Analysis of the Bacterial Community and Fatty Acid Composition in the Bacteriome of the Lac Insect Llaveia axin axin
Abstract
1. Introduction
2. Materials and Methods
2.1. Neuro-Fuzzy Controlled Biosystem for Artificial Breeding of L. axin axin
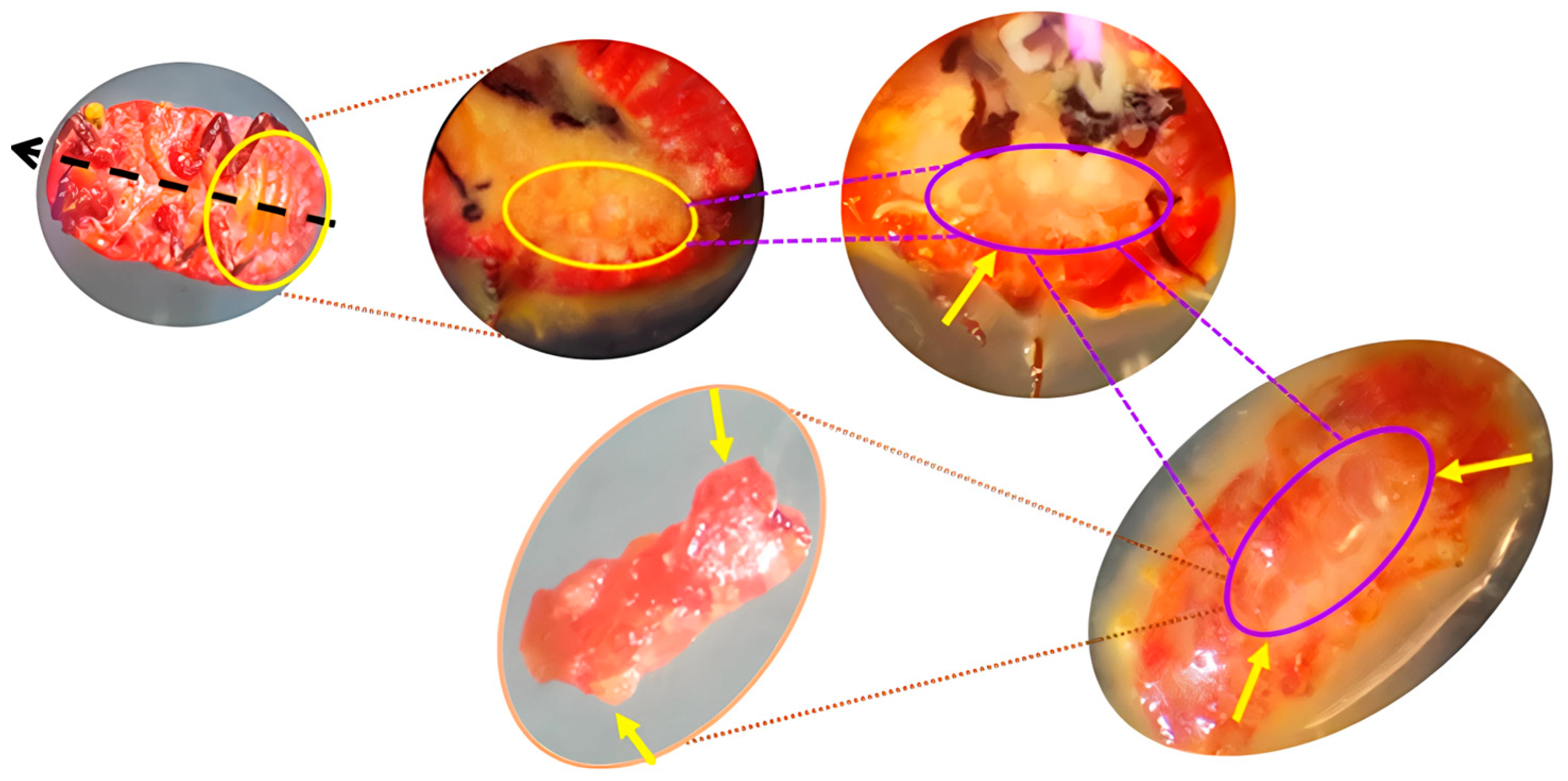
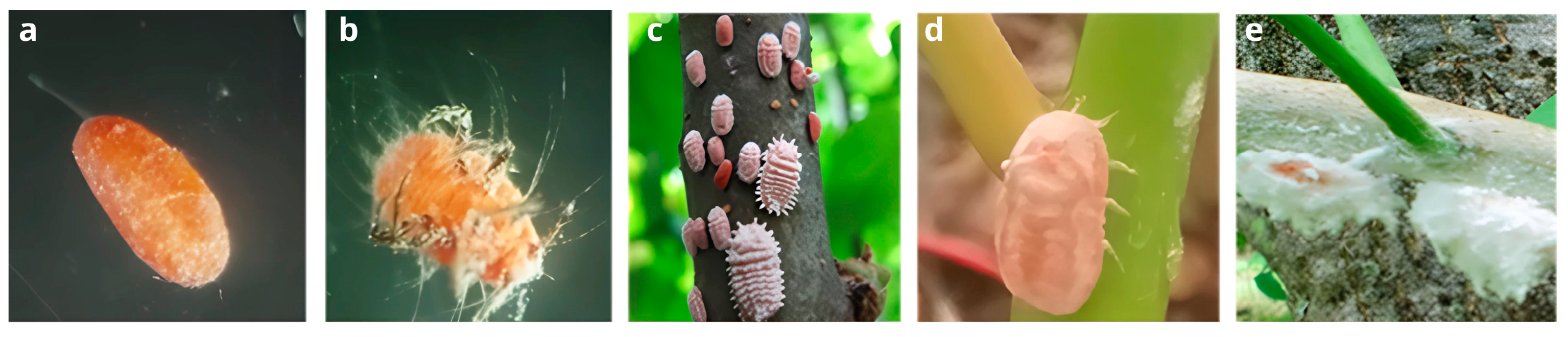
2.2. Sample Processing and Bacteriome Extraction in the Lac Insect L. axin axin
2.3. Culture-Independent Characterization of Bacteria Associated with the Bacteriomes of L. axin axin
2.4. Genomic Characterization of Symbiotic Bacteria in the Lac Insect L. axin axin
2.5. Fatty Acid Profile of the Bacteriome in the Lac Insect L. axin axin
3. Results
3.1. Artificial Breeding of L. axin axin Using a Neuro-Fuzzy Controlled Biosystem
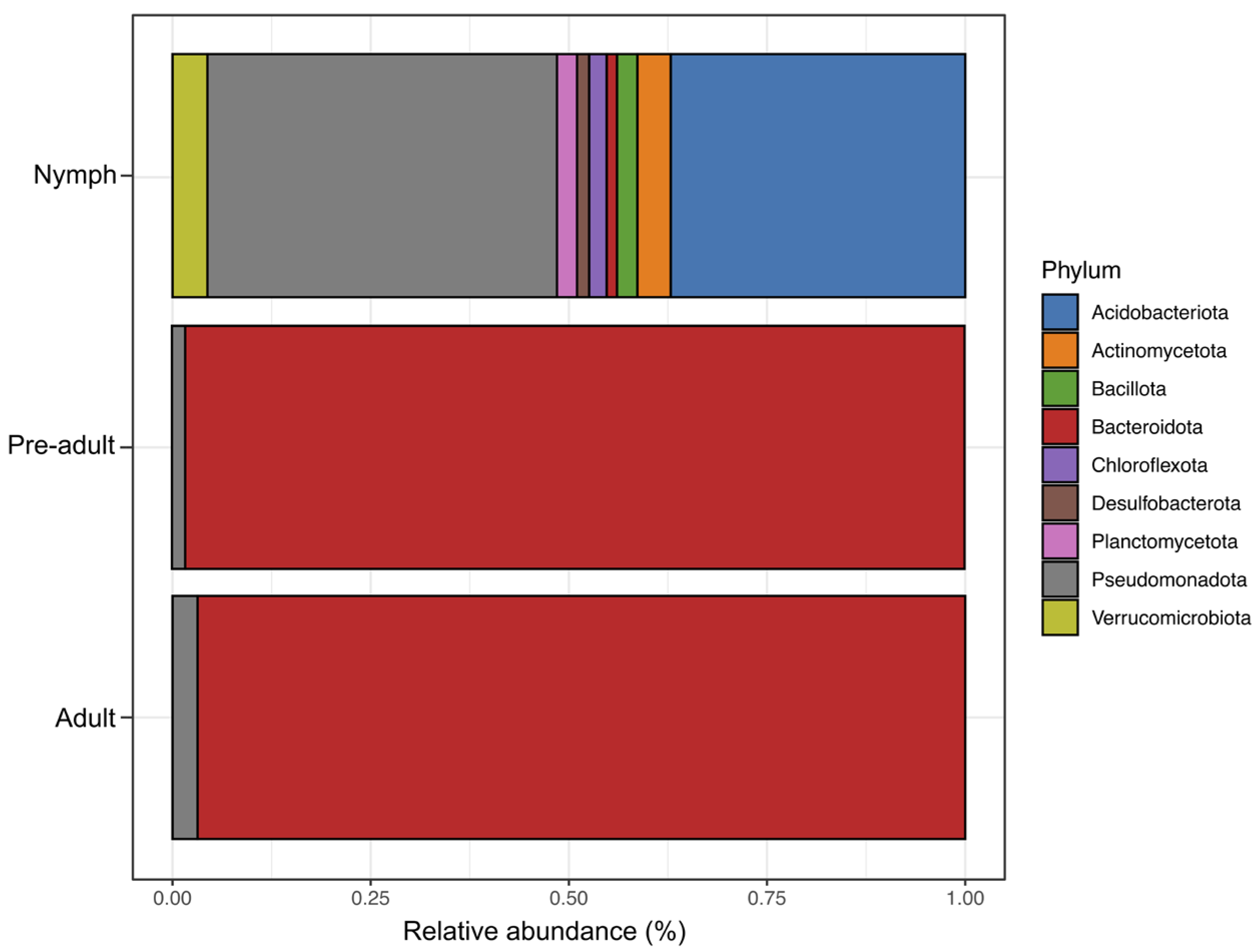
3.2. 16S rRNA Gene-Based Profiling of Bacterial Communities Associated with L. axin axin
3.3. ARDRA-Based Genomic Profiling and Diversity Indices of Cultivable Bacteria
3.4. Molecular Characterization and Taxonomic Classification of Cultivable Bacteria from L. axin axin
3.5. Fatty Acid Composition of L. axin axin Across Developmental Stages and Its Bacteriome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Douglas, A.E. Multiorganismal Insects: Diversity and Function of Resident Microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Somani, J.; Roy, S.; Babu, A.; Pandey, A.K. Insect Microbial Symbionts: Ecology, Interactions, and Biological Significance. Microorganisms 2023, 11, 2665. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, M.M.; Engl, T.; Kaltenpoth, M. Microbial Symbionts Expanding or Constraining Abiotic Niche Space in Insects. Curr. Opin. Insect Sci. 2020, 39, 14–20. [Google Scholar] [CrossRef]
- Rupawate, P.S.; Roylawar, P.; Khandagale, K.; Gawande, S.; Ade, A.B.; Jaiswal, D.K.; Borgave, S. Role of Gut Symbionts of Insect Pests: A Novel Target for Insect-Pest Control. Front. Microbiol. 2023, 14, 1146390. [Google Scholar] [CrossRef] [PubMed]
- Visser, B.; Scheifler, M. Insect Lipid Metabolism in the Presence of Symbiotic and Pathogenic Viruses and Bacteria. Adv. Exp. Med. Biol. 2024, 1493, 1–25. [Google Scholar] [CrossRef]
- Attardo, G.M.; Benoit, J.B.; Michalkova, V.; Kondragunta, A.; Baumann, A.A.; Weiss, B.L.; Malacrida, A.; Scolari, F.; Aksoy, S. Lipid Metabolism Dysfunction Following Symbiont Elimination Is Linked to Altered Kennedy Pathway Homeostasis. iScience 2023, 26, 107108. [Google Scholar] [CrossRef]
- Williams, M.; MacVean, C. Ethnococcidology: Use of the Giant Margarodids, by Indigenous Peoples of Mesoamerica in Their Culture, Medicine and Arts. Isr. J. Entomol. 1995, 29, 147–148. [Google Scholar]
- Rosenblueth, M.; Martínez-Romero, J.; Tabita Ramírez-Puebla, S.; Vera-Ponce De León, A.; Rosas-Pérez, T.; Bustamante-Brito, R.; Rincón-Rosales, R.; Martínez-Romero, E. Endosymbiotic Microorganisms of Scale Insects. TIP Rev. Espec. Cienc. Químico-Biológicas 2018, 21, 53–69. [Google Scholar] [CrossRef]
- Suazo-Ortuño, I.; Del Val-De Gortari, E.; Benítez-Malvido, J. Rediscovering an Extraordinary Vanishing Bug: Llaveia axin axin. Rev. Mex. Biodivers. 2013, 84, 338–346. [Google Scholar] [CrossRef]
- Rincón-Rosales, R.; Gutiérrez-Miceli, F.A. Biological Characteristics of Acaciella angustissima (Mill.) Britton & Rose in Its Natural Habitat and Assessment of Its Bark Potential in Chiapas, México. Agrociencia 2008, 42, 129–137. [Google Scholar]
- Ferrarini, M.G.; Dell’Aglio, E.; Vallier, A.; Balmand, S.; Vincent-Monégat, C.; Hughes, S.; Gillet, B.; Parisot, N.; Zaidman-Rémy, A.; Vieira, C.; et al. Efficient Compartmentalization in Insect Bacteriomes Protects Symbiotic Bacteria from Host Immune System. Microbiome 2022, 10, 156. [Google Scholar] [CrossRef]
- Gil, R.; Latorre, A. Unity Makes Strength: A Review on Mutualistic Symbiosis in Representative Insect Clades. Life 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Pérez, T.; Rosenblueth, M.; Rincón-Rosales, R.; Mora, J.; Martínez-Romero, E. Genome Sequence of “Candidatus Walczuchella Monophlebidarum” the Flavobacterial Endosymbiont of Llaveia axin axin (Hemiptera: Coccoidea: Monophlebidae). Genome Biol. Evol. 2014, 6, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Ponce-de-Leon, M.; Tamarit, D.; Calle-Espinosa, J.; Mori, M.; Latorre, A.; Montero, F.; Pereto, J. A Genome-Scale Study of Metabolic Complementation in Endosymbiotic Consortia: The Case of the Cedar Aphid 2017. bioRxiv 2017. [Google Scholar] [CrossRef]
- Sloan, D.B.; Nakabachi, A.; Richards, S.; Qu, J.; Murali, S.C.; Gibbs, R.A.; Moran, N.A. Parallel Histories of Horizontal Gene Transfer Facilitated Extreme Reduction of Endosymbiont Genomes in Sap-Feeding Insects. Mol. Biol. Evol. 2014, 31, 857–871. [Google Scholar] [CrossRef]
- Majumder, R.; Sutcliffe, B.; Taylor, P.W.; Chapman, T.A. Next-Generation Sequencing Reveals Relationship between the Larval Microbiome and Food Substrate in the Polyphagous Queensland Fruit Fly. Sci. Rep. 2019, 9, 14292. [Google Scholar] [CrossRef]
- Ramírez-Puebla, S.T.; Ormeño-Orrillo, E.; Vera-Ponce de León, A.; Lozano, L.; Sanchez-Flores, A.; Rosenblueth, M.; Martínez-Romero, E. Genomes of Candidatus Wolbachia bourtzisii wDacA and Candidatus Wolbachia pipientis wDacB from the Cochineal Insect Dactylopius Coccus (Hemiptera: Dactylopiidae). G3 Genes Genomes Genet. 2016, 6, 3343–3394. [Google Scholar] [CrossRef]
- Wrońska, A.K.; Kaczmarek, A.; Boguś, M.I.; Kuna, A. Lipids as a Key Element of Insect Defense Systems. Front. Genet. 2023, 14, 1183659. [Google Scholar] [CrossRef]
- Fan, Y.; Thompson, J.W.; Dubois, L.G.; Moseley, M.A.; Wernegreen, J.J. Proteomic Analysis of an Unculturable Bacterial Endosymbiont (Blochmannia) Reveals High Abundance of Chaperonins and Biosynthetic Enzymes. J. Proteome Res. 2013, 12, 704–718. [Google Scholar] [CrossRef]
- Silva, F.J.; Domínguez-Santos, R.; Latorre, A.; García-Ferris, C. Comparative Transcriptomics of Fat Bodies between Symbiotic and Quasi-Aposymbiotic Adult Females of Blattella germanica with Emphasis on the Metabolic Integration with Its Endosymbiont Blattabacterium and Its Immune System. Int. J. Mol. Sci. 2024, 25, 4228. [Google Scholar] [CrossRef]
- Morales-Mancilla, J.A.; Guerra-Crespo, H.; Cossío-Martínez, A.G.; Suárez-Ruiz, F.d.J.; Gómez-Pérez, J.; Orozco-Torres, J.A. Neurofuzzy Model for the Control of Ambient Temperature and Soil Moisture to Cultivate the insect NIIJ (Llaveia axin). Rev. Tecnol. Digit. 2015, 5, 115–127. [Google Scholar]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S RRNA Sequence Formation and Detection in Sanger and 454-Pyrosequenced PCR Amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- R Core Team. R Core Team R: A Language and Environment for Statistical Computing; Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Petrovski, S.; Seviour, R.J.; Tillett, D. Genome Sequence and Characterization of the Tsukamurella Bacteriophage TPA2. Appl. Environ. Microbiol. 2011, 77, 1389–1398. [Google Scholar] [CrossRef]
- Koskey, G.; Mburu, S.W.; Kimiti, J.M.; Ombori, O.; Maingi, J.M.; Njeru, E.M. Genetic Characterization and Diversity of Rhizobium Isolated from Root Nodules of Mid-Altitude Climbing Bean (Phaseolus vulgaris L.) Varieties. Front. Microbiol. 2018, 9, 968. [Google Scholar] [CrossRef]
- Ranjbar, R.; Tabatabaee, A.; Behzadi, P.; Kheiri, R. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) Genotyping of Escherichia coli Strains Isolated from Different Animal Stool Specimens. J. Pathol. Iran. J. Pathol. 2017, 12, 25–34. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S RRNA Genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Xu, C.W.; Wang, M.M.; Li, T.; Zhao, Z.W. Quantitatively Evaluating Mistaken Clone Assignments by RFLP Analysis of 16S RRNA Genes: A Case Study. Can. J. Microbiol. 2008, 54, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Yang, X.; Zhang, T.; Chen, J.; Xue, D. Molecular Characterization of the Bacterial Composition in Two Waste Silk Refining Systems. World J. Microbiol. Biotechnol. 2011, 27, 2335–2341. [Google Scholar] [CrossRef]
- Adeleke, B.S. 16S RRNA Gene Sequencing Data of Plant Growth-Promoting Jute-Associated Endophytic and Rhizobacteria from Coastal-Environment of Ondo State, Nigeria. Data Brief. 2024, 54, 110286. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Deibel, D.; Parrish, C.C.; Grønkjær, P.; Munk, P.; Gissel Nielsen, T. Lipid Class and Fatty Acid Content of the Leptocephalus Larva of Tropical Eels. Lipids 2012, 47, 623–634. [Google Scholar] [CrossRef]
- Gołȩbiowski, M.; Cerkowniak, M.; Urbanek, A.; Słocińska, M.; Rosiński, G.; Stepnowski, P. Adipokinetic Hormone Induces Changes in the Fat Body Lipid Composition of the Beetle Zophobas Atratus. Peptides 2014, 58, 65–73. [Google Scholar] [CrossRef]
- Rodríguez-Hernandez, L.; Rodríguez-Mendiola, M.A.; Arias-Castro, C.; Gutiérrez-Miceli, F.A. Effect of Plant Growth Regulators on Fatty Acids Composition in Jatropha curcas L. Callus Culture. J. Oleo Sci. 2015, 64, 325–330. [Google Scholar] [CrossRef]
- Sabri, A.; Leroy, P.; Haubruge, E.; Hance, T.; Frère, I.; Destain, J.; Thonart, P. Isolation, Pure Culture and Characterization of Serratia symbiotica Sp. Nov., the R-Type of Secondary Endosymbiont of the Black Bean Aphid Aphis fabae. Int. J. Syst. Evol. Microbiol. 2011, 61, 2081–2088. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Hosokawa, T.; Fukatsu, T. Insect-Microbe Mutualism without Vertical Transmission: A Stinkbug Acquires a Beneficial Gut Symbiont from the Environment Every Generation. Appl. Environ. Microbiol. 2007, 73, 4308–4316. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and Evolution of Heritable Bacterial Symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Andongma, A.A.; Wan, L.; Dong, Y.C.; Wang, Y.L.; He, J.; Niu, C.Y. Assessment of the Bacteria Community Structure across Life Stages of the Chinese Citrus Fly, Bactrocera minax (Diptera: Tephritidae). BMC Microbiol. 2019, 19, 285. [Google Scholar] [CrossRef]
- Kang, W.N.; Jin, L.; Fu, K.Y.; Guo, W.C.; Li, G.Q. A Switch of Microbial Flora Coupled with Ontogenetic Niche Shift in Leptinotarsa decemlineata. Arch. Insect Biochem. Physiol. 2021, 107, e21782. [Google Scholar] [CrossRef]
- Sun, T.; Wang, X.Q.; Zhao, Z.L.; Yu, S.H.; Yang, P.; Chen, X.M. A Lethal Fungus Infects the Chinese White Wax Scale Insect and Causes Dramatic Changes in the Host Microbiota. Sci. Rep. 2018, 8, 5324. [Google Scholar] [CrossRef]
- Dhami, M.K.; Buckley, T.R.; Beggs, J.R.; Taylor, M.W. Primary Symbiont of the Ancient Scale Insect Family Coelostomidiidae Exhibits Strict Cophylogenetic Patterns. Symbiosis 2013, 61, 77–91. [Google Scholar] [CrossRef]
- Szklarzewicz, T.; Kalandyk-Kołodziejczyk, M.; Michalik, K.; Jankowska, W.; Michalik, A. Symbiotic Microorganisms in Puto superbus (Leonardi, 1907) (Insecta, Hemiptera, Coccomorpha: Putoidae). Protoplasma 2018, 255, 129–138. [Google Scholar] [CrossRef]
- Toh, H.; Weiss, B.L.; Perkin, S.A.H.; Yamashita, A.; Oshima, K.; Hattori, M.; Aksoy, S. Massive Genome Erosion and Functional Adaptations Provide Insights into the Symbiotic Lifestyle of Sodalis glossinidius in the Tsetse Host. Genome Res. 2006, 16, 149–156. [Google Scholar] [CrossRef]
- Morrow, J.L.; Hall, A.A.G.; Riegler, M. Symbionts in Waiting: The Dynamics of Incipient Endosymbiont Complementation and Replacement in Minimal Bacterial Communities of Psyllids. Microbiome 2017, 5, 58. [Google Scholar] [CrossRef]
- Lefèvre, C.; Charles, H.; Vallier, A.; Delobel, B.; Farrell, B.; Heddi, A. Endosymbiont Phylogenesis in the Dryophthoridae Weevils: Evidence for Bacterial Replacement. Mol. Biol. Evol. 2004, 21, 965–973. [Google Scholar] [CrossRef]
- Conord, C.; Despres, L.; Vallier, A.; Balmand, S.; Miquel, C.; Zundel, S.; Lemperiere, G.; Heddi, A. Long-Term Evolutionary Stability of Bacterial Endosymbiosis in Curculionoidea: Additional Evidence of Symbiont Replacement in the Dryophthoridae Family. Mol. Biol. Evol. 2008, 25, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.; Miller, D.; Brewster, C.; Husseneder, C.; Dickerman, A. Diversity of Gut Bacteria of Reticulitermes Flavipes as Examined by 16S RRNA Gene Sequencing and Amplified RDNA Restriction Analysis. Curr. Microbiol. 2007, 55, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Pidiyar, V.J.; Jangid, K.; Patole, M.S.; Shouche, Y.S. Studies on Cultured and Uncultured Microbiota of Wild Culex Quinquefasciatus Mosquito Midgut Based on 16s Ribosomal RNA Gene Analysis. Am. J. Trop. Med. Hyg. 2004, 70, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.F.; Ling, X.F.; Lu, Q.; Wang, W.W.; Zhang, X.; Feng, Y.; Chen, X.M.; Chen, H. Identification of the Key Pathways and Genes Involved in the Wax Biosynthesis of the Chinese White Wax Scale Insect (Ericerus pela Chavannes) by Integrated Weighted Gene Coexpression Network Analysis. Genes 2022, 13, 1364. [Google Scholar] [CrossRef]
- Fu, Z.; Shi, Y.; Yu, S.; Zhao, Q.; Mo, H.; Yang, P. Variation of Gene Expression of Fatty Acid Acyl CoA Reductase Associated with Wax Secretion of a Scale Insect, Ericerus pela, and Identification of Its Regulation Factors through the Accessible Chromatin Analyses and Yeast One-Hybrid. Arch. Insect Biochem. Physiol. 2024, 15, e22101. [Google Scholar] [CrossRef]
- Kanyile, S.N.; Engl, T.; Heddi, A.; Kaltenpoth, M. Endosymbiosis Allows Sitophilus Oryzae to Persist in Dry Conditions. Front. Microbiol. 2023, 14, 1199370. [Google Scholar] [CrossRef]
- Kiefer, J.S.T.; Bauer, E.; Okude, G.; Fukatsu, T.; Kaltenpoth, M.; Engl, T. Cuticle Supplementation and Nitrogen Recycling by a Dual Bacterial Symbiosis in a Family of Xylophagous Beetles. ISME J. 2023, 17, 1029–1039. [Google Scholar] [CrossRef]
- Cilia, G.; Fratini, F.; Tafi, E.; Mancini, S.; Turchi, B.; Sagona, S.; Cerri, D.; Felicioli, A.; Nanetti, A. Changes of Western Honey Bee Apis Mellifera ligustica (Spinola, 1806) Ventriculus Microbial Profile Related to Their in-Hive Tasks. J. Apic. Res. 2021, 60, 198–202. [Google Scholar] [CrossRef]
- Moyano, A.; Palladini, A.; Díaz, V.; Abraham, S.; Castillo, G.; Giudice, A.; Araóz, V.C.; Fernandez, P.; Van Nieuwenhove, G.; Rull, J. Gut Bacteria Symbiosis Affects Cuticular Hydrocarbon Profile and Mating Success in Wild Ceratitis capitata (Diptera: Tephritidae) Males. Aust. Entomol. 2025, 64, e70002. [Google Scholar] [CrossRef]
- Fladerer, J.; Grollitsch, S.; Bucar, F. Three Cuticular Amides in the Tripartite Symbiosis of Leafcutter Ants. Arch. Insect Biochem. Physiol. 2023, 114, 1–13. [Google Scholar] [CrossRef]
- Oakeson, K.F.; Gil, R.; Clayton, A.L.; Dunn, D.M.; von Niederhausern, A.C.; Hamil, C.; Aoyagi, A.; Duval, B.; Baca, A.; Silva, F.J.; et al. Genome Degeneration and Adaptation in a Nascent Stage of Symbiosis. Genome Biol. Evol. 2014, 6, 76–93. [Google Scholar] [CrossRef]
- Sabree, Z.L.; Kambhampati, S.; Moran, N.A. Nitrogen Recycling and Nutritional Provisioning by Blattabacterium, the Cockroach Endosymbiont. Proc. Natl. Acad. Sci. USA 2009, 106, 19521–19526. [Google Scholar] [CrossRef]
- Hansen, A.K.; Moran, N.A. Aphid Genome Expression Reveals Host–Symbiont Cooperation in the Production of Amino Acids. Proc. Natl. Acad. Sci. USA 2011, 108, 2849–2854. [Google Scholar] [CrossRef]
- Ruiu, L. Insect Pathogenic Bacteria in Integrated Pest Management. Insects 2015, 6, 352–367. [Google Scholar] [CrossRef]
- Ugwu, J.A.; Liu, M.; Sun, H.; Asiegbu, F.O. Microbiome of the Larvae of Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) from Maize Plants. J. Appl. Entomol. 2020, 144, 764–776. [Google Scholar] [CrossRef]


| Developmental Stage | Number of Reads (n) | Abundance (d) | Shannon–Weaver Diversity Index (H′) |
|---|---|---|---|
| First-instar nymphs | 27,847 | 7.2304 | 1.8842 |
| Pre-adult females | 149,875 | 6.2093 | 2.0202 |
| Adult females | 174,001 | 1.5745 | 0.8101 |
| Sample Source | No. of Isolates | No. of Groups ARDRA Profiles a | Relative Abundance (%) | Shannon–Weaver Index b | |
|---|---|---|---|---|---|
| Richness (d) | Diversity (H′) | ||||
| First-instar nymphs | 8 | 4 | 40 | 1.44 | 1.32 |
| Pre-adult females | 6 | 3 | 30 | 1.24 | 1.05 |
| Adult females | 6 | 2 | 30 | 0.62 | 0.67 |
| Total | 20 | 9 | 100 | ||
| Strain | Closest NCBI Match a/Identity (%) | Accession Number | Developmental Stage | Phylum |
|---|---|---|---|---|
| NI-01 | Staphylococcus warneri (98%) | MH223592.1 | First-instar nymph | Bacillota |
| NI-02 | Pseudomonas oryzihabitans (99%) | MH223593.1 | First-instar nymph | Pseudomonadota |
| NI-03 | Acinetobacter pittii (98%) | MH223594.1 | First-instar nymph | Pseudomonadota |
| NI-04 | Moraxella osloensis (98%) | MH223595.1 | First-instar nymph | Pseudomonadota |
| PA-01 | Methylobacterium hispanicum (99%) | PV888580 | Pre-adult female | Pseudomonadota |
| PA-04 | Microbacterium marinilacus (90%) | PV888581 | Pre-adult female | Actinobacteriota |
| PA-05 | Bacillus sp. (97%) | PV888582 | Pre-adult female | Bacillota |
| AD-03 | Bacillus sp. (94%) | PV888584 | Adult female | Bacillota |
| AD-04 | Microbacterium marinilacus (93%) | PV888583 | Adult female | Actinobacteriota |
| Fatty Acid | Eggs | First-Instar Nymphs | Pre-Adult Females | Adult Females | Axe Wax |
|---|---|---|---|---|---|
| Relative Abundance (%) | |||||
| Palmitic (C16:0) | 2.918 | 21.78 | 4.53 | 4.15 | 0.389 |
| Stearic (C18:0) | 43.82 | 31.90 | 43.08 | 49.68 | 40.99 |
| Oleic (C18:1) | 13.74 | 8.52 | 21.62 | 15.22 | 17.41 |
| Linoleic (C18:2) | 16.14 | 13.52 | 19.20 | 16.08 | 19.79 |
| Arachidic (C20:0) | 17.97 | 5.42 | 10.84 | 13.29 | 13.66 |
| Behenic (C22:0) | 2.32 | 3.22 | ND | ND | 0.561 |
| Butyric (C4:0) | ND a | ND | ND | ND | 0.258 |
| Valeric (C5:0) | ND | ND | 0.157 | 0.277 | ND |
| Undecanoic | ND | ND | ND | ND | 6.924 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rincón-Rosales, R.; Díaz-Hernández, M.; Manzano-Gómez, L.A.; Rincón-Molina, F.A.; Ruíz-Valdiviezo, V.M.; Gen-Jiménez, A.; Villalobos-Maldonado, J.J.; Maldonado-Gómez, J.C.; Rincón-Molina, C.I. Analysis of the Bacterial Community and Fatty Acid Composition in the Bacteriome of the Lac Insect Llaveia axin axin. Microorganisms 2025, 13, 1930. https://doi.org/10.3390/microorganisms13081930
Rincón-Rosales R, Díaz-Hernández M, Manzano-Gómez LA, Rincón-Molina FA, Ruíz-Valdiviezo VM, Gen-Jiménez A, Villalobos-Maldonado JJ, Maldonado-Gómez JC, Rincón-Molina CI. Analysis of the Bacterial Community and Fatty Acid Composition in the Bacteriome of the Lac Insect Llaveia axin axin. Microorganisms. 2025; 13(8):1930. https://doi.org/10.3390/microorganisms13081930
Chicago/Turabian StyleRincón-Rosales, Reiner, Miriam Díaz-Hernández, Luis Alberto Manzano-Gómez, Francisco Alexander Rincón-Molina, Víctor Manuel Ruíz-Valdiviezo, Adriana Gen-Jiménez, Juan José Villalobos-Maldonado, Julio César Maldonado-Gómez, and Clara Ivette Rincón-Molina. 2025. "Analysis of the Bacterial Community and Fatty Acid Composition in the Bacteriome of the Lac Insect Llaveia axin axin" Microorganisms 13, no. 8: 1930. https://doi.org/10.3390/microorganisms13081930
APA StyleRincón-Rosales, R., Díaz-Hernández, M., Manzano-Gómez, L. A., Rincón-Molina, F. A., Ruíz-Valdiviezo, V. M., Gen-Jiménez, A., Villalobos-Maldonado, J. J., Maldonado-Gómez, J. C., & Rincón-Molina, C. I. (2025). Analysis of the Bacterial Community and Fatty Acid Composition in the Bacteriome of the Lac Insect Llaveia axin axin. Microorganisms, 13(8), 1930. https://doi.org/10.3390/microorganisms13081930





