Short-Term Maize Rotation Suppresses Verticillium Wilt and Restructures Soil Microbiomes in Xinjiang Cotton Fields
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Site Selection and Verticillium Wilt Assessment
2.2. Soil Sampling and Analysis
2.3. DNA Extraction and High-Throughput Sequencing
2.4. Bioinformatics and Statistical Analyses
3. Results
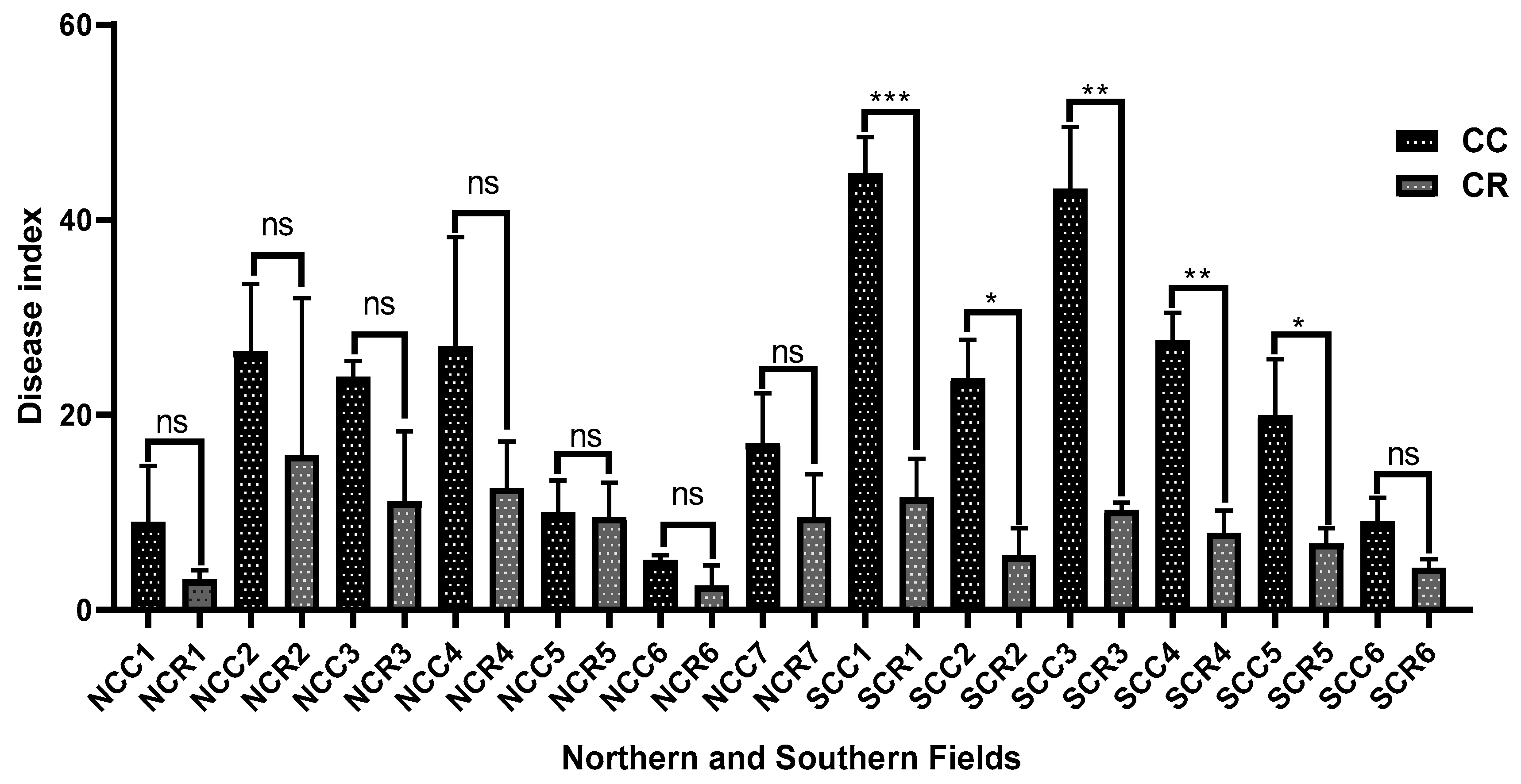
3.1. Comparative Analysis of Verticillium Wilt Suppression
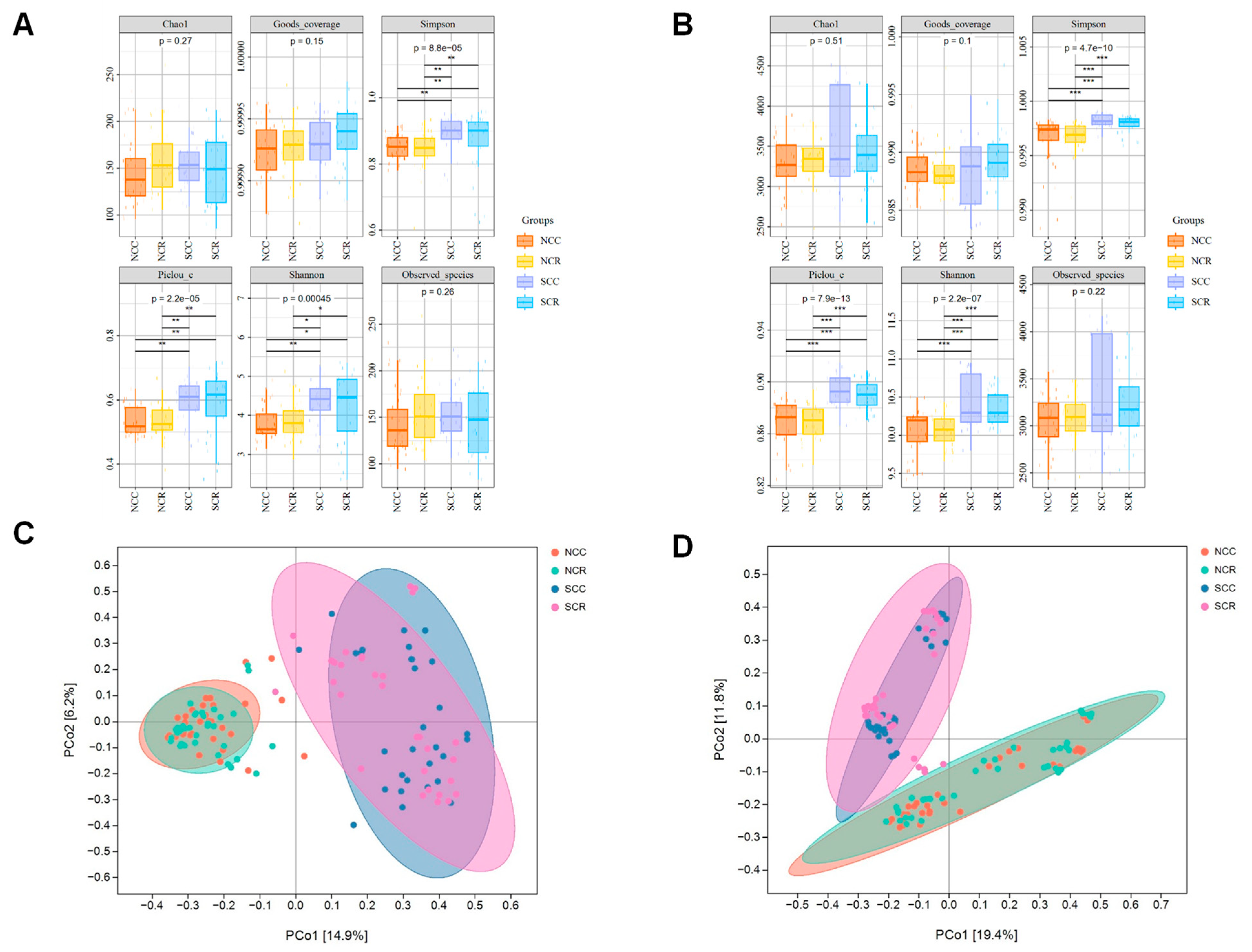
3.2. Soil Microbiome Diversity Under Short-Term Rotation
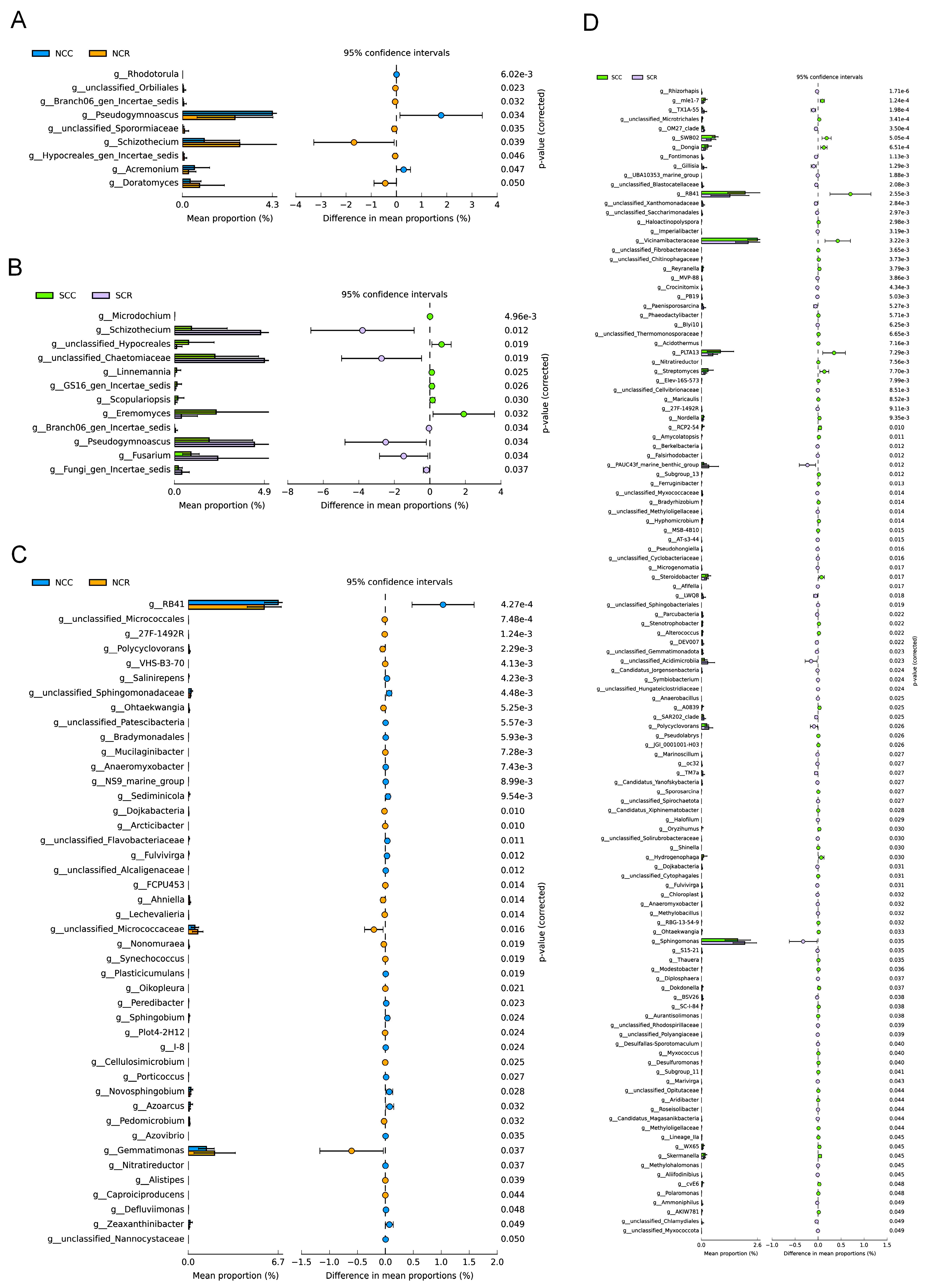
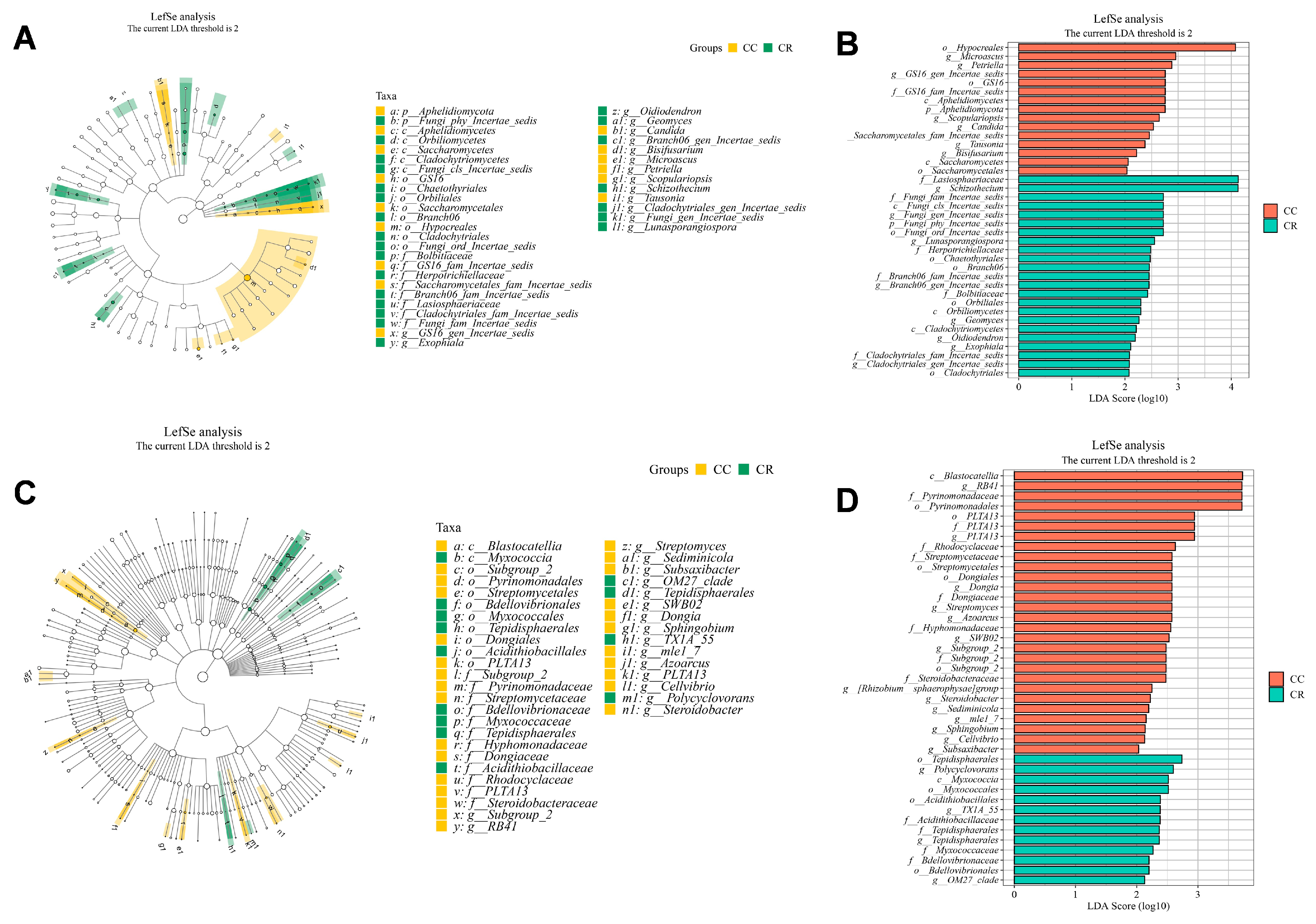
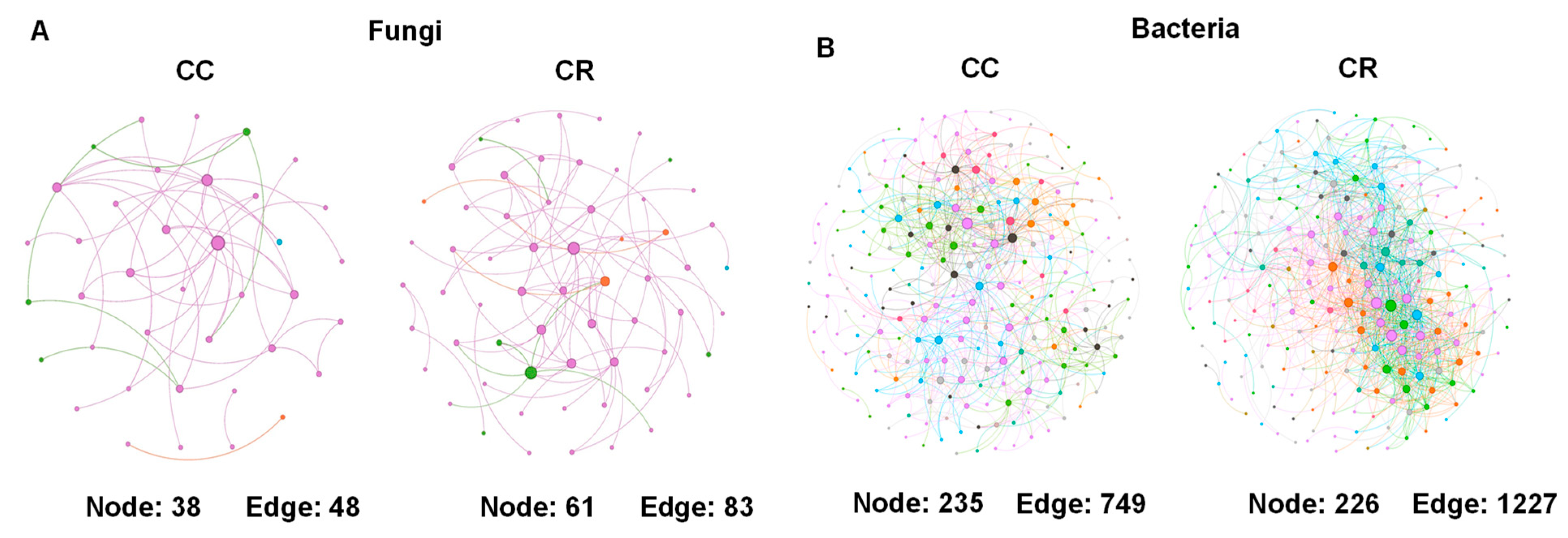
3.3. Short Rotation-Induced Shifts in Microbiome Composition and Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NCC | North Cotton continuous cropping |
| NCR | North Cotton–Maize rotation |
| SCC | South Cotton continuous cropping |
| SCR | South Cotton–Maize rotation |
References
- Zhou, Y.; Li, F.; Xin, Q.; Li, Y.; Lin, Z. Historical variability of cotton yield and response to climate and agronomic management in Xinjiang, China. Sci. Total Environ. 2024, 912, 169327. [Google Scholar] [CrossRef]
- Li, X.-G.; Ding, C.-F.; Zhang, T.-L.; Wang, X.-X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Liu, H.; Pan, F.; Han, X.; Song, F.; Zhang, Z.; Yan, J.; Xu, Y. Response of Soil Fungal Community Structure to Long-Term Continuous Soybean Cropping. Front. Microbiol. 2019, 9, 3316. [Google Scholar] [CrossRef]
- Xi, H.; Shen, J.; Qu, Z.; Yang, D.; Liu, S.; Nie, X.; Zhu, L. Effects of Long-term Cotton Continuous Cropping on Soil Microbiome. Sci. Rep. 2019, 9, 18297. [Google Scholar] [CrossRef]
- Kuo, S.-Y.; Hu, C.-C.; Huang, Y.-W.; Lee, C.-W.; Luo, M.-J.; Tu, C.-W.; Lee, S.-C.; Lin, N.-S.; Hsu, Y.-H. Argonaute 5 family proteins play crucial roles in the defence against Cymbidium mosaic virus and Odontoglossum ringspot virus in Phalaenopsis aphrodite subsp. formosana. Mol. Plant Pathol. 2021, 22, 627–643. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Wang, Y.; Qin, J.; Wei, M.; Liao, X.; Shang, W.; Chen, J.; Subbarao, K.V.; Hu, X. Verticillium dahliae Elicitor VdSP8 Enhances Disease Resistance Through Increasing Lignin Biosynthesis in Cotton. Plant Cell Environ. 2025, 48, 728–745. [Google Scholar] [CrossRef]
- Zhou, Y.; Luo, W.; Li, M.; Wang, Q.; Liu, Y.; Guo, H. Summer Rice–Winter Potato Rotation Suppresses Various Soil-Borne Plant Fungal Pathogens. Agronomy 2023, 13, 2143. [Google Scholar] [CrossRef]
- De Haro, S. The Invisibility of Diffeomorphisms. Found. Phys. 2017, 47, 1464–1497. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, Y.; Ma, Y.; Hou, T.; Wang, J.; Che, Q.; Chen, B.; Wang, Q.; Feng, G. Soil Bacterial Diversity and Community Structure of Cotton Rhizosphere under Mulched Drip-Irrigation in Arid and Semi-arid Regions of Northwest China. Microb. Ecol. 2025, 88, 39. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Wood, S.A.; Bueno de Mesquita, C.P. How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef]
- Wang, M.; Ge, A.-H.; Ma, X.; Wang, X.; Xie, Q.; Wang, L.; Song, X.; Jiang, M.; Yang, W.; Murray, J.D.; et al. Dynamic root microbiome sustains soybean productivity under unbalanced fertilization. Nat. Commun. 2024, 15, 1668. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, J.; Yu, Z.; Yao, Q.; Li, Y.; Liang, A.; Zhang, W.; Mi, G.; Jin, J.; Liu, X.; et al. Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 2020, 197, 104503. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, S.; Li, K.; Qiao, J.; Guo, Y.; Liu, Z.; Guo, X. Responses of soil bacterial and fungal communities to the long-term monoculture of grapevine. Appl. Microbio. Biot. 2021, 105, 7035–7050. [Google Scholar] [CrossRef]
- Mohanan, A.; Nigam, R.; Yeliya, P.; Daniel, G.R.; Devi, N.O.; Sahu, R.; Mohapatra, D.; Sahoo, L. The Role of Plant Microbiomes in Suppressing Soilborne Pathogens: A Review. J. Adv. Microbiol. 2025, 25, 160–178. [Google Scholar] [CrossRef]
- Xiao, Z.; Lu, C.; Wu, Z.; Li, X.; Ding, K.; Zhu, Z.; Han, R.; Zhao, J.; Ge, T.; Li, G.; et al. Continuous cropping disorders of eggplants (Solanum melongena L.) and tomatoes (Solanum lycopersicum L.) in suburban agriculture: Microbial structure and assembly processes. Sci. Total Environ. 2024, 909, 168558. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, X.; Yang, Q.; Chi, X.; Pan, L.; Chen, N.; Yang, Z.; Wang, T.; Wang, M.; Yu, S. Dynamic succession of soil bacterial community during continuous cropping of peanut (Arachis hypogaea L.). PLoS ONE 2014, 9, e101355. [Google Scholar] [CrossRef] [PubMed]
- Larkin, R.P.; Griffin, T.S.; Honeycutt, C.W. Rotation and Cover Crop Effects on Soilborne Potato Diseases, Tuber Yield, and Soil Microbial Communities. Plant Dis. 2010, 94, 1491–1502. [Google Scholar] [CrossRef]
- Trivedi, P.; Rochester, I.J.; Trivedi, C.; Van Nostrand, J.D.; Zhou, J.; Karunaratne, S.; Anderson, I.C.; Singh, B.K. Soil aggregate size mediates the impacts of cropping regimes on soil carbon and microbial communities. Soil Biol. Biochem. 2015, 91, 169–181. [Google Scholar] [CrossRef]
- Passaris, N.; Flower, K.C.; Ward, P.R.; Cordingley, N. Effect of crop rotation diversity and windrow burning of residue on soil chemical composition under long-term no-tillage. Soil Tillage Res. 2021, 213, 105153. [Google Scholar] [CrossRef]
- Wheeler, T.A.; Bordovsky, J.P.; Keeling, J.W.; Mullinix, B.G. Effects of Crop Rotation, Cultivar, and Irrigation and Nitrogen Rate on Verticillium Wilt in Cotton. Plant Dis. 2012, 96, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Lu, X.; Wang, A.; Xue, C.; Zhao, M.; Zhang, J. The effects and interrelationships of intercropping on Cotton Verticillium wilt and soil microbial communities. BMC Microbiol. 2023, 23, 41. [Google Scholar] [CrossRef]
- dos Santos, L.F.C.; Ruiz-Sánchez, E.; Andueza-Noh, R.H.; Garruña-Hernández, R.; Latournerie-Moreno, L.; Mijangos-Cortés, J.O. Leaf damage by Spodoptera frugiperda J. E. Smith (Lepidoptera: Noctuidae) and its relation to leaf morphological traits in maize landraces and commercial cultivars. J. Plant Dis. Prot. 2020, 127, 103–109. [Google Scholar] [CrossRef]
- Toepfer, S.; Fallet, P.; Kajuga, J.; Bazagwira, D.; Mukundwa, I.P.; Szalai, M.; Turlings, T.C.J. Streamlining leaf damage rating scales for the fall armyworm on maize. J. Pest Sci. 2021, 94, 1075–1089. [Google Scholar] [CrossRef]
- Edwards, J.A.; Santos-Medellín, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.-I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Guimerà, R.; Nunes Amaral, L.A. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Dong, J.; Horvath, S. Understanding network concepts in modules. BMC Syst. Biol. 2007, 1, 24. [Google Scholar] [CrossRef]
- Xi, H.; Zhang, X.; Qu, Z.; Yang, D.; Alariqi, M.; Yang, Z.; Nie, X.; Zhu, L. Effects of cotton–maize rotation on soil microbiome structure. Mol. Plant Pathol. 2021, 22, 673–682. [Google Scholar] [CrossRef]
- Larkin, R.P. Soil health paradigms and implications for disease management. Annu. Rev. Phytopathol. 2015, 53, 199–221. [Google Scholar] [CrossRef]
- Rotili, D.H.; Alvarez Prado, S.; Barattini, A.; Lamattina, I.; Saks, M.G.; Gregorini, M.; Garcia, F.O.; Andrade, J.F. Medium-term fertilization strategies on extensive grain cropping systems under water table influence. Agric. Syst. 2023, 210, 103715. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, Z.; Liu, J.; Li, X.; Wang, X.; Dai, C.; Zhang, T.; Carrión, V.J.; Wei, Z.; Cao, F.; et al. Crop rotation and native microbiome inoculation restore soil capacity to suppress a root disease. Nat. Commun. 2023, 14, 8126. [Google Scholar] [CrossRef]
- Huang, J.; Liu, X.; Liu, J.; Zhang, Z.; Zhang, W.; Qi, Y.; Li, W.; Chen, Y. Changes of soil bacterial community, network structure, and carbon, nitrogen and sulfur functional genes under different land use types. Catena 2023, 231, 107385. [Google Scholar] [CrossRef]
- Kumar, U.; Cheng, M.; Islam, M.J.; Maniruzzaman, M.; Nasreen, S.S.; Haque, M.E.; Rahman, M.T.; Jahiruddin, M.; Bell, R.W.; Jahangir, M.M.R. Long-term Conservation Agriculture increases sulfur pools in soils together with increased soil organic carbon compared to conventional practices. Soil Tillage Res. 2022, 223, 105474. [Google Scholar] [CrossRef]
- Neupane, A.; Bulbul, I.; Wang, Z.; Lehman, R.M.; Nafziger, E.; Marzano, S.-Y.L. Long term crop rotation effect on subsequent soybean yield explained by soil and root-associated microbiomes and soil health indicators. Sci. Rep. 2021, 11, 9200. [Google Scholar] [CrossRef]
- Wang, B.; Shang, N.; Feng, X.; Hu, Z.; Li, P.; Chen, Y.; Hu, B.; Ding, M.; Xu, J. Understanding the microbiome–crop rotation nexus in karst agricultural systems: Insights from Southwestern China. Front. Microbiol. 2025, 16, 1503636. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Liu, Y.; Chen, Y.; Yang, S. Altitude Distribution Patterns and Driving Factors of Rhizosphere Soil Microbial Diversity in the Mountainous and Hilly Region of Southwest, China. Agronomy 2024, 14, 2441. [Google Scholar] [CrossRef]
- Yang, X.; Hu, H.-W.; Yang, G.-W.; Cui, Z.-L.; Chen, Y.-L. Crop rotational diversity enhances soil microbiome network complexity and multifunctionality. Geoderma 2023, 436, 116562. [Google Scholar] [CrossRef]
- Niazmoradi, M.; Kazemi, H.; Gherekhloo, J.; Soltani, A.; Kamkar, B. Health assessment of wheat agroecosystems in Iran. Sci. Rep. 2025, 15, 18133. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Evans, B.; Bainard, L.D. Pulse Frequency in Crop Rotations Alters Soil Microbial Community Networks and the Relative Abundance of Fungal Plant Pathogens. Front. Microbiol. 2021, 12, 667394. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, F.H.; Tie, Z.; Zhang, X.; Ma, Y.; Yu, Y.; Zhao, S.; Zhang, X.; Xi, H. Short-Term Maize Rotation Suppresses Verticillium Wilt and Restructures Soil Microbiomes in Xinjiang Cotton Fields. Microorganisms 2025, 13, 1968. https://doi.org/10.3390/microorganisms13091968
Khan FH, Tie Z, Zhang X, Ma Y, Yu Y, Zhao S, Zhang X, Xi H. Short-Term Maize Rotation Suppresses Verticillium Wilt and Restructures Soil Microbiomes in Xinjiang Cotton Fields. Microorganisms. 2025; 13(9):1968. https://doi.org/10.3390/microorganisms13091968
Chicago/Turabian StyleKhan, Faisal Hayat, Zhanjiang Tie, Xueqin Zhang, Yanjun Ma, Yu Yu, Sifeng Zhao, Xuekun Zhang, and Hui Xi. 2025. "Short-Term Maize Rotation Suppresses Verticillium Wilt and Restructures Soil Microbiomes in Xinjiang Cotton Fields" Microorganisms 13, no. 9: 1968. https://doi.org/10.3390/microorganisms13091968
APA StyleKhan, F. H., Tie, Z., Zhang, X., Ma, Y., Yu, Y., Zhao, S., Zhang, X., & Xi, H. (2025). Short-Term Maize Rotation Suppresses Verticillium Wilt and Restructures Soil Microbiomes in Xinjiang Cotton Fields. Microorganisms, 13(9), 1968. https://doi.org/10.3390/microorganisms13091968






