Desulfitobacterium elongatum sp. nov. NIT-TF6 Isolated from Trichloroethene-Dechlorinating Culture with Formate
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Growth Conditions
2.2. Physiological, Morphological, and Biochemical Analyses
2.3. Sanger Sequencing of 16S rRNA Gene and Genome Pyrosequencing
2.4. Phylogenetic Analysis Based on 16S rRNA Gene and Core Genome
2.5. TB Tool II for Genome Analysis
2.6. GC-FID Analysis for Dechlorination Experiments
2.7. GC-MS for Detection of 13C-Labeled Acetate
3. Results and Discussion
3.1. Isolation and Morphology of Strain NIT-TF6
3.2. Physiological Characteristics of Strain NIT-TF6
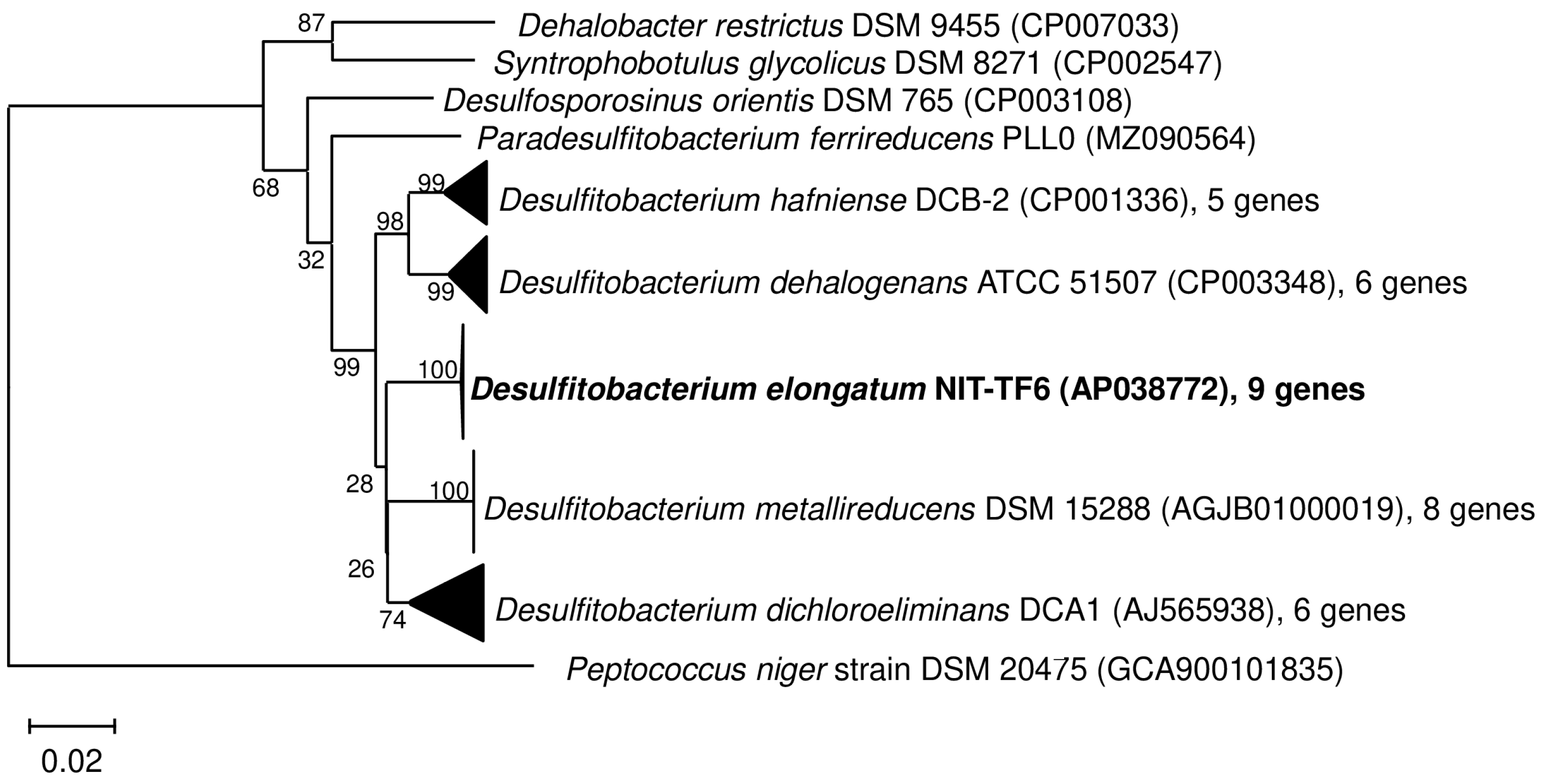
3.3. Phylogenetic Analysis
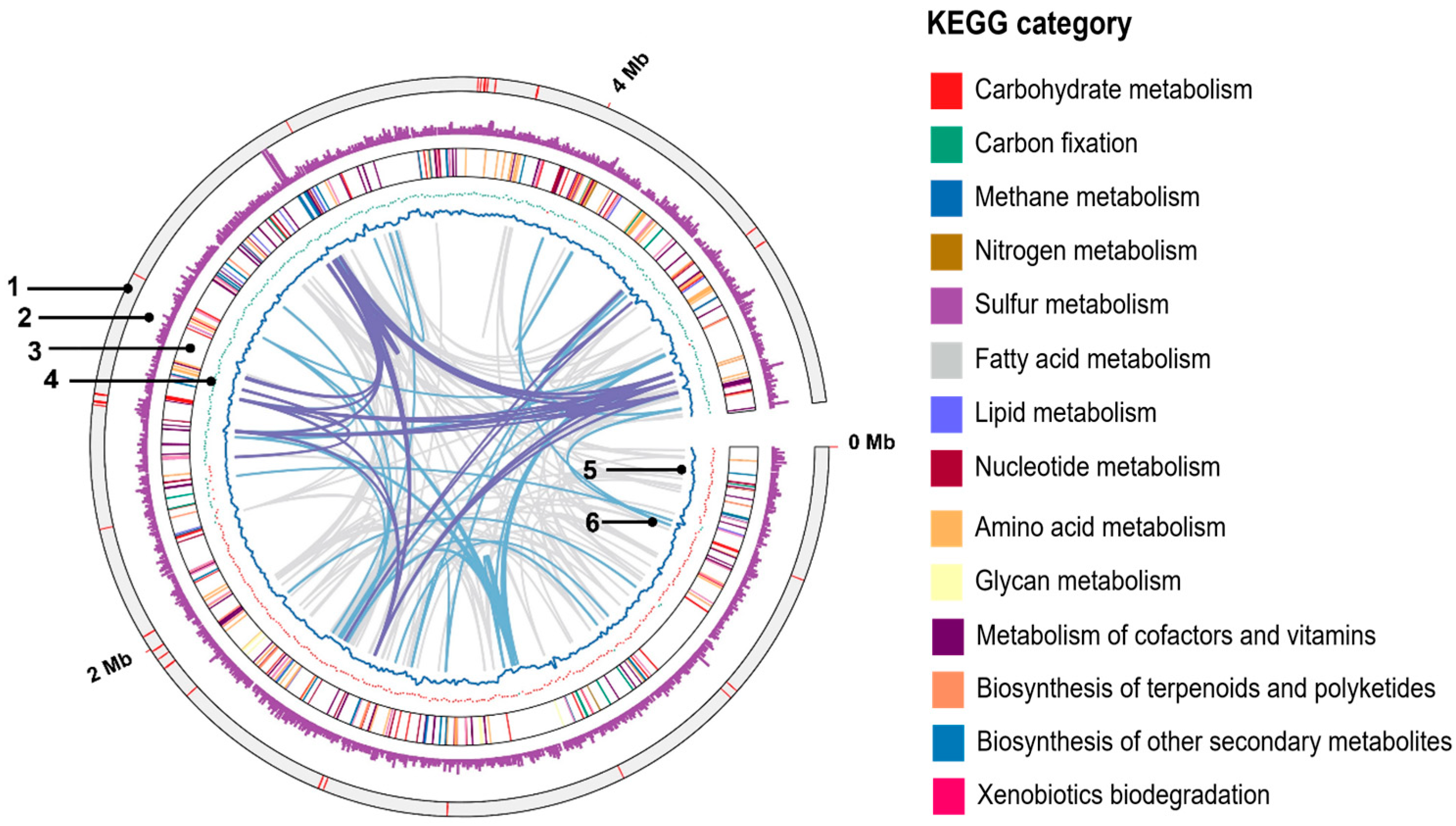
3.4. General Genomic Feature
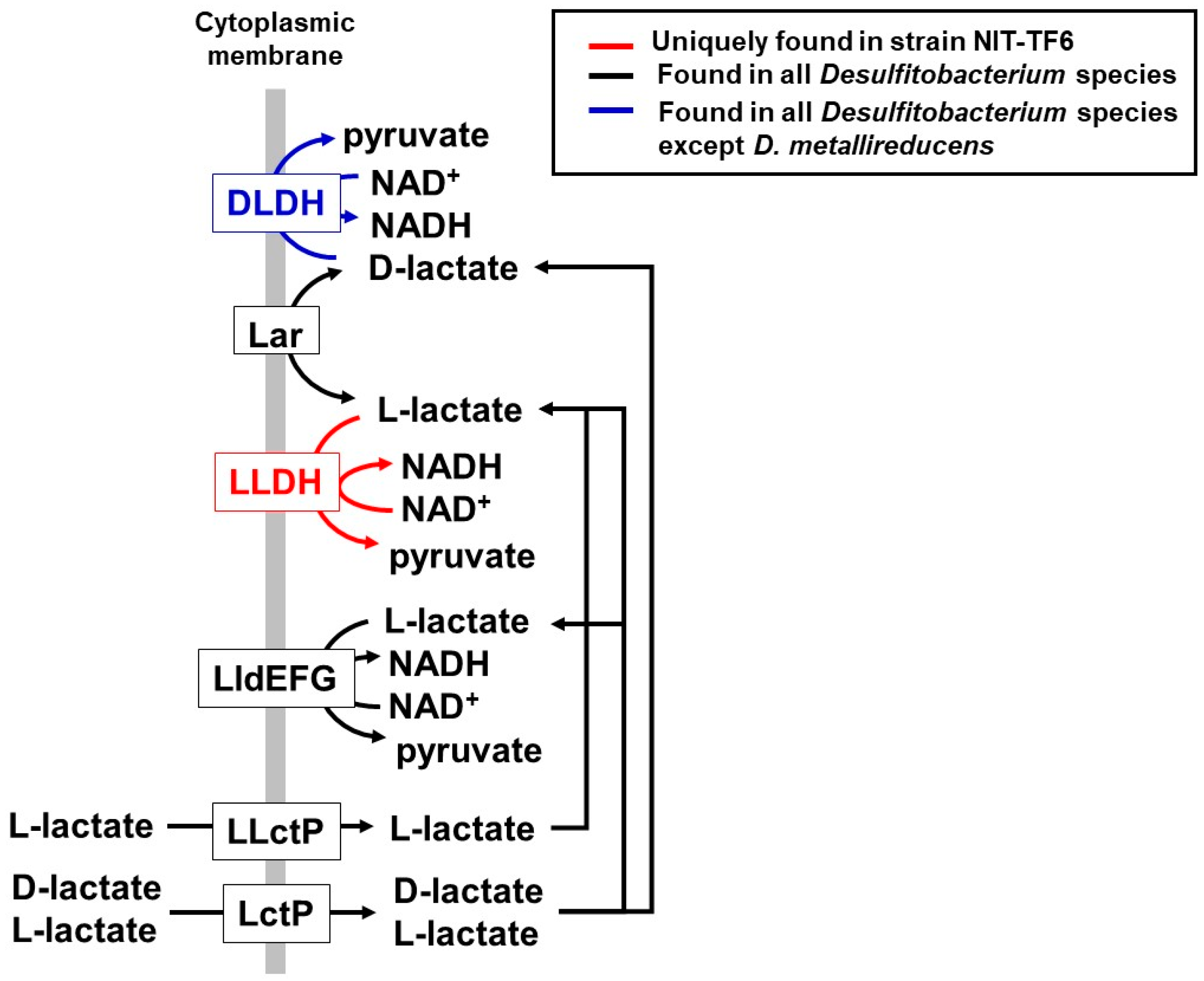
3.5. Metabolic Potential and Unique Characteristics Based on the Genome
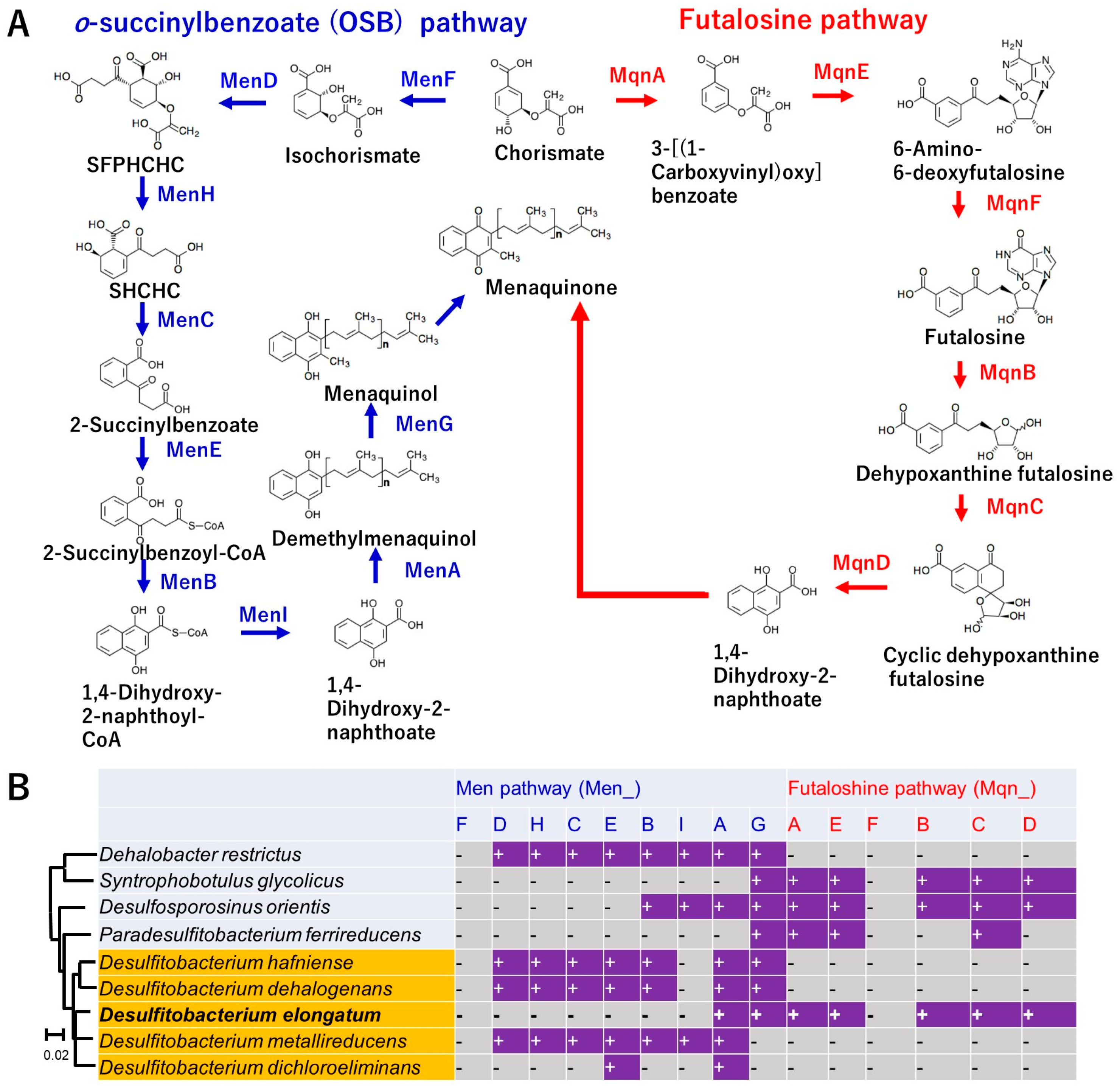
3.6. Unique Menaquinone Synthesis Pathway Based on the Genome
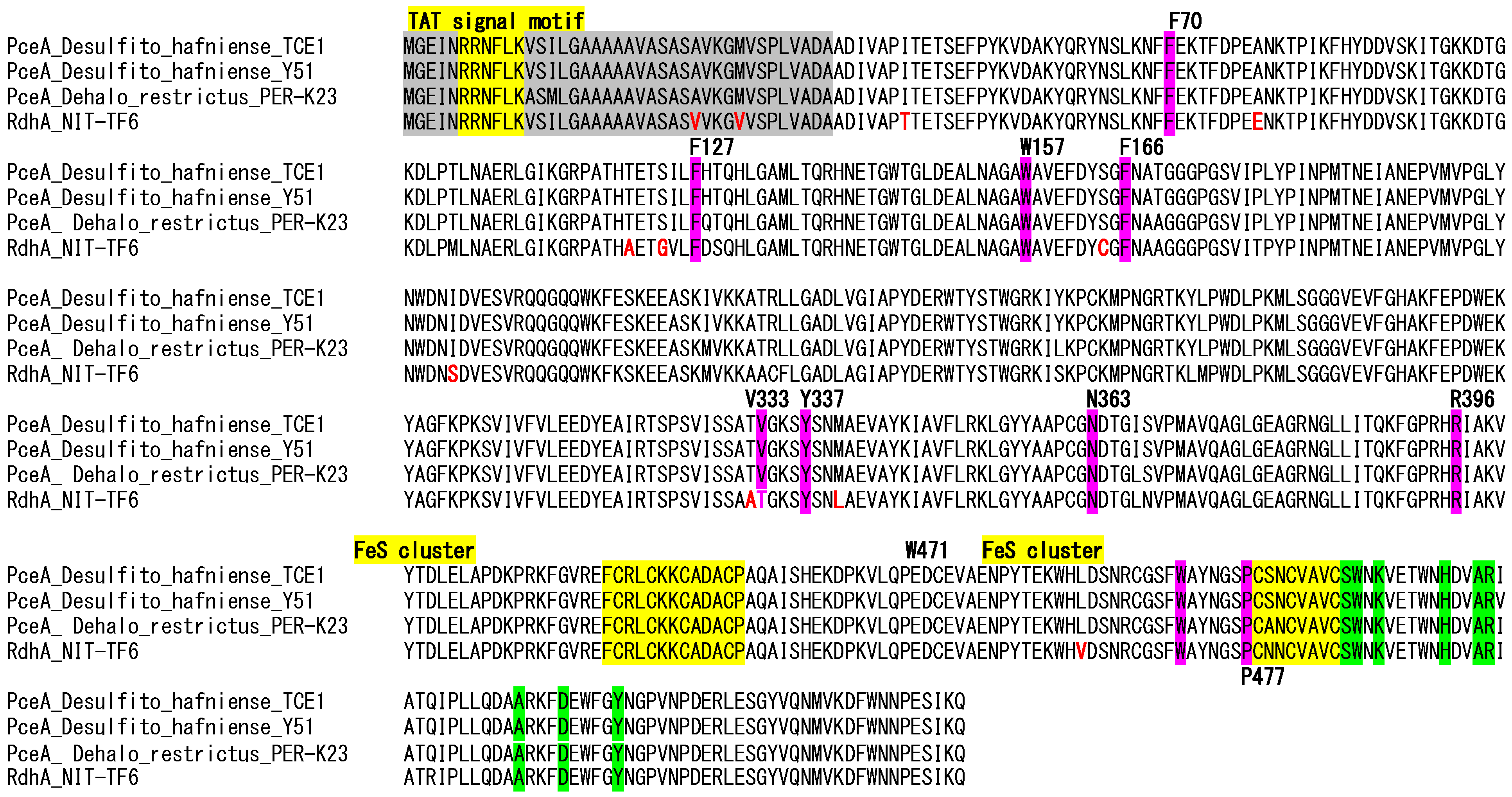
3.7. The RdhA Gene Cluster and Mechanism for the Inability to Dechlorinate Chloroethene
3.8. Role of Strain NIT-TF6 in the TCE-Dechlorinating Culture
4. Conclusions
Description of Desulfitobacterium elongatum sp. nov.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christiansen, N.; Ahring, B.K. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int. J. Syst. Bacteriol. 1996, 46, 442–448. [Google Scholar] [CrossRef]
- Utkin, I.; Woese, C.; Wiegel, J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int. J. Syst. Bacteriol. 1994, 44, 612–619. [Google Scholar] [CrossRef]
- Finneran, K.T.; Forbush, H.M.; Gaw VanPraagh, C.V.; Lovley, D.R. Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Int. J. Syst. Evol. Microbiol. 2002, 52, 1929–1935. [Google Scholar] [CrossRef]
- De Wildeman, S.; Diekert, G.; Van Langenhove, H.; Verstraete, W. Stereoselective microbial dehalorespiration with vicinal dichlorinated alkanes. Appl. Environ. Microbiol. 2003, 69, 5643–5647. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, G.; Yao, S.; Zhuang, L. Paradesulfitobacterium ferrireducens gen. nov., sp. nov., a Fe(III)-reducing bacterium from petroleum-contaminated soil and reclassification of Desulfitobacterium aromaticivorans as Paradesulfitobacterium aromaticivorans comb. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 005025. [Google Scholar] [CrossRef]
- Niggemyer, A.; Spring, S.; Stackebrandt, E.; Rosenzweig, R.F. Isolation and Characterization of a Novel As(V)-Reducing Bacterium: Implications for Arsenic Mobilization and the Genus Desulfitobacterium. Appl. Environ. Microbiol. 2001, 67, 5568–5580. [Google Scholar] [CrossRef] [PubMed]
- Villemur, R.; Lanthier, M.; Beaudet, R.; Lepine, F. The Desulfitobacterium genus. FEMS Microbiol. Rev. 2006, 30, 706–733. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ge, R.; Shi, C.; Wan, Z.; Zheng, D.; Huang, W.; Wu, Y.; Yang, K.; Li, G.; Zhang, F. Reductive dechlorination of trichloroethene at concentrations approaching saturation by a Desulfitobacterium-containing community. J. Hazard. Mater. 2025, 486, 137005. [Google Scholar] [CrossRef]
- Tomita, R.; Yoshida, N.; Meng, L. Formate: A promising electron donor to enhance trichloroethene-to-ethene dechlorination in Dehalococcoides-augmented groundwater ecosystems with minimal bacterial growth. Chemosphere 2022, 307, 136080. [Google Scholar] [CrossRef]
- Maymó-Gatell, X.; Chien, Y.T.; Gossett, J.M.; Zinder, S.H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 1997, 276, 1568–1571. [Google Scholar] [CrossRef]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Muller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 2, 625–635. [Google Scholar]
- Yan, J.; Wang, J.; Solis, V.; Jin, H.; Chourey, K.; Li, X.; Yang, Y.; Yin, Y.; Hettich, R.L.; Löffler, F.E. Respiratory vinyl chloride reductive dechlorination to ethene in TceA-expressing Dehalococcoides mccartyi. Environ. Sci. Technol. 2021, 55, 4831–4841. [Google Scholar] [CrossRef]
- Nonaka, H.; Keresztes, G.; Shinoda, Y.; Ikenaga, Y.; Abe, M.; Naito, K.; Inatomi, K.; Furukawa, K.; Inui, M.; Yukawa, H. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 2006, 188, 2262–2274. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, S.; Liu, N.; Du, Y.; Ding, G. Interspecies transfer of biosynthetic cobalamin for complete dechlorination of trichloroethene by Dehalococcoides mccartyi. Water Sci. Technol. 2022, 85, 1335–1350. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Harzman, C.; Davis, J.K.; Hutcheson, R.; Broderick, J.B.; Marsh, T.L.; Tiedje, J. Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol. 2012, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Asai, M.; Yoshida, N.; Kusakabe, T.; Ismaeil, M.; Nishiuchi, T.; Katayama, A. Dehalococcoides mccartyi NIT01, a novel isolate, dechlorinates high concentrations of chloroethenes by expressing at least six different reductive dehalogenases. Environ. Res. 2022, 207, 112150. [Google Scholar] [CrossRef]
- Ismaeil, M.; Yoshida, N.; Katayama, A. Identification of Multiple Dehalogenase Genes Involved in Tetrachloroethene-to-Ethene Dechlorination in a Dehalococcoides-Dominated Enrichment Culture. Biomed Res. Int. 2017, 2017, 9191086. [Google Scholar] [CrossRef]
- Yoshida, N.; Asahi, K.; Sakakibara, Y.; Miyake, K.; Katayama, A. Isolation and quantitative detection of tetrachloroethene (PCE)-dechlorinating bacteria in unsaturated subsurface soils contaminated with chloroethenes. J. Biosci. Bioeng. 2007, 104, 91–97. [Google Scholar] [CrossRef]
- Xie, L.; Yoshida, N.; Ishii, S.; Meng, L. Isolation and polyphasic characterization of Desulfuromonas versatilis sp. Nov., an electrogenic bacteria capable of versatile metabolism isolated from a graphene oxide-reducing enrichment culture. Microorganisms 2021, 9, 1953. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.; Gerdes, S.; Parello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Yoshida, N.; Ye, L.; Baba, D.; Katayama, A. A novel Dehalobacter species is involved in extensive 4,5,6,7-Tetrachlorophthalide dichlorination. Appl. Envirom. Microbiol. 2009, 75, 2400–2405. [Google Scholar] [CrossRef]
- Rodriguez-R., L.M.; Konstantinidis, K.T. Bypassing Cultivation To Identify Bacterial Species Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. Microbe 2014, 9, 111–118. [Google Scholar]
- Pohanka, M. D-lactic acid as a metabolite: Toxicology, diagnosis, and detection. Biomed Res. Int. 2020, 2020, 3419034. [Google Scholar] [CrossRef]
- Alfán-Guzmán, R.; Ertan, H.; Manefield, M.; Lee, M. Isolation and characterization of Dehalobacter sp. strain TeCB1 including identification of TcbA: A novel tetra-and trichlorobenzene reductive dehalogenase. Front. Microbiol. 2017, 8, 558. [Google Scholar] [CrossRef] [PubMed]

| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| Size (µm) | 1.6 − 6 × 0.25 − 0.5 | 2.5 − 4 × 0.5 − 0.7 | 2 − 5 × 0.5 − 0.7 | 3.3 − 6 × 0.6 − 0.7 | 2 − 5 × 0.5 |
| Spores | + | − | ND | + | − |
| Cell morphology | Straight Rod | Straight or curved rod | Curved rod | Straight rod | Curved rod |
| e− donor for thiosulfate respiration | |||||
| H2+acetate | + | ND | + | ND | − |
| Formate | − | + | + | − | + |
| Acetate | − | − | − | ND | − |
| Citrate | − | ND | ND | ND | − |
| Fumarate | − | ND | ND | ND | ND |
| Malate | + | ND | ND | ND | + |
| Pyruvate | + | + | ND | + | − |
| Lactate | + | + | + | + | + |
| Propionate | − | − | ND | − | − |
| Butyrate | − | + | ND | − | + |
| Methanol | − | − | ND | ND | ND |
| Ethanol | − | + | ND | ND | + |
| Propanol | − | ND | ND | ND | − |
| Benzoate | − | ND | ND | − | − |
| e− acceptors for pyruvate oxidization | |||||
| Thiosulfate | + | + | + | + | + |
| Sulfate | − | − | − | − | − |
| Sulfite | − | + | + | + | − |
| Nitrate | − | + | + | + | − |
| Nitrite | − | ND | − | ND | − |
| Fe(III)-citrate | + | ND | ND | ND | + |
| O2 (air) | − | − | ND | ND | − |
| Features | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Size (Mb) | 4.8 | 4.3 | 3.6 | 5.2 | 3.1 |
| GC (%) | 43.2 | 45 | 44.2 | 47.5 | 41.9 |
| CDSs | 4655 | 4403 | 3544 | 5247 | 3138 |
| 16S rRNAs | 9 | 6 | 6 | 5 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharjee, U.; Tomita, R.; Xie, L.; Yoshida, N. Desulfitobacterium elongatum sp. nov. NIT-TF6 Isolated from Trichloroethene-Dechlorinating Culture with Formate. Microorganisms 2025, 13, 1863. https://doi.org/10.3390/microorganisms13081863
Bhattacharjee U, Tomita R, Xie L, Yoshida N. Desulfitobacterium elongatum sp. nov. NIT-TF6 Isolated from Trichloroethene-Dechlorinating Culture with Formate. Microorganisms. 2025; 13(8):1863. https://doi.org/10.3390/microorganisms13081863
Chicago/Turabian StyleBhattacharjee, Udaratta, Ryuya Tomita, Li Xie, and Naoko Yoshida. 2025. "Desulfitobacterium elongatum sp. nov. NIT-TF6 Isolated from Trichloroethene-Dechlorinating Culture with Formate" Microorganisms 13, no. 8: 1863. https://doi.org/10.3390/microorganisms13081863
APA StyleBhattacharjee, U., Tomita, R., Xie, L., & Yoshida, N. (2025). Desulfitobacterium elongatum sp. nov. NIT-TF6 Isolated from Trichloroethene-Dechlorinating Culture with Formate. Microorganisms, 13(8), 1863. https://doi.org/10.3390/microorganisms13081863






