy
2.13. Western Blot Assays
2.14. Protease Activity
2.15. Statistical Analyses
3. Results
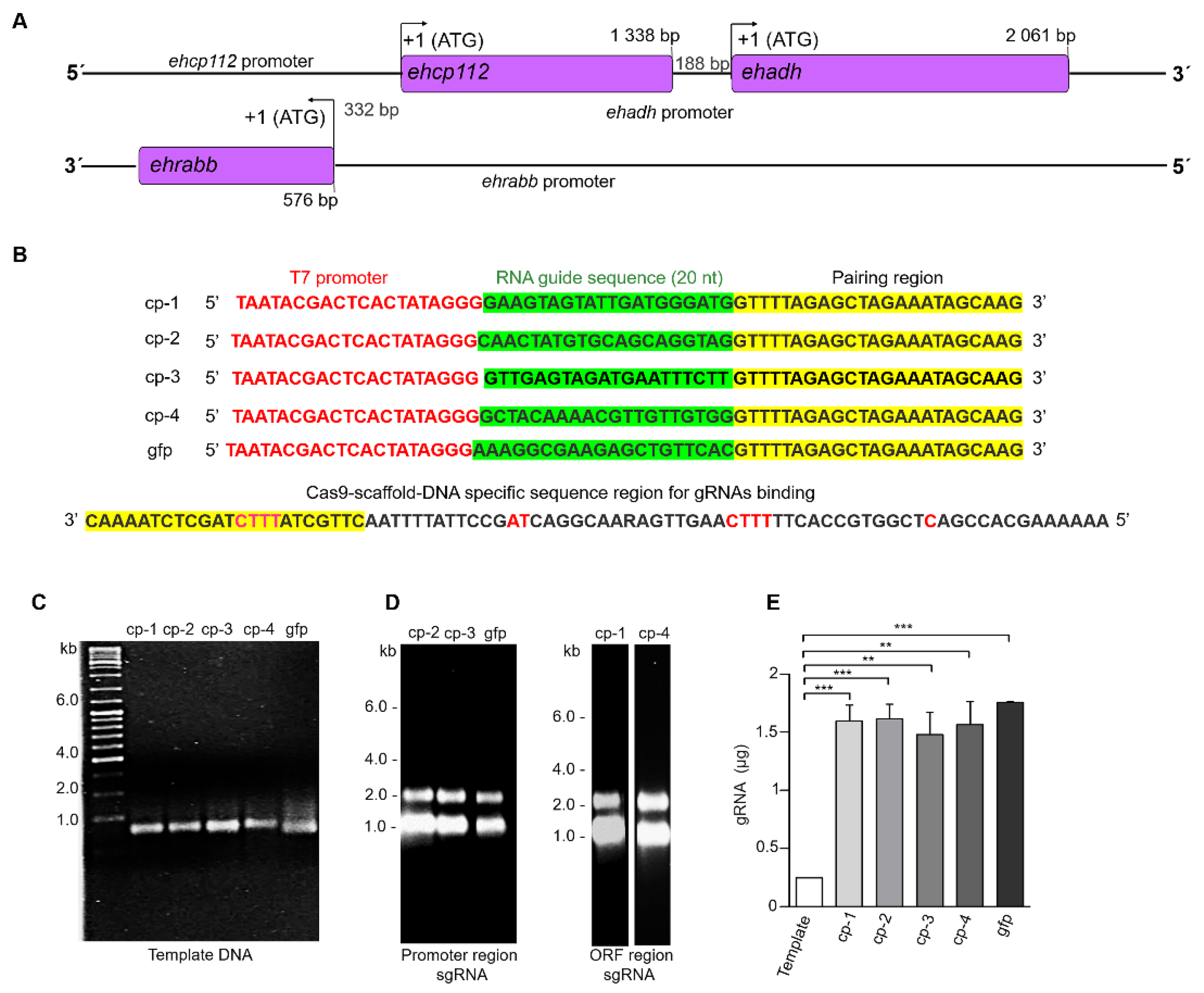
3.1. sgRNA Design and Production
3.2. The ehcp112 Gene Was In Vitro Edited Using the CRISPR-Cas9 System
3.3. The RNPs Formed by the Cas9 Enzyme and cp-gRNAs Are Efficiently Introduced into the Cytoplasm and Nucleus of E. histolytica Trophozoites
3.4. Transmission Electron Microscopy (TEM) Evidenced That the RNPs Reached the Chromatin in E. histolytica Trophozoites
3.5. EhCP112-KO Trophozoites Grew at a Similar Rate than the HM1:IMSS Strain, but They Presented Changes in Their Morphology
3.6. The ehcp112 Gene Sequence Is Altered and the Gene Is Not Expressed in the EhCP112-KO Trophozoites
3.7. The EhCP112 Protein Is Expressed in HM1:IMSS but Not in EhCP112-KO Trophozoites
3.8. The ehcp112 Gene Knockout Augments the Enzymatic Activity of EhCP112-KO Trophozoite Extracts but Its Activity Diminished in the Secretion Products
3.9. The ehcp112 Gene Knockout Affects Other Genes of the V1 Locus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chou, A.; Austin, R.L. Entamoeba histolytica Infection. In StatPearls Internet; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar] [PubMed]
- Mathur, S.; Kaushik, S.; Kothari, S.L.; Srivastava, V.K. Role of various virulence factors involved in the pathogenesis of Entamoeba histolytica. Exp. Parasitol. 2024, 266, 108841. [Google Scholar] [CrossRef] [PubMed]
- Orozco, M.E.; Guarneros, G.; Martínez Palomo, A. Clones of E. histolytica deficient in phagocytosis present a deficiency in virulence. Arch. Investig. Médica 1982, 13 (Suppl. 3), 137–143. [Google Scholar]
- Zhang, H.; Pompey, J.M.; Singh, U. RNA interference in Entamoeba histolytica: Implications for parasite biology and gene silencing. Future Microbiol. 2011, 6, 103–117. [Google Scholar] [CrossRef]
- Khalil, M.I.; Foda, B.M.; Suresh, S.; Singh, U. Technical advances in trigger-induced RNA interference gene silencing in the parasite Entamoeba histolytica. Int. J. Parasitol. 2016, 46, 205–212. [Google Scholar] [CrossRef]
- Bhattacharya, S. Episomal and chromosomal DNA replication and recombination in Entamoeba histolytica. Front. Mol. Biosci. 2023, 10, 1212082. [Google Scholar] [CrossRef]
- Kangussu-Marcolino, M.M.; Morgado, P.; Manna, D.; Yee, H.; Singh, U. Development of a CRISPR/Cas9 system in Entamoeba histolytica: Proof of concept. Int. J. Parasitol. 2021, 51, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Hildebrandt, F.; Matias, A.N.; Treeck, M. A CRISPR view on genetic screens in Toxoplasma gondii. Curr. Opin. Microbiol. 2025, 83, 102577. [Google Scholar] [CrossRef] [PubMed]
- Di Cristina, M.; Carruthers, V.B. New and emerging uses of CRISPR/Cas9 to genetically manipulate apicomplexan parasites. Parasitology 2018, 145, 1119–1126. [Google Scholar] [CrossRef]
- Zakharova, A.; Saura, A.; Butenko, A.; Podešvová, L.; Warmusová, S.; Kostygov, A.Y.; Nenarokova, A.; Lukeš, J.; Opperdoes, F.R.; Yurchenko, V. A New Model Trypanosomatid, Novymonas esmeraldas: Genomic perception of its ‘Candidatus Pandoraea novymonadis’ endosymbiont. mBio 2021, 12. [Google Scholar] [CrossRef]
- Pawlowic, M.C.; Vinayak, S.; Sateriale, A.; Brooks, C.F.; Striepen, B. Generating and maintaining transgenic Cryptosporidium parvum parasites. Curr. Protoc. Microbiol. 2017, 46. [Google Scholar] [CrossRef]
- Sollelis, L.; Ghorbal, M.; MacPherson, C.R.; Martins, R.M.; Kuk, N.; Crobu, L.; Bastien, P.; Scherf, A.; Lopez-Rubio, J.-J.; Sterkers, Y. First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell. Microbiol. 2015, 17, 1405–1412. [Google Scholar] [CrossRef]
- Janssen, B.D.; Chen, Y.P.; Molgora, B.M.; Wang, S.E.; Simoes-Barbosa, A.; Johnson, P.J. CRISPR/Cas9-mediated gene modification and gene knock out in the human-infective parasite Trichomonas vaginalis. Sci. Rep. 2018, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, Y.; Zhang, T. Computational approaches for effective CRISPR guide RNA design and evaluation. Comput. Struct. Biotechnol. J. 2020, 18, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Bañuelos, C.; García-Rivera, G.; López-Reyes, I.; Mendoza, L.; González-Robles, A.; Herranz, S.; Vincent, O.; Orozco, E. EhADH112 is a Bro1 domain-containing protein involved in the Entamoeba histolytica multivesicular bodies pathway. J. Biomed. Biotechnol. 2012, 2012, 657942. [Google Scholar] [CrossRef]
- Rodríguez, M.A.; García-Pérez, R.M.; García-Rivera, G.; López-Reyes, I.; Mendoza, L.; Ortiz-Navarrete, V.; Orozco, E. An Entamoeba histolytica Rab-like encoding gene and protein: Function and cellular location. Mol. Biochem. Parasitol. 2000, 108, 199–206. [Google Scholar] [CrossRef]
- Galindo, A.; Javier-Reyna, R.; García-Rivera, G.; Bañuelos, C.; Chávez-Munguía, B.; Salazar-Villatoro, L.; Orozco, E. EhVps23, an ESCRT-I member, is a key factor in secretion, motility, phagocytosis and tissue invasion by Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2022, 12, 835654. [Google Scholar] [CrossRef]
- Ocádiz, R.; Orozco, E.; Carrillo, E.; Quintas, L.I.; Ortega-López, J.; García-Pérez, R.M.; Sánchez, T.; Castillo-Juárez, B.A.; García-Rivera, G.; Rodríguez, M.A. EhCP112 is an Entamoeba histolytica secreted cysteine protease that may be involved in the parasite-virulence. Cell. Microbiol. 2005, 7, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Rigothier, M.C.; García-Rivera, G.; Guaderrama, M.; Orozco, E. Purification and functional characterization of the 112 kDa adhesin of Entamoeba histolytica. Arch. Med. Res. 1992, 23, 239–241. [Google Scholar] [PubMed]
- Garcia-Rivera, G.; Rodriguez, M.A.; Ocadiz, R.; Martinez-Lopez, M.C.; Arroyo, R.; Gonzalez-Robles, A.; Orozco, E. Entamoeba histolytica: A novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol. Microbiol. 1999, 33, 556–568. [Google Scholar] [CrossRef]
- Yang, N.; Matthew, M.A.; Yao, C. Roles of cysteine proteases in biology and pathogenesis of parasites. Microorganisms 2023, 11, 1397. [Google Scholar] [CrossRef]
- Bruchhaus, I.; Loftus, B.J.; Hall, N.; Tannich, E. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2003, 2, 501–509. [Google Scholar] [CrossRef]
- Brás, X.P.; Zimorski, V.; Bolte, K.; Maier, U.G.; Martin, W.F.; Gould, S.B. Knockout of the abundant Trichomonas vaginalis hydrogenosomal membrane protein TvHMP23 increases hydrogenosome size but induces no compensatory up-regulation of paralogous copies. FEBS Lett. 2013, 587, 1333–1339. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Dolatshad, H.; Valletta, S.; Boultwood, J. Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Arch. Toxicol. 2015, 89, 1023–1034. [Google Scholar] [CrossRef]
- Jackson, R.N.; van Erp, P.B.; Sternberg, S.H.; Wiedenheft, B. Conformational regulation of CRISPR-associated nucleases. Curr. Opin. Microbiol. 2017, 37, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Grzybek, M.; Golonko, A.; Górska, A.; Szczepaniak, K.; Strachecka, A.; Lass, A.; Lisowski, P. The CRISPR/Cas9 system sheds new lights on the biology of protozoan parasites. Appl. Microbiol. Biotechnol. 2018, 102, 4629–4640. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Jinek, M. In vitro enzymology of Cas9. Methods Enzymol. 2014, 546, 1–20. [Google Scholar] [CrossRef]
- Mehravar, M.; Shirazi, A.; Mehrazar, M.M.; Nazari, M. In vitro pre-validation of gene editing by CRISPR/Cas9 ribonucleoprotein. Avicenna J. Med. Biotechnol. 2019, 11, 259–263. [Google Scholar] [PubMed]
- Solis, C.F.; Guillén, N. Silencing genes by RNA interference in the protozoan parasite Entamoeba histolytic. In RNAi: Design and Application; Humana Press: Totowa, NJ, USA, 2008; pp. 113–128. [Google Scholar] [CrossRef]
- Javier-Reyna, R.; Montaño, S.; García-Rivera, G.; Rodríguez, M.A.; González-Robles, A.; Orozco, E. EhRabB mobilises the EhCPADH complex through the actin cytoskeleton during phagocytosis of Entamoeba histolytica. Cell. Microbiol. 2019, 21, e13071. [Google Scholar] [CrossRef] [PubMed]
- Orozco, E.; Guarneros, G.; Martinez-Palomo, A.; Sánchez, T. Entamoeba histolytica. Phagocytosis as a virulence factor. J. Exp. Med. 1983, 158, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Ocádiz-Ruiz, R.; Fonseca, W.; Martínez, M.B.; Ocádiz-Quintanar, R.; Orozco, E.; Rodríguez, M.A. Effect of the silencing of the Ehcp112 gene on the in vitro virulence of Entamoeba histolytica. Parasites Vectors 2013, 6, 248. [Google Scholar] [CrossRef] [PubMed]
- Azuara-Liceaga, E.; Flores-Soto, E.; López-Camarillo, C.; Orozco, E. Entamoeba histolytica: Structural and functional analysis of the Ehadh112 gene promoter. Exp. Parasitol. 2005, 110, 280–285. [Google Scholar] [CrossRef] [PubMed]








| gRNA | Sequence (PAM “NGG”) | Location | % GC | Off Target | Score |
|---|---|---|---|---|---|
| cp-1 | GAAGTAGTATTGATGGGATGGGG | ORF-985 bp | 40% | no | 0.62 |
| cp-2 | CAACTATGTGCAGCAGGTAGTGG | Promoter 662 bp reverse | 50% | no | 0.61 |
| cp-3 | GTTGAGTAGATGAATTTCTTTGG | Promoter 502 bp | 30% | no | 0.59 |
| cp-4 | GCTACAAAACGTTGTTGTGGTGG | ORF-449 bp | 45% | no | 0.58 |
| cp-5 | AATGGCGGTACTTCATTCCATGG | ORF-337 bp reverse | 45% | no | 0.57 |
| cp-6 | GCAGGATCTGACTTTCTCATTGG | Promoter 244 bp | 45% | no | 0.56 |
| cp-7 | GAAGTACCGCCATTACCTTCTGG | ORF-364 bp | 50% | no | 0.56 |
| cp-8 | GGGTGTTGTTGCATGGCTCTAGG | Promoter 539 bp reverse | 55% | no | 0.56 |
| cp-9 | GTTCTGAAGTGCATAGGTAAAGG | Promoter 281 bp | 40% | no | 0.56 |
| cp-10 | AAATTCATGGGCTAGTGGATGGG | Promoter 480 bp | 40% | no | 0.56 |
| Primers | Sequence | Assay |
|---|---|---|
| cp-1 Forward | 5′ CAAAACGTTGTTGTGGTGGTC 3′ | PCR |
| cp-1 Reverse | 5′ TGATTGTAGAATTGGACATAGGTTG 3′ | PCR |
| cp-2 Forward | 5′ TGCCTTTACCTATGCACTTCAGA 3′ | PCR |
| cp-2 Reverse | 5′ TGCCACTCTAAGTCGTTGGAC 3′ | PCR |
| cp-3 Forward | 5′ AACACCTTCACCAGTTTTGGC 3′ | PCR |
| cp-3 Reverse | 5′ CCCATCCACTAGCCCATGAA 3′ | PCR |
| cp-4 Forward | 5′ CAGCGATTGTTGTCGCTTTTT 3′ | PCR |
| cp-4 Reverse | 5′ CCCATCCACTAGCCCATGAA 3′ | PCR |
| cp112-Forward | 5′ GGAGGTTGTTGGGCAGTTTC 3′ | RT-qPCR |
| cp112-Reverse | 5′ CTTCACCCCATCCACTAGCC 3′ | RT-qPCR |
| Eh40s-Forward | 5′ ATTCGGAAATAGAAGAGGAGG 3′ | RT-qPCR |
| Eh40s-Reverse | 5′ ACTAATCTTCCAAGCTTGGT 3′ | RT-qPCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reyes, L.V.; García-Rivera, G.; Javier-Reyna, R.; Morales-Rios, E.; Tinajero, S.; Bañuelos, C.; Talamás-Lara, D.; Orozco, E. The CRISPR-Cas9 System in Entamoeba histolytica Trophozoites: ehcp112 Gene Knockout and Effects on Other Genes in the V1 Virulence Locus. Microorganisms 2025, 13, 2219. https://doi.org/10.3390/microorganisms13092219
Reyes LV, García-Rivera G, Javier-Reyna R, Morales-Rios E, Tinajero S, Bañuelos C, Talamás-Lara D, Orozco E. The CRISPR-Cas9 System in Entamoeba histolytica Trophozoites: ehcp112 Gene Knockout and Effects on Other Genes in the V1 Virulence Locus. Microorganisms. 2025; 13(9):2219. https://doi.org/10.3390/microorganisms13092219
Chicago/Turabian StyleReyes, Luz Virginia, Guillermina García-Rivera, Rosario Javier-Reyna, Edgar Morales-Rios, Sergio Tinajero, Cecilia Bañuelos, Daniel Talamás-Lara, and Esther Orozco. 2025. "The CRISPR-Cas9 System in Entamoeba histolytica Trophozoites: ehcp112 Gene Knockout and Effects on Other Genes in the V1 Virulence Locus" Microorganisms 13, no. 9: 2219. https://doi.org/10.3390/microorganisms13092219
APA StyleReyes, L. V., García-Rivera, G., Javier-Reyna, R., Morales-Rios, E., Tinajero, S., Bañuelos, C., Talamás-Lara, D., & Orozco, E. (2025). The CRISPR-Cas9 System in Entamoeba histolytica Trophozoites: ehcp112 Gene Knockout and Effects on Other Genes in the V1 Virulence Locus. Microorganisms, 13(9), 2219. https://doi.org/10.3390/microorganisms13092219








