Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis?
Abstract
1. Introduction
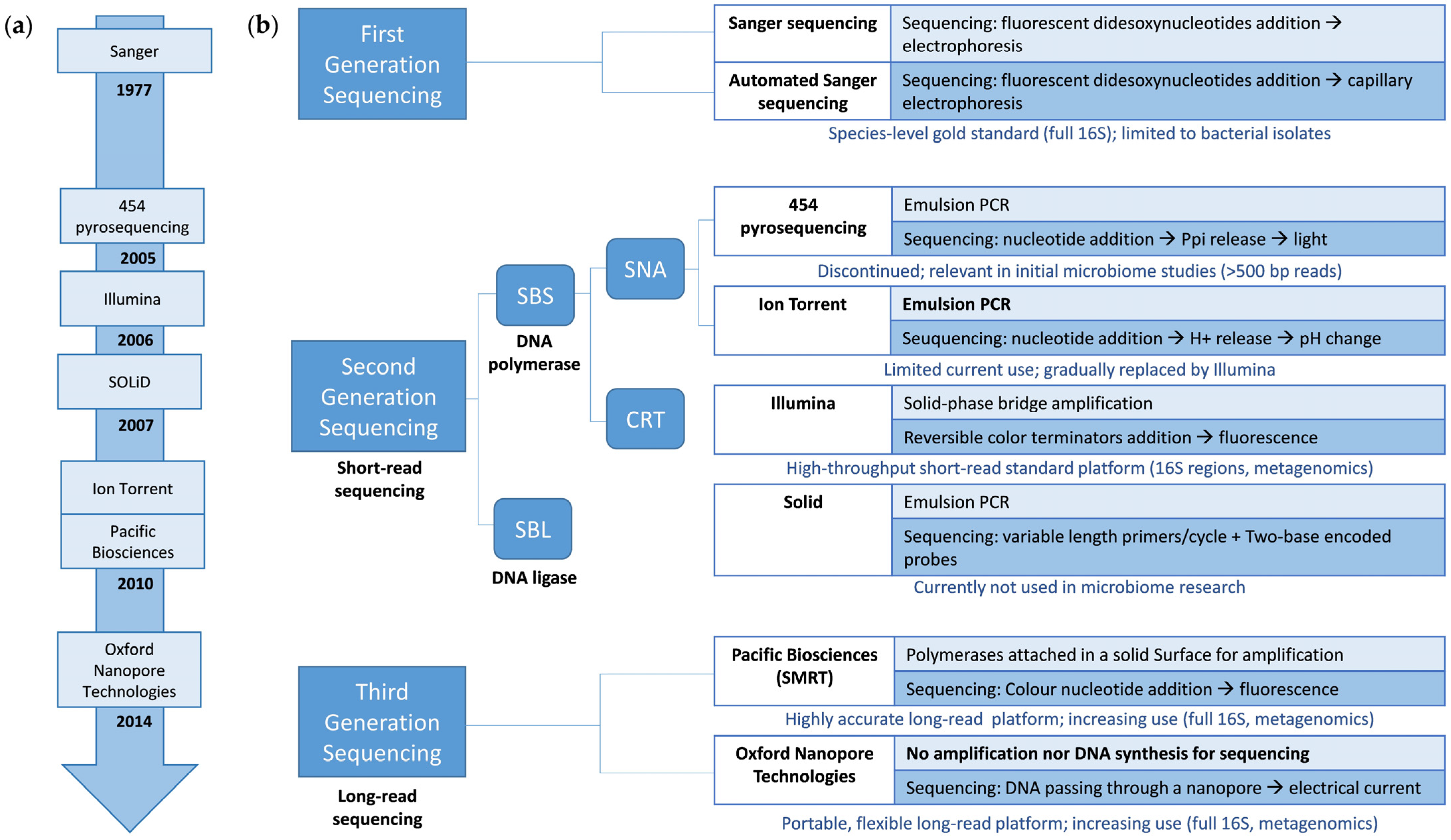
2. Evolution of Sequencing Technologies for Microbiome Analysis in Ecosystems
3. The Early Development of Microbiome Sequencing: From Culture to First-Generation Sequencing
4. A Revolution in Microbiome Analysis: The Advent of Next-Generation Sequencing
4.1. Sequencing by Synthesis: Single-Nucleotide Addition (454 and Ion Torrent)
4.1.1. 454 Pyrosequencing
4.1.2. Ion Torrent
4.2. Sequencing by Synthesis: Cyclic Reversible Termination (Illumina)
4.3. Sequencing by Ligation (SOLiD)
5. New Era in Microbiome Analysis: The Development of Third-Generation Sequencing
5.1. Pacific Biosciences
5.2. Oxford Nanopore Technologies
6. Long-Read Sequencing Technologies: New Perspectives in the Analysis of Microbiome
6.1. Predominance of Host DNA: A Challenge in Microbiome Analysis
6.2. Towards a More Accurate Taxonomic Identification in Microbial Communities
6.2.1. Bacteria
6.2.2. Fungi
6.2.3. Virome
6.3. Complete and Accurate Assembly of Microbial Genomes
6.4. Microbial Epigenome Profiling
7. Desirable Characteristics for Microbiome Sequencing Methods
7.1. Read Length
7.2. Accuracy
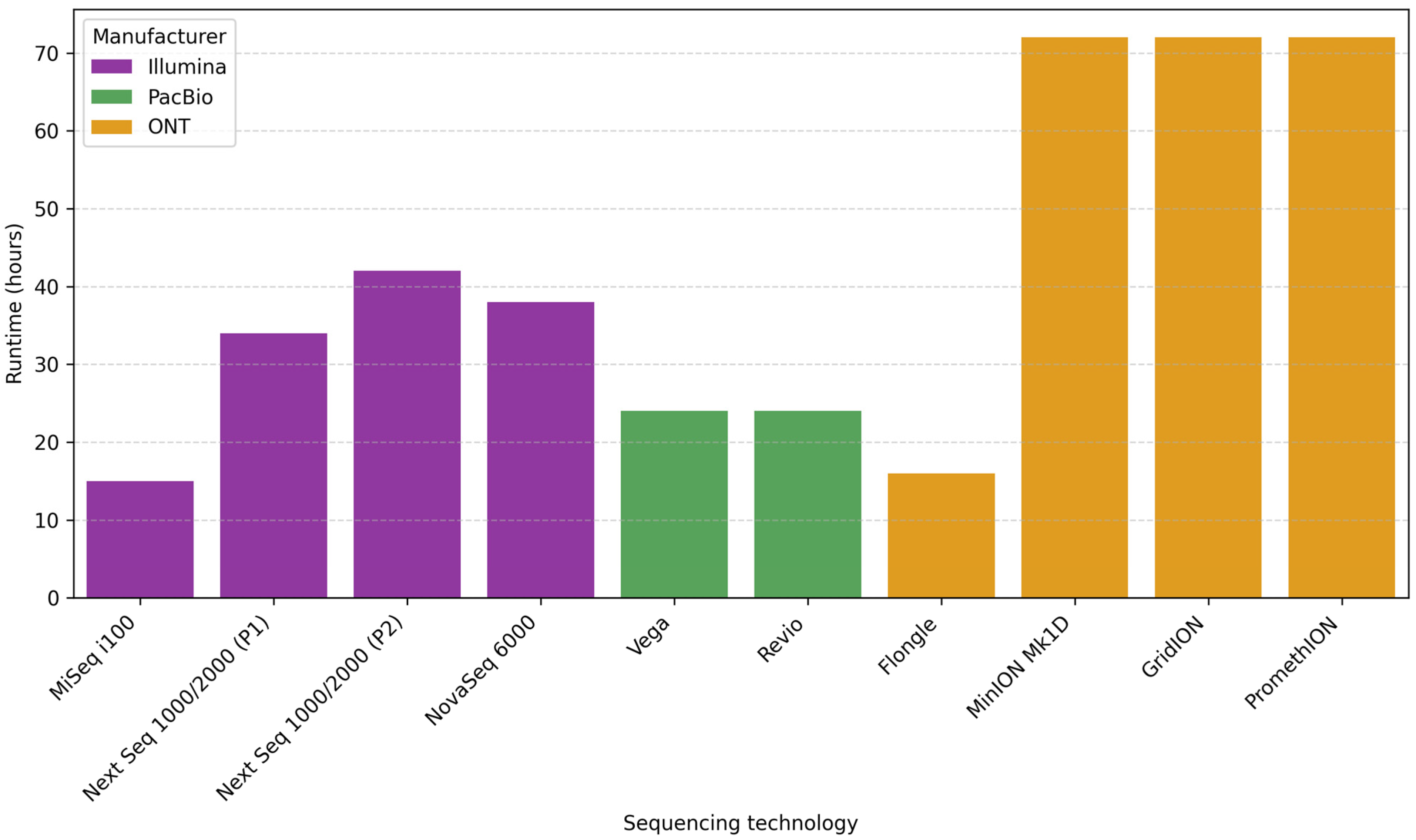
7.3. Runtime
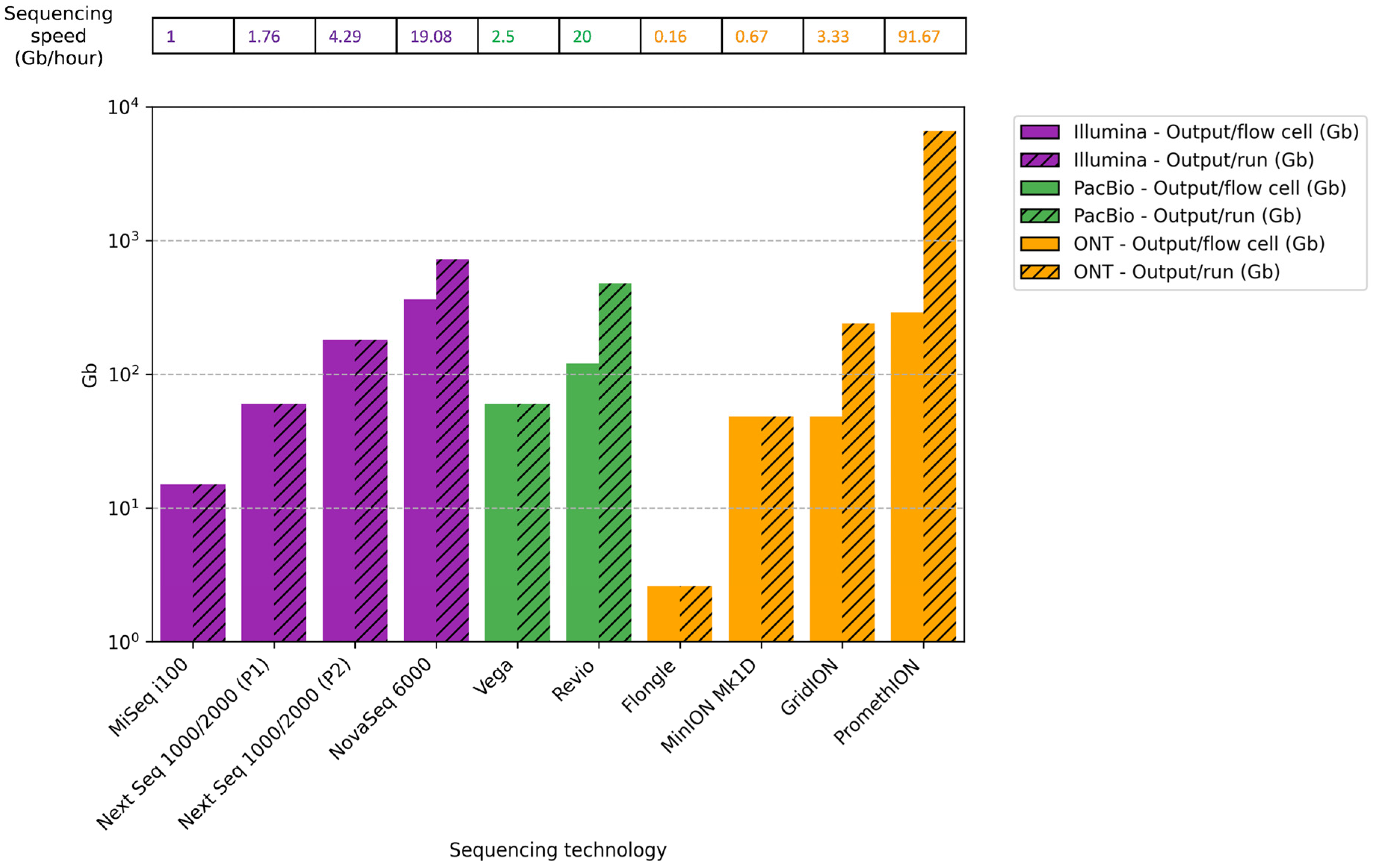
7.4. Sequencing Output per Cell
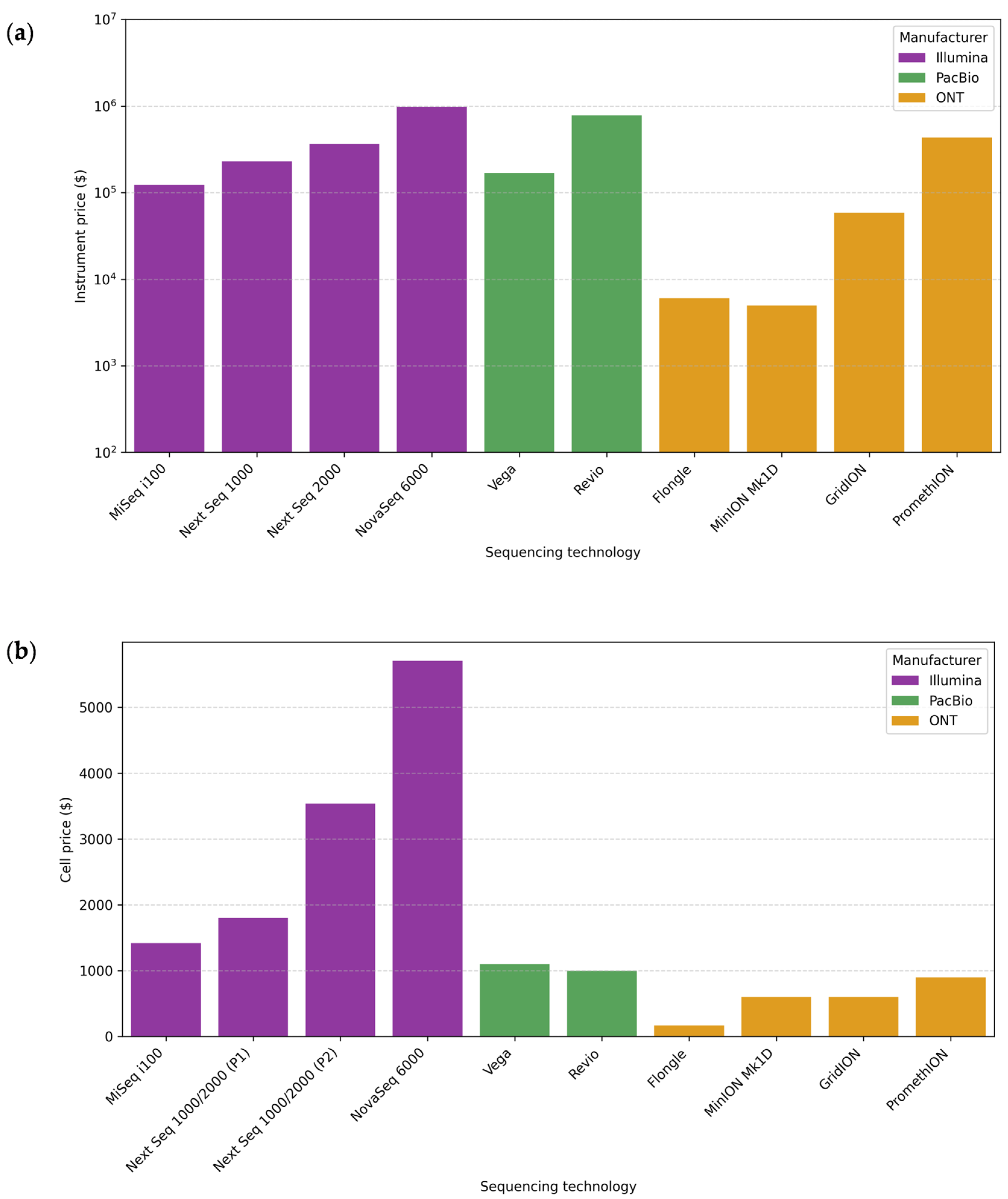
7.5. Cost
7.6. Equipment Portability
7.7. Bioinformatic Tools for Sequencing Data
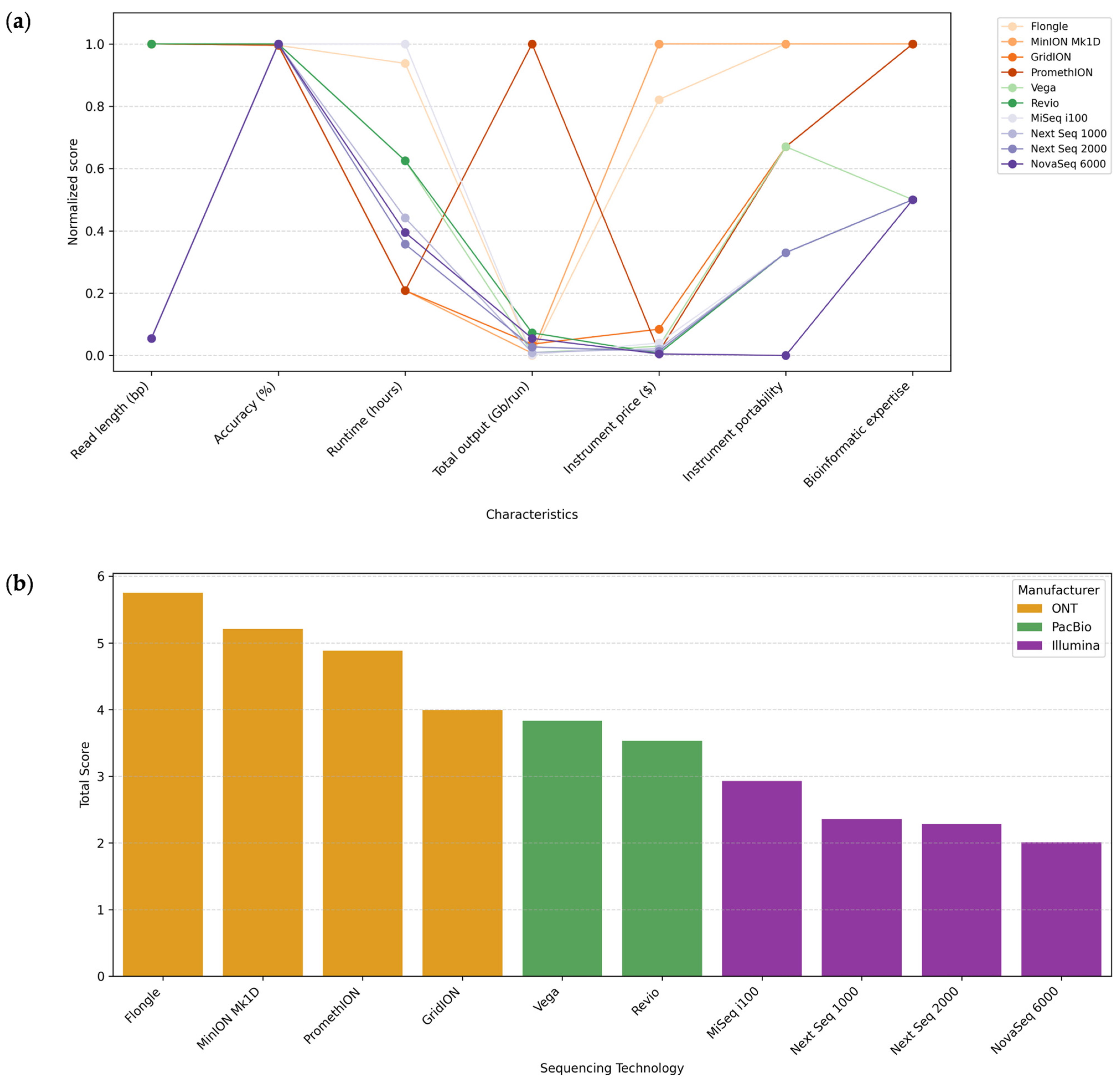
7.8. General Comparison
8. ONT Applications: Portable, Affordable, Fast, and Real-Time Sequencing
9. Additional Key Aspects When Using Long-Read Sequencing for Microbiome Analysis in Ecosystems
9.1. Current Perspectives of Amplicon and Shotgun Sequencing Approach
9.1.1. 16S Ribosomal RNA (16S) Gene Amplicons
| Target Region | Primer Pairs | Amplicon Length (~) | Primer Specificity | Accurate Taxonomic Resolution | Other Remarks |
|---|---|---|---|---|---|
| 16S-ITS-23S rRNA operon | 27F, 519F, 2241R and 2428R [6,196] | 4500 bp [196] | Universal [6,196] | Species and strain levels [6,196] | It is especially useful for distinguishing Escherichia coli and Shigella spp.; limitations in detecting archaeal taxa; emerging method; requires long-read sequencing [6,196] |
| V1–V9 (16S rRNA) | 27F-1492R [6] | 1465 bp [53] | Universal [188] | Species and strain levels [53,205] | Better taxonomic resolution than 16S regions; 27 F primer has limited amplification for Bifidobacterium [206]; requires long-read sequencing [53] |
| V1–V2 | 27F-338R [207] | 310 bp [53] | Universal [188] | Genus level; good for archaea [127,193] | Low sensitivity for Bifidobacterium [190], Verrucomicrobia [191], and Proteobacteria [53]; suitable for low-bacterial biomass samples [208]; recommended region for sputum microbiome analysis; commonly used with Illumina [202] |
| V1–V3 | 27F-534R [209] | 507 bp [53] | Universal [188] | Genus level; informative at species level [53] | Good sensitivity for Escherichia/Shigella. Poor for Bacteroides intestinalis [53] and Verrucomicrobia [191]; used in HMP (454) [189]; recommended region for plant [210] and skin microbiome analyses [211]; suitable for long-read sequencing platforms |
| V3 | 338F-533R [212]; ARC344F-519R [193,213] | 200 bp [212,213] | Bacteria: 338F-533R [212]; archaea: ARC344F-519R [193,213] | Genus level; ARC344F-519R good for archaea [193,212,213] | ARC344F-519R is considered the best choice for archaea community profiling [193] |
| V4 | 515F-806R [214] | 291 bp [53] | Universal [188] | Genus level [53] | Susceptible to human DNA amplification [208]; recommended region for diverse microbial communities [18]; used in EMP; commonly used with Illumina [215]; reduced bias against the SAR11 bacterial clade with 806RB primer [216] |
| V3–V4 | 341F-785R [192] | 464 bp [192] | Bacteria [192] | Genus level [192] | Fails to detect Chloroflexi and Elusimicrobia [192]; widely used region for human-associated, soil, and plant microbiome analysis; commonly used with Illumina [188] |
| V3–V5 | 357F-926R [53] | 569 bp [53] | Bacteria [188] | Genus level [53] | Susceptible to human DNA amplification [208]; good sensitivity for Klebsiella and poor for Actinobacteria [53]; used in HMP (454) [189] and MetaHit [217]; suitable for long-read sequencing platforms |
| V4–V5 | 515F-944R [188] | 429 bp [188] | Bacteria [188]; 515F-Y/926R universal [194] | Genus level [218]; 515F-Y/926R good for archaea [194] | Low sensitivity for Bacteroidota, with few overlaps with other primer pairs [188]; 515F-Y/926R primer pair has reduced bias against environmental archaea Crenarchaeota/Thaumarchaeota [194]; 515F-Y/926R is widely used in marine microbiome studies and tested in temperate water microbiomes [194,219] |
| V6–V9 | 968F/1492R [53] | 524 bp [53] | Bacteria [188] | Genus level [53] | Good sensitivity for Clostridium and Staphylococcus [53]; suitable for long-read sequencing platforms |
9.1.2. Metagenomic Shotgun Sequencing
9.2. Current Perspectives on Sequencing Depth
9.3. The Emergence of Microbiome Databases Specific to Ecosystems
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 16S | 16S ribosomal RNA gene |
| ASV | Amplicon Sequence Variant |
| bp | Base pair |
| CCD | Charge-coupled device |
| CMOS | Integrated complementary metal-oxide semiconductor |
| CRT | Cyclic reversible termination |
| EMP | Earth Microbiome Project |
| ES-DBs | Ecosystem-specific databases |
| ETS | External transcribed spacer |
| Gb | Gigabases |
| HiFi | High fidelity |
| HMP | Human Microbiome Project |
| HPRC | Human Pangenome Reference Consortium |
| IGS | Intergenic spacer |
| Indel | Insertions and deletions |
| ISFET | Ion-sensitive field-effect transistor |
| ISS | International Space Station |
| ITS | Internal transcribed spacer |
| Kb | Kilobases |
| M | Millions |
| Mb | Megabases |
| MAG | Metagenome-assembled genome |
| MeDIP-seq | Immunoprecipitation sequencing of methylated DNA |
| MIDAS 3 | Microbial Database for Activated Sludge |
| NGS | Next-generation sequencing |
| NHGR | National Human Genome Research Institute |
| NIH | National Institutes of Health |
| ONT | Oxford Nanopore Technologies |
| OTU | Operational Taxonomic Unit |
| PacBio | Pacific Biosciences |
| PPi | Inorganic pyrophosphate |
| SBL | Sequencing by ligation |
| SBS | Sequencing by synthesis |
| SMRT | Single-molecule real-time sequencing |
| SNA | Single-nucleotide addition |
| SNP | Single-nucleotide polymorphism |
| T2T | Telomere-to-Telomere Consortium |
| WHO | World Health Organization |
| WGBS | Whole-genome bisulfite sequencing |
| ZMD | Zero-mode waveguide |
References
- Seitz, T.J.; Schütte, U.M.E.; Drown, D.M. Soil Disturbance Affects Plant Productivity via Soil Microbial Community Shifts. Front. Microbiol. 2021, 12, 619711. [Google Scholar] [CrossRef]
- Klinsawat, W.; Uthaipaisanwong, P.; Jenjaroenpun, P.; Sripiboon, S.; Wongsurawat, T.; Kusonmano, K. Microbiome Variations among Age Classes and Diets of Captive Asian Elephants (Elephas maximus) in Thailand Using Full-Length 16S rRNA Nanopore Sequencing. Sci. Rep. 2023, 13, 17685. [Google Scholar] [CrossRef]
- Zarantonello, G.; Cuenca, A. Nanopore-Enabled Microbiome Analysis: Investigating Environmental and Host-Associated Samples in Rainbow Trout Aquaculture. Curr. Protoc. 2024, 4, e1069. [Google Scholar] [CrossRef]
- Esberg, A.; Fries, N.; Haworth, S.; Johansson, I. Saliva Microbiome Profiling by Full-Gene 16S rRNA Oxford Nanopore Technology versus Illumina MiSeq Sequencing. Npj Biofilms Microbiomes 2024, 10, 149. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, D.; Rillig, M.C.; Yang, Y.; Chu, H.; Chen, Q.; Penuelas, J.; Cui, H.; Gillings, M. Ecosystem Microbiome Science. mLife 2023, 2, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Albertsen, M.; Anslan, S.; Callahan, B. Perspectives and Benefits of High-Throughput Long-Read Sequencing in Microbial Ecology. Appl. Environ. Microbiol. 2021, 87, e00626-21. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Li, X.; Wu, Z.; Nie, C.; Cheng, Z.; Sun, Y.; Liu, L.; Zhang, T. Strategies and Tools in Illumina and Nanopore-integrated Metagenomic Analysis of Microbiome Data. iMeta 2023, 2, e72. [Google Scholar] [CrossRef] [PubMed]
- Tomasulo, A.; Simionati, B.; Facchin, S. Microbiome One Health Model for a Healthy Ecosystem. Sci. One Health 2024, 3, 100065. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO) Tripartite and UNEP Support OHHLEP’s Definition of “One Health”. Available online: https://www.who.int/news/item/01-12-2021-tripartite-and-unep-support-ohhlep-s-definition-of-one-health (accessed on 16 April 2025).
- Ma, L.; Zhao, H.; Wu, L.B.; Cheng, Z.; Liu, C. Impact of the Microbiome on Human, Animal, and Environmental Health from a One Health Perspective. Sci. One Health 2023, 2, 100037. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar]
- Rappé, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA Sequencing with Chain-Terminating Inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Prober, J.M.; Trainor, G.L.; Dam, R.J.; Hobbs, F.W.; Robertson, C.W.; Zagursky, R.J.; Cocuzza, A.J.; Jensen, M.A.; Baumeister, K. A System for Rapid DNA Sequencing with Fluorescent Chain-Terminating Dideoxynucleotides. Science 1987, 238, 336–341. [Google Scholar] [CrossRef]
- Smith, L.M.; Sanders, J.Z.; Kaiser, R.J.; Hughes, P.; Dodd, C.; Connell, C.R.; Heiner, C.; Kent, S.B.; Hood, L.E. Fluorescence Detection in Automated DNA Sequence Analysis. Nature 1986, 321, 674–679. [Google Scholar] [CrossRef]
- Singh, A.P. Genomic Techniques Used to Investigate the Human Gut Microbiota. In Human Microbiome; IntechOpen: London, UK, 2021; ISBN 978-1-78984-849-6. [Google Scholar]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation Sequencing Systems. BioMed Res. Int. 2012, 2012, 251364. [Google Scholar] [CrossRef]
- Kuczynski, J.; Lauber, C.L.; Walters, W.A.; Parfrey, L.W.; Clemente, J.C.; Gevers, D.; Knight, R. Experimental and Analytical Tools for Studying the Human Microbiome. Nat. Rev. Genet. 2012, 13, 47–58. [Google Scholar] [CrossRef]
- Collins, F.S.; Morgan, M.; Patrinos, A. The Human Genome Project: Lessons from Large-Scale Biology. Science 2003, 300, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Metzker, M.L. Emerging Technologies in DNA Sequencing. Genome Res. 2005, 15, 1767–1776. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Fan, D.; Wang, J.; Zhou, X.; Teng, X.; Rao, C. High Throughput Construction of Species Characterized Bacterial Biobank for Functional Bacteria Screening: Demonstration with GABA-Producing Bacteria. Front. Microbiol. 2025, 16, 1545877. [Google Scholar] [CrossRef] [PubMed]
- National Human Genome Research Institute (NHGRI). DNA Sequencing Costs: Data. Available online: https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data (accessed on 7 June 2025).
- National Human Genome Research Institute (NHGRI). The Cost of Sequencing a Human Genome. Available online: https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost (accessed on 7 June 2025).
- Mardis, E.R. A Decade’s Perspective on DNA Sequencing Technology. Nature 2011, 470, 198–203. [Google Scholar] [CrossRef]
- Feng, X.; Ding, W.; Xiong, L.; Guo, L.; Sun, J.; Xiao, P. Recent Advancements in Intestinal Microbiota Analyses: A Review for Non-Microbiologists. Curr. Med. Sci. 2018, 38, 949–961. [Google Scholar] [CrossRef]
- Methé, B.A.; Nelson, K.E.; Pop, M.; Creasy, H.H.; Giglio, M.G.; Huttenhower, C.; Gevers, D.; Petrosino, J.F.; Abubucker, S.; Badger, J.H.; et al. A Framework for Human Microbiome Research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C.; et al. The NIH Human Microbiome Project. Genome Res. 2009, 19, 2317–2323. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd_Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of Age: Ten Years of next-Generation Sequencing Technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing Technologies-The next Generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Nyrén, P.; Pettersson, B.; Uhlén, M. Solid Phase DNA Minisequencing by an Enzymatic Luminometric Inorganic Pyrophosphate Detection Assay. Anal. Biochem. 1993, 208, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z.; et al. Genome Sequencing in Microfabricated High-Density Picolitre Reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, Function and Diversity of the Healthy Human Microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Granberg, F.; Vicente-Rubiano, M.; Rubio-Guerri, C.; Karlsson, O.E.; Kukielka, D.; Belák, S.; Sánchez-Vizcaíno, J.M. Metagenomic Detection of Viral Pathogens in Spanish Honeybees: Co-Infection by Aphid Lethal Paralysis, Israel Acute Paralysis and Lake Sinai Viruses. PLoS ONE 2013, 8, e57459. [Google Scholar] [CrossRef]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An Integrated Semiconductor Device Enabling Non-Optical Genome Sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef]
- Martín, J.M.V.; Ortigosa, F.; Cañas, R.A. Métodos de secuenciación: Segunda generación. Encuentros Biol. 2020, 13, 17–23. [Google Scholar]
- Merriman, B.; Ion Torrent R&D Team; Rothberg, J.M. Progress in Ion Torrent Semiconductor Chip Based Sequencing. Electrophoresis 2012, 33, 3397–3417. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Krishnamurthy, K.; Goldstein, D.Y. P744: Comparative Analysis of Ion Torrent Sequencing Platforms: Unveiling Enhanced Performance and Precision with the Genexus Integrated Sequencer in Clinical Applications. Genet. Med. Open 2024, 2, 101648. [Google Scholar] [CrossRef]
- Conrads, G.; Abdelbary, M.M.H. Challenges of Next-Generation Sequencing Targeting Anaerobes. Anaerobe 2019, 58, 47–52. [Google Scholar] [CrossRef]
- Salipante, S.J.; Kawashima, T.; Rosenthal, C.; Hoogestraat, D.R.; Cummings, L.A.; Sengupta, D.J.; Harkins, T.T.; Cookson, B.T.; Hoffman, N.G. Performance Comparison of Illumina and Ion Torrent Next-Generation Sequencing Platforms for 16S rRNA-Based Bacterial Community Profiling. Appl. Environ. Microbiol. 2014, 80, 7583–7591. [Google Scholar] [CrossRef] [PubMed]
- Onywera, H.; Meiring, T.L. Comparative Analyses of Ion Torrent V4 and Illumina V3-V4 16S rRNA Gene Metabarcoding Methods for Characterization of Cervical Microbiota: Taxonomic and Functional Profiling. Sci. Afr. 2020, 7, e00278. [Google Scholar] [CrossRef]
- Pylro, V.S.; Roesch, L.F.W.; Morais, D.K.; Clark, I.M.; Hirsch, P.R.; Tótola, M.R. Data Analysis for 16S Microbial Profiling from Different Benchtop Sequencing Platforms. J. Microbiol. Methods 2014, 107, 30–37. [Google Scholar] [CrossRef]
- Loman, N.J.; Misra, R.V.; Dallman, T.J.; Constantinidou, C.; Gharbia, S.E.; Wain, J.; Pallen, M.J. Performance Comparison of Benchtop High-Throughput Sequencing Platforms. Nat. Biotechnol. 2012, 30, 434–439. [Google Scholar] [CrossRef]
- Torrell, H.; Cereto-Massagué, A.; Kazakova, P.; García, L.; Palacios, H.; Canela, N. Multiomic Approach to Analyze Infant Gut Microbiota: Experimental and Analytical Method Optimization. Biomolecules 2021, 11, 999. [Google Scholar] [CrossRef]
- Terrazzan, A.C.; Procianoy, R.S.; Roesch, L.F.W.; Corso, A.L.; Dobbler, P.T.; Silveira, R.C. Meconium Microbiome and Its Relation to Neonatal Growth and Head Circumference Catch-up in Preterm Infants. PLoS ONE 2020, 15, e0238632. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A Practical Guide to Amplicon and Metagenomic Analysis of Microbiome Data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Landegren, U.; Kaiser, R.; Sanders, J.; Hood, L. A Ligase-Mediated Gene Detection Technique. Science 1988, 241, 1077–1080. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Förster-Fromme, K.; Damms-Machado, A.; Scheurenbrand, T.; Biskup, S.; Huson, D.H.; Bischoff, S.C. Analysis of the Intestinal Microbiota Using SOLiD 16S rRNA Gene Sequencing and SOLiD Shotgun Sequencing. BMC Genom. 2013, 14, S16. [Google Scholar] [CrossRef]
- McCarroll, S.A.; Altshuler, D.M. Copy-Number Variation and Association Studies of Human Disease. Nat. Genet. 2007, 39, S37–S42. [Google Scholar] [CrossRef]
- Stankiewicz, P.; Lupski, J.R. Structural Variation in the Human Genome and Its Role in Disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef]
- Nossa, C.W.; Oberdorf, W.; Yang, L.; Aas, J.; Paster, B.; Desantis, T.; Brodie, E.; Malamud, D.; Poles, M.; Pei, Z. Design of 16S rRNA Gene Primers for 454 Pyrosequencing of the Human Foregut Microbiome. World J. Gastroenterol. WJG 2010, 16, 4135. [Google Scholar] [CrossRef]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium Acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.-Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA Gene Sequencing for Species and Strain-Level Microbiome Analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Marx, V. Method of the Year: Long-Read Sequencing. Nat. Methods 2023, 20, 6–11. [Google Scholar] [CrossRef]
- Formenti, G.; Theissinger, K.; Fernandes, C.; Bista, I.; Bombarely, A.; Bleidorn, C.; Ciofi, C.; Crottini, A.; Godoy, J.A.; Höglund, J.; et al. The Era of Reference Genomes in Conservation Genomics. Trends Ecol. Evol. 2022, 37, 197–202. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The Complete Sequence of a Human Genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Wang, T.; Antonacci-Fulton, L.; Howe, K.; Lawson, H.A.; Lucas, J.K.; Phillippy, A.M.; Popejoy, A.B.; Asri, M.; Carson, C.; Chaisson, M.J.P.; et al. The Human Pangenome Project: A Global Resource to Map Genomic Diversity. Nature 2022, 604, 437–446. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D.; et al. Accurate Circular Consensus Long-Read Sequencing Improves Variant Detection and Assembly of a Human Genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Pacific Biosciences. Sequencing Systems. Available online: https://www.pacb.com/sequencing-systems/ (accessed on 17 June 2025).
- Chin, C.-S.; Sorenson, J.; Harris, J.B.; Robins, W.P.; Charles, R.C.; Jean-Charles, R.R.; Bullard, J.; Webster, D.R.; Kasarskis, A.; Peluso, P.; et al. The Origin of the Haitian Cholera Outbreak Strain. N. Engl. J. Med. 2011, 364, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Su, L.; Wang, Y.; Deng, S. Improved High-Throughput Sequencing of the Human Oral Microbiome: From Illumina to PacBio. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 6678872. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Akeson, M.; Branton, D. Three Decades of Nanopore Sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Oxford Nanopore Technologies. Welcome to Oxford Nanopore Technologies. Available online: https://nanoporetech.com/ (accessed on 12 June 2025).
- Leggett, R.M.; Clark, M.D. A World of Opportunities with Nanopore Sequencing. J. Exp. Bot. 2017, 68, 5419–5429. [Google Scholar] [CrossRef]
- Lin, B.; Hui, J.; Mao, H. Nanopore Technology and Its Applications in Gene Sequencing. Biosensors 2021, 11, 214. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Kit 14 Sequencing and Duplex Basecalling. Available online: https://nanoporetech.com/document/kit-14-device-and-informatics (accessed on 17 June 2025).
- Stoeck, T.; Katzenmeier, S.N.; Breiner, H.-W.; Rubel, V. Nanopore Duplex Sequencing as an Alternative to Illumina MiSeq Sequencing for eDNA-Based Biomonitoring of Coastal Aquaculture Impacts. Metabarcoding Metagenom. 2024, 8, e121817. [Google Scholar] [CrossRef]
- Chen, B.; Zhou, Y.; Duan, L.; Gong, X.; Liu, X.; Pan, K.; Zeng, D.; Ni, X.; Zeng, Y. Complete Genome Analysis of Bacillus velezensis TS5 and Its Potential as a Probiotic Strain in Mice. Front. Microbiol. 2023, 14, 1322910. [Google Scholar] [CrossRef]
- Cha, T.; Kim, H.H.; Keum, J.; Kwak, M.-J.; Park, J.Y.; Hoh, J.K.; Kim, C.-R.; Jeon, B.-H.; Park, H.-K. Gut Microbiome Profiling of Neonates Using Nanopore MinION and Illumina MiSeq Sequencing. Front. Microbiol. 2023, 14, 1148466. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun Metagenomics, from Sampling to Analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Marques, J.; Hout, A.; Ferreira, R.M.; Weber, M.; Pinto-Ribeiro, I.; van Doorn, L.-J.; Knetsch, C.W.; Figueiredo, C. Impact of Host DNA and Sequencing Depth on the Taxonomic Resolution of Whole Metagenome Sequencing for Microbiome Analysis. Front. Microbiol. 2019, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Notario, E.; Visci, G.; Fosso, B.; Gissi, C.; Tanaskovic, N.; Rescigno, M.; Marzano, M.; Pesole, G. Amplicon-Based Microbiome Profiling: From Second- to Third-Generation Sequencing for Higher Taxonomic Resolution. Genes 2023, 14, 1567. [Google Scholar] [CrossRef]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic Fungi: Past, Present, and Future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Dai, M.; Xu, J.; Chen, L.; Zhang, F.; Zhao, N.; Wang, J. Profiling of Human Gut Virome with Oxford Nanopore Technology. Med. Microecol. 2020, 4, 100012. [Google Scholar] [CrossRef]
- Lee, C.Z.; Zoqratt, M.Z.H.M.; Phipps, M.E.; Barr, J.J.; Lal, S.K.; Ayub, Q.; Rahman, S. The Gut Virome in Two Indigenous Populations from Malaysia. Sci. Rep. 2022, 12, 1824. [Google Scholar] [CrossRef]
- Zhao, L.; Shi, Y.; Lau, H.C.-H.; Liu, W.; Luo, G.; Wang, G.; Liu, C.; Pan, Y.; Zhou, Q.; Ding, Y.; et al. Uncovering 1058 Novel Human Enteric DNA Viruses Through Deep Long-Read Third-Generation Sequencing and Their Clinical Impact. Gastroenterology 2022, 163, 699–711. [Google Scholar] [CrossRef]
- Singleton, C.M.; Petriglieri, F.; Kristensen, J.M.; Kirkegaard, R.H.; Michaelsen, T.Y.; Andersen, M.H.; Kondrotaite, Z.; Karst, S.M.; Dueholm, M.S.; Nielsen, P.H.; et al. Connecting Structure to Function with the Recovery of over 1000 High-Quality Metagenome-Assembled Genomes from Activated Sludge Using Long-Read Sequencing. Nat. Commun. 2021, 12, 2009. [Google Scholar] [CrossRef]
- Fang, G.; Munera, D.; Friedman, D.I.; Mandlik, A.; Chao, M.C.; Banerjee, O.; Feng, Z.; Losic, B.; Mahajan, M.C.; Jabado, O.J.; et al. Genome-Wide Mapping of Methylated Adenine Residues in Pathogenic Escherichia coli Using Single-Molecule Real-Time Sequencing. Nat. Biotechnol. 2012, 30, 1232–1239. [Google Scholar] [CrossRef] [PubMed]
- Tourancheau, A.; Mead, E.A.; Zhang, X.-S.; Fang, G. Discovering Multiple Types of DNA Methylation from Bacteria and Microbiome Using Nanopore Sequencing. Nat. Methods 2021, 18, 491–498. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, Functions and Dynamics in the Expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Cleveland, K.; Schnoll-Sussman, F.; McClure, B.; Bigg, M.; Thakkar, P.; Schultz, N.; Shah, M.A.; Betel, D. Identification of Low Abundance Microbiome in Clinical Samples Using Whole Genome Sequencing. Genome Biol. 2015, 16, 265. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Adaptive Sampling. Available online: https://nanoporetech.com/document/adaptive-sampling (accessed on 7 June 2025).
- Zhang, W.; Fan, X.; Shi, H.; Li, J.; Zhang, M.; Zhao, J.; Su, X. Comprehensive Assessment of 16S rRNA Gene Amplicon Sequencing for Microbiome Profiling across Multiple Habitats. Microbiol. Spectr. 2023, 11, e00563-23. [Google Scholar] [CrossRef]
- Djemiel, C.; Maron, P.-A.; Terrat, S.; Dequiedt, S.; Cottin, A.; Ranjard, L. Inferring Microbiota Functions from Taxonomic Genes: A Review. GigaScience 2022, 11, giab090. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Wu, C.-C.; Li, Y.-E.; Chen, C.-L.; Lin, C.-R.; Ni, Y.-H. Full-Length 16S rRNA Sequencing Reveals Gut Microbiome Signatures Predictive of MASLD in Children with Obesity. BMC Microbiol. 2025, 25, 146. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome Diversity: High-Throughput Sequencing and Identification of Fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, H.; Hildebrand, F.; et al. Shotgun Metagenomes and Multiple Primer Pair-Barcode Combinations of Amplicons Reveal Biases in Metabarcoding Analyses of Fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Wurzbacher, C.; Larsson, E.; Bengtsson-Palme, J.; Van den Wyngaert, S.; Svantesson, S.; Kristiansson, E.; Kagami, M.; Nilsson, R.H. Introducing Ribosomal Tandem Repeat Barcoding for Fungi. Mol. Ecol. Resour. 2019, 19, 118–127. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Puusepp, R.; Nilsson, R.H.; James, T.Y. Novel Soil-Inhabiting Clades Fill Gaps in the Fungal Tree of Life. Microbiome 2017, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Jamy, M.; Foster, R.; Barbera, P.; Czech, L.; Kozlov, A.; Stamatakis, A.; Bending, G.; Hilton, S.; Bass, D.; Burki, F. Long-Read Metabarcoding of the Eukaryotic rDNA Operon to Phylogenetically and Taxonomically Resolve Environmental Diversity. Mol. Ecol. Resour. 2020, 20, 429–443. [Google Scholar] [CrossRef] [PubMed]
- D’Andreano, S.; Cuscó, A.; Francino, O. Rapid and Real-Time Identification of Fungi up to Species Level with Long Amplicon Nanopore Sequencing from Clinical Samples. Biol. Methods Protoc. 2020, 6, bpaa026. [Google Scholar] [CrossRef]
- Hoyles, L.; McCartney, A.L.; Neve, H.; Gibson, G.R.; Sanderson, J.D.; Heller, K.J.; van Sinderen, D. Characterization of Virus-like Particles Associated with the Human Faecal and Caecal Microbiota. Res. Microbiol. 2014, 165, 803–812. [Google Scholar] [CrossRef]
- Kim, M.-S.; Park, E.-J.; Roh, S.W.; Bae, J.-W. Diversity and Abundance of Single-Stranded DNA Viruses in Human Feces. Appl. Environ. Microbiol. 2011, 77, 8062–8070. [Google Scholar] [CrossRef]
- Sereika, M.; Kirkegaard, R.H.; Karst, S.M.; Michaelsen, T.Y.; Sørensen, E.A.; Wollenberg, R.D.; Albertsen, M. Oxford Nanopore R10.4 Long-Read Sequencing Enables the Generation of near-Finished Bacterial Genomes from Pure Cultures and Metagenomes without Short-Read or Reference Polishing. Nat. Methods 2022, 19, 823–826. [Google Scholar] [CrossRef]
- Moss, E.L.; Maghini, D.G.; Bhatt, A.S. Complete, Closed Bacterial Genomes from Microbiomes Using Nanopore Sequencing. Nat. Biotechnol. 2020, 38, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Pinto, Y.; Bhatt, A.S. Sequencing-Based Analysis of Microbiomes. Nat. Rev. Genet. 2024, 25, 829–845. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, H.; Portik, D.; Li, H. Metagenome Assembly of High-Fidelity Long Reads with Hifiasm-Meta. Nat. Methods 2022, 19, 671–674. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus Long-Read Assemblies for Bacterial Genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef] [PubMed]
- Delahaye, C.; Nicolas, J. Sequencing DNA with Nanopores: Troubles and Biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, F.B.; Maghini, D.; Oduaran, O.H.; Brewster, R.; Hulley, M.R.; Sahibdeen, V.; Norris, S.A.; Tollman, S.; Kahn, K.; Wagner, R.G.; et al. Short- and Long-Read Metagenomics of Urban and Rural South African Gut Microbiomes Reveal a Transitional Composition and Undescribed Taxa. Nat. Commun. 2022, 13, 926. [Google Scholar] [CrossRef]
- Datema, E.; Hulzink, R.J.M.; Blommers, L.; Valle-Inclan, J.E.; Van Orsouw, N.; Wittenberg, A.H.J.; De Vos, M. The Megabase-Sized Fungal Genome of Rhizoctonia solani Assembled from Nanopore Reads Only. BioRxiv 2016, 084772. [Google Scholar] [CrossRef]
- Luo, Y.; Jang, J.H.; Balkey, M.; Hoffmann, M. 217 Closed Salmonella Reference Genomes Using PacBio Sequencing. BMC Genom. Data 2025, 26, 15. [Google Scholar] [CrossRef]
- Sydenham, T.V.; Overballe-Petersen, S.; Hasman, H.; Wexler, H.; Kemp, M.; Justesen, U.S. Complete Hybrid Genome Assembly of Clinical Multidrug-Resistant Bacteroides Fragilis Isolates Enables Comprehensive Identification of Antimicrobial-Resistance Genes and Plasmids. Microb. Genom. 2019, 5, e000312. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Bollas, A.; Wang, Y.; Au, K.F. Nanopore Sequencing Technology, Bioinformatics and Applications. Nat. Biotechnol. 2021, 39, 1348–1365. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Casadesús, J. The Bacterial Epigenome. Nat. Rev. Microbiol. 2020, 18, 7–20. [Google Scholar] [CrossRef]
- Won, C.; Yim, S.S. Emerging Methylation-Based Approaches in Microbiome Engineering. Biotechnol. Biofuels Bioprod. 2024, 17, 96. [Google Scholar] [CrossRef]
- Beaulaurier, J.; Zhang, X.-S.; Zhu, S.; Sebra, R.; Rosenbluh, C.; Deikus, G.; Shen, N.; Munera, D.; Waldor, M.K.; Chess, A.; et al. Single Molecule-Level Detection and Long Read-Based Phasing of Epigenetic Variations in Bacterial Methylomes. Nat. Commun. 2015, 6, 7438. [Google Scholar] [CrossRef] [PubMed]
- Mattei, A.L.; Bailly, N.; Meissner, A. DNA Methylation: A Historical Perspective. Trends Genet. 2022, 38, 676–707. [Google Scholar] [CrossRef]
- Casadesús, J.; Low, D. Epigenetic Gene Regulation in the Bacterial World. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef]
- Messer, W.; Bellekes, U.; Lother, H. Effect of Dam Methylation on the Activity of the E. Coli Replication Origin, oriC. EMBO J. 1985, 4, 1327–1332. [Google Scholar] [CrossRef]
- Stephens, C.; Reisenauer, A.; Wright, R.; Shapiro, L. A Cell Cycle-Regulated Bacterial DNA Methyltransferase Is Essential for Viability. Proc. Natl. Acad. Sci. USA 1996, 93, 1210–1214. [Google Scholar] [CrossRef]
- Robbins-Manke, J.L.; Zdraveski, Z.Z.; Marinus, M.; Essigmann, J.M. Analysis of Global Gene Expression and Double-Strand-Break Formation in DNA Adenine Methyltransferase- and Mismatch Repair-Deficient Escherichia coli. J. Bacteriol. 2005, 187, 7027–7037. [Google Scholar] [CrossRef]
- Seib, K.L.; Jen, F.E.-C.; Scott, A.L.; Tan, A.; Jennings, M.P. Phase Variation of DNA Methyltransferases and the Regulation of Virulence and Immune Evasion in the Pathogenic Neisseria. Pathog. Dis. 2017, 75, ftx080. [Google Scholar] [CrossRef]
- Hajkova, P.; El-Maarri, O.; Engemann, S.; Oswald, J.; Olek, A.; Walter, J. DNA-Methylation Analysis by the Bisulfite-Assisted Genomic Sequencing Method. In DNA Methylation Protocols; Mills, K.I., Ramsahoye, B.H., Eds.; Springer: New York, NY, USA; Totowa, NJ, USA, 2002; pp. 143–154. ISBN 978-1-59259-182-4. [Google Scholar]
- Shiratori, H.; Feinweber, C.; Knothe, C.; Lötsch, J.; Thomas, D.; Geisslinger, G.; Parnham, M.J.; Resch, E. High-Throughput Analysis of Global DNA Methylation Using Methyl-Sensitive Digestion. PLoS ONE 2016, 11, e0163184. [Google Scholar] [CrossRef]
- Bonora, G.; Rubbi, L.; Morselli, M.; Ma, F.; Chronis, C.; Plath, K.; Pellegrini, M. DNA Methylation Estimation Using Methylation-Sensitive Restriction Enzyme Bisulfite Sequencing (MREBS). PLoS ONE 2019, 14, e0214368. [Google Scholar] [CrossRef]
- Li, D.; Zhang, B.; Xing, X.; Wang, T. Combining MeDIP-Seq and MRE-Seq to Investigate Genome-Wide CpG Methylation. Methods 2015, 72, 29–40. [Google Scholar] [CrossRef]
- Jensen, T.Ø.; Tellgren-Roth, C.; Redl, S.; Maury, J.; Jacobsen, S.A.B.; Pedersen, L.E.; Nielsen, A.T. Genome-Wide Systematic Identification of Methyltransferase Recognition and Modification Patterns. Nat. Commun. 2019, 10, 3311. [Google Scholar] [CrossRef]
- MacKenzie, M.; Argyropoulos, C. An Introduction to Nanopore Sequencing: Past, Present, and Future Considerations. Micromachines 2023, 14, 459. [Google Scholar] [CrossRef]
- Lu, B.; Guo, Z.; Liu, X.; Ni, Y.; Xu, L.; Huang, J.; Li, T.; Feng, T.; Li, R.; Deng, X. Comprehensive Comparison of the Third-Generation Sequencing Tools for Bacterial 6mA Profiling. Nat. Commun. 2025, 16, 3982. [Google Scholar] [CrossRef] [PubMed]
- Gouil, Q.; Keniry, A. Latest Techniques to Study DNA Methylation. Essays Biochem. 2019, 63, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Arnold, B.J.; Huang, I.-T.; Hanage, W.P. Horizontal Gene Transfer and Adaptive Evolution in Bacteria. Nat. Rev. Microbiol. 2022, 20, 206–218. [Google Scholar] [CrossRef]
- Barreto, H.C.; Gordo, I. Intrahost Evolution of the Gut Microbiota. Nat. Rev. Microbiol. 2023, 21, 590–603. [Google Scholar] [CrossRef]
- Bharti, R.; Grimm, D.G. Current Challenges and Best-Practice Protocols for Microbiome Analysis. Brief. Bioinform. 2021, 22, 178–193. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to Limit False Positives in Environmental DNA and Metabarcoding? Mol. Ecol. Resour. 2016, 16, 604–607. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Wong, J.; Heiner, C.; Oh, S.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-Throughput Amplicon Sequencing of the Full-Length 16S rRNA Gene with Single-Nucleotide Resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Ma, S.; Cao, J.; Liao, H.; Huang, Q.; Chen, W. The Newest Oxford Nanopore R10.4.1 Full-Length 16S rRNA Sequencing Enables the Accurate Resolution of Species-Level Microbial Community Profiling. Appl. Environ. Microbiol. 2023, 89, e00605–e00623. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Application Note: Oxford Nanopore Sequencing Provides Superior MAG Recovery and Strain-Level Resolution from a Complex Microbiome. Available online: https://nanoporetech.com/resource-centre/oxford-nanopore-sequencing-provides-superior-metagenome-assembled-genome-recovery-and-strain-level-resolution-from-a-complex-microbiome (accessed on 7 June 2025).
- Balvočiūtė, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How Do These Taxonomies Compare? BMC Genom. 2017, 18, 114. [Google Scholar] [CrossRef]
- Buetas, E.; Jordán-López, M.; López-Roldán, A.; D’Auria, G.; Martínez-Priego, L.; De Marco, G.; Carda-Diéguez, M.; Mira, A. Full-Length 16S rRNA Gene Sequencing by PacBio Improves Taxonomic Resolution in Human Microbiome Samples. BMC Genom. 2024, 25, 310. [Google Scholar] [CrossRef]
- Lu, J.; Salzberg, S.L. Ultrafast and Accurate 16S rRNA Microbial Community Analysis Using Kraken 2. Microbiome 2020, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, M.; Kakinuma, K.; Kawaguchi, R. Phylogenetic Analysis of Salmonella, Shigella, and Escherichia coli Strains on the Basis of the gyrB Gene Sequence. J. Clin. Microbiol. 2002, 40, 2779–2785. [Google Scholar] [CrossRef]
- Halimeh, F.B.; Rafei, R.; Osman, M.; Kassem, I.I.; Diene, S.M.; Dabboussi, F.; Rolain, J.-M.; Hamze, M. Historical, Current, and Emerging Tools for Identification and Serotyping of Shigella. Braz. J. Microbiol. 2021, 52, 2043–2055. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic Insights That Advance the Species Definition for Prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Chen, T.; Misir, M.; Lin, Y. Deep Learning for Predicting 16S rRNA Gene Copy Number. Sci. Rep. 2024, 14, 14282. [Google Scholar] [CrossRef]
- Edgar, R. Taxonomy Annotation and Guide Tree Errors in 16S rRNA Databases. PeerJ 2018, 6, e5030. [Google Scholar] [CrossRef]
- Jia, H.; Tan, S.; Zhang, Y.E. Chasing Sequencing Perfection: Marching Toward Higher Accuracy and Lower Costs. Genom. Proteom. Bioinform. 2024, 22, qzae024. [Google Scholar] [CrossRef] [PubMed]
- Stoler, N.; Nekrutenko, A. Sequencing Error Profiles of Illumina Sequencing Instruments. NAR Genom. Bioinform. 2021, 3, lqab019. [Google Scholar] [CrossRef]
- Illumina. Sequencing Technology|Sequencing by Synthesis. Available online: https://www.illumina.com/science/technology/next-generation-sequencing/sequencing-technology.html (accessed on 17 June 2025).
- Illumina. Sequencing Platforms|Illumina NGS Platforms. Available online: https://www.illumina.com/systems/sequencing-platforms.html (accessed on 7 June 2025).
- Dohm, J.C.; Peters, P.; Stralis-Pavese, N.; Himmelbauer, H. Benchmarking of Long-Read Correction Methods. NAR Genom. Bioinform. 2020, 2, lqaa037. [Google Scholar] [CrossRef]
- Cuber, P.; Chooneea, D.; Geeves, C.; Salatino, S.; Creedy, T.J.; Griffin, C.; Sivess, L.; Barnes, I.; Price, B.; Misra, R. Comparing the Accuracy and Efficiency of Third Generation Sequencing Technologies, Oxford Nanopore Technologies, and Pacific Biosciences, for DNA Barcode Sequencing Applications. Ecol. Genet. Genom. 2023, 28, 100181. [Google Scholar] [CrossRef]
- Pacific Biosciences. PacBio Revio|Long-Read Sequencing at Scale. Available online: https://www.pacb.com/revio/ (accessed on 11 June 2025).
- Pacific Biosciences. Microbial Genomics. Available online: https://www.pacb.com/microbial-genomics/ (accessed on 7 June 2025).
- Oxford Nanopore Technologies. Oxford Nanopore Flow Cells and Sequencing Devices. Available online: https://nanoporetech.com/products/sequence (accessed on 26 May 2025).
- Hall, C.L.; Zascavage, R.R.; Sedlazeck, F.J.; Planz, J.V. Potential Applications of Nanopore Sequencing for Forensic Analysis. Forensic Sci. Rev. 2020, 32, 23–54. [Google Scholar]
- Oxford Nanopore Technologies. Nanopore Store: Flow Cells. Available online: https://store.nanoporetech.com/eu/flow-cells.html (accessed on 13 June 2025).
- Pacific Biosciences. Vega Benchtop System. Available online: https://www.pacb.com/vega/ (accessed on 11 June 2025).
- Xu, W.; Chen, T.; Pei, Y.; Guo, H.; Li, Z.; Yang, Y.; Zhang, F.; Yu, J.; Li, X.; Yang, Y.; et al. Characterization of Shallow Whole-Metagenome Shotgun Sequencing as a High-Accuracy and Low-Cost Method by Complicated Mock Microbiomes. Front. Microbiol. 2021, 12, 678319. [Google Scholar] [CrossRef] [PubMed]
- Diao, Z.; Han, D.; Zhang, R.; Li, J. Metagenomics Next-Generation Sequencing Tests Take the Stage in the Diagnosis of Lower Respiratory Tract Infections. J. Adv. Res. 2022, 38, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Lao, H.-Y.; Ng, T.T.-L.; Wong, R.Y.-L.; Wong, C.S.-T.; Lee, L.-K.; Wong, D.S.-H.; Chan, C.T.-M.; Jim, S.H.-C.; Leung, J.S.-L.; Lo, H.W.-H.; et al. The Clinical Utility of Two High-Throughput 16S rRNA Gene Sequencing Workflows for Taxonomic Assignment of Unidentifiable Bacterial Pathogens in Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2022, 60, e01769-21. [Google Scholar] [CrossRef]
- Petrone, J.R.; Rios Glusberger, P.; George, C.D.; Milletich, P.L.; Ahrens, A.P.; Roesch, L.F.W.; Triplett, E.W. RESCUE: A Validated Nanopore Pipeline to Classify Bacteria through Long-Read, 16S-ITS-23S rRNA Sequencing. Front. Microbiol. 2023, 14, 1201064. [Google Scholar] [CrossRef]
- Quick, J.; Loman, N.J.; Duraffour, S.; Simpson, J.T.; Severi, E.; Cowley, L.; Bore, J.A.; Koundouno, R.; Dudas, G.; Mikhail, A.; et al. Real-Time, Portable Genome Sequencing for Ebola Surveillance. Nature 2016, 530, 228–232. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and Clustering Orders of Magnitude Faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- One Codex. A Fast, Easy-to-Use Platform for Microbiome Sequencing and Analysis. Available online: https://onecodex.com/ (accessed on 1 June 2025).
- EzBiome. Empowering Microbiome Innovation & Discovery. Available online: https://ezbiome.com/ (accessed on 1 June 2025).
- Pacific Biosciences. HiFi-16S-Workflow. Available online: https://github.com/PacificBiosciences/HiFi-16S-workflow (accessed on 1 June 2025).
- Pacific Biosciences. Pb-Metagenomics-Tools. Available online: https://github.com/PacificBiosciences/pb-metagenomics-tools (accessed on 1 June 2025).
- BugSeq. Rapid and Accurate Analysis for Microbiology Labs. Available online: https://bugseq.com (accessed on 1 June 2025).
- Kolmogorov, M.; Bickhart, D.M.; Behsaz, B.; Gurevich, A.; Rayko, M.; Shin, S.B.; Kuhn, K.; Yuan, J.; Polevikov, E.; Smith, T.P.L.; et al. metaFlye: Scalable Long-Read Metagenome Assembly Using Repeat Graphs. Nat. Methods 2020, 17, 1103–1110. [Google Scholar] [CrossRef]
- Benoit, G.; Raguideau, S.; James, R.; Phillippy, A.M.; Chikhi, R.; Quince, C. High-Quality Metagenome Assembly from Long Accurate Reads with metaMDBG. Nat. Biotechnol. 2024, 42, 1378–1383. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Getting Started Guide: Microbial Sequencing. Available online: https://nanoporetech.com/resource-centre/a-guide-to-microbial-sequencing-with-oxford-nanopore (accessed on 7 June 2025).
- Oxford Nanopore Technologies. Rapid Sequencing DNA-16S Barcoding Kit 24 V14 (SQK-16S114.24). Available online: https://nanoporetech.com/document/rapid-sequencing-DNA-16s-barcoding-kit-v14-sqk-16114-24 (accessed on 7 June 2025).
- Rodríguez-Pérez, H.; Ciuffreda, L.; Flores, C. NanoCLUST: A Species-Level Analysis of 16S rRNA Nanopore Sequencing Data. Bioinformatics 2021, 37, 1600–1601. [Google Scholar] [CrossRef] [PubMed]
- Curry, K.D.; Wang, Q.; Nute, M.G.; Tyshaieva, A.; Reeves, E.; Soriano, S.; Wu, Q.; Graeber, E.; Finzer, P.; Mendling, W.; et al. Emu: Species-Level Microbial Community Profiling of Full-Length 16S rRNA Oxford Nanopore Sequencing Data. Nat. Methods 2022, 19, 845–853. [Google Scholar] [CrossRef]
- Gowers, G.-O.F.; Vince, O.; Charles, J.-H.; Klarenberg, I.; Ellis, T.; Edwards, A. Entirely Off-Grid and Solar-Powered DNA Sequencing of Microbial Communities during an Ice Cap Traverse Expedition. Genes 2019, 10, 902. [Google Scholar] [CrossRef]
- Castro-Wallace, S.L.; Chiu, C.Y.; John, K.K.; Stahl, S.E.; Rubins, K.H.; McIntyre, A.B.R.; Dworkin, J.P.; Lupisella, M.L.; Smith, D.J.; Botkin, D.J.; et al. Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Sci. Rep. 2017, 7, 18022. [Google Scholar] [CrossRef]
- Drown, D.M.; Lekanoff, R.M.; Khalsa, N.S.; Smith, H.H.; Drown, D.M. Introducing DNA Sequencing to the Next Generation on a Research Vessel Sailing the Bering Sea Through a Storm. Preprints 2019. [Google Scholar] [CrossRef]
- Blanco, M.B.; Greene, L.K.; Rasambainarivo, F.; Toomey, E.; Williams, R.C.; Andrianandrasana, L.; Larsen, P.A.; Yoder, A.D. Next-Generation Technologies Applied to Age-Old Challenges in Madagascar. Conserv. Genet. 2020, 21, 785–793. [Google Scholar] [CrossRef]
- Loit, K.; Adamson, K.; Bahram, M.; Puusepp, R.; Anslan, S.; Kiiker, R.; Drenkhan, R.; Tedersoo, L. Relative Performance of MinION (Oxford Nanopore Technologies) versus Sequel (Pacific Biosciences) Third-Generation Sequencing Instruments in Identification of Agricultural and Forest Fungal Pathogens. Appl. Environ. Microbiol. 2019, 85, e01368-19. [Google Scholar] [CrossRef]
- Charalampous, T.; Alcolea-Medina, A.; Snell, L.B.; Alder, C.; Tan, M.; Williams, T.G.S.; Al-Yaakoubi, N.; Humayun, G.; Meadows, C.I.S.; Wyncoll, D.L.A.; et al. Routine Metagenomics Service for ICU Patients with Respiratory Infection. Am. J. Respir. Crit. Care Med. 2024, 209, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Buytaers, F.E.; Verhaegen, B.; Van Nieuwenhuysen, T.; Roosens, N.H.C.; Vanneste, K.; Marchal, K.; De Keersmaecker, S.C.J. Strain-Level Characterization of Foodborne Pathogens without Culture Enrichment for Outbreak Investigation Using Shotgun Metagenomics Facilitated with Nanopore Adaptive Sampling. Front. Microbiol. 2024, 15, 1300814. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Ziemski, M.; Robeson, M.S.; Kaehler, B.D. Measuring the Microbiome: Best Practices for Developing and Benchmarking Microbiomics Methods. Comput. Struct. Biotechnol. J. 2020, 18, 4048–4062. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Mazel, F.; Doebeli, M.; Parfrey, L.W. A Census-Based Estimate of Earth’s Bacterial and Archaeal Diversity. PLoS Biol. 2019, 17, e3000106. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Status of the Microbial Census. Microbiol. Mol. Biol. Rev. 2004, 68, 686–691. [Google Scholar] [CrossRef]
- Pel, J.; Leung, A.; Choi, W.W.Y.; Despotovic, M.; Ung, W.L.; Shibahara, G.; Gelinas, L.; Marziali, A. Rapid and Highly-Specific Generation of Targeted DNA Sequencing Libraries Enabled by Linking Capture Probes with Universal Primers. PLoS ONE 2018, 13, e0208283. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing. mSphere 2021, 6, e01202-20. [Google Scholar] [CrossRef]
- Hao, Y.; Pei, Z.; Brown, S.M. Bioinformatics in Microbiome Analysis. In Methods in Microbiology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 44, pp. 1–18. ISBN 978-0-12-813714-7. [Google Scholar]
- Hayashi, H.; Sakamoto, M.; Benno, Y. Evaluation of Three Different Forward Primers by Terminal Restriction Fragment Length Polymorphism Analysis for Determination of Fecal Bifidobacterium Spp. in Healthy Subjects. Microbiol. Immunol. 2004, 48, 1–6. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Bates, S.T.; Eilers, K.G.; Lauber, C.L.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. The Under-Recognized Dominance of Verrucomicrobia in Soil Bacterial Communities. Soil Biol. Biochem. 2011, 43, 1450–1455. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Yu, Z.; García-González, R.; Schanbacher, F.L.; Morrison, M. Evaluations of Different Hypervariable Regions of Archaeal 16S rRNA Genes in Profiling of Methanogens by Archaea-Specific PCR and Denaturing Gradient Gel Electrophoresis. Appl. Environ. Microbiol. 2008, 74, 889–893. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every Base Matters: Assessing Small Subunit rRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Fraher, M.H.; O’Toole, P.W.; Quigley, E.M.M. Techniques Used to Characterize the Gut Microbiota: A Guide for the Clinician. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 312–322. [Google Scholar] [CrossRef]
- Srinivas, M.; Walsh, C.J.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; van Sinderen, D.; Kenny, J.G. Evaluating the Efficiency of 16S-ITS-23S Operon Sequencing for Species Level Resolution in Microbial Communities. Sci. Rep. 2025, 15, 2822. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Rogers, Y.-H.; Venter, J.C. Massively Parallel Sequencing. Nature 2005, 437, 326–327. [Google Scholar] [CrossRef]
- Sharpton, T.J. An Introduction to the Analysis of Shotgun Metagenomic Data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef]
- Hong, S.; Bunge, J.; Leslin, C.; Jeon, S.; Epstein, S.S. Polymerase Chain Reaction Primers Miss Half of rRNA Microbial Diversity. ISME J. 2009, 3, 1365–1373. [Google Scholar] [CrossRef]
- Logares, R.; Sunagawa, S.; Salazar, G.; Cornejo-Castillo, F.M.; Ferrera, I.; Sarmento, H.; Hingamp, P.; Ogata, H.; de Vargas, C.; Lima-Mendez, G.; et al. Metagenomic 16S rDNA Illumina Tags Are a Powerful Alternative to Amplicon Sequencing to Explore Diversity and Structure of Microbial Communities. Environ. Microbiol. 2014, 16, 2659–2671. [Google Scholar] [CrossRef]
- López-Aladid, R.; Fernández-Barat, L.; Alcaraz-Serrano, V.; Bueno-Freire, L.; Vázquez, N.; Pastor-Ibáñez, R.; Palomeque, A.; Oscanoa, P.; Torres, A. Determining the Most Accurate 16S rRNA Hypervariable Region for Taxonomic Identification from Respiratory Samples. Sci. Rep. 2023, 13, 3974. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M. Accounting for 16S rRNA Copy Number Prediction Uncertainty and Its Implications in Bacterial Diversity Analyses. ISME Commun. 2023, 3, 59. [Google Scholar] [CrossRef]
- Acinas, S.G.; Marcelino, L.A.; Klepac-Ceraj, V.; Polz, M.F. Divergence and Redundancy of 16S rRNA Sequences in Genomes with Multiple Rrn Operons. J. Bacteriol. 2004, 186, 2629–2635. [Google Scholar] [CrossRef]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-Length 16S rRNA Gene Amplicon Analysis of Human Gut Microbiota Using MinIONTM Nanopore Sequencing Confers Species-Level Resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef]
- Kai, S.; Matsuo, Y.; Nakagawa, S.; Kryukov, K.; Matsukawa, S.; Tanaka, H.; Iwai, T.; Imanishi, T.; Hirota, K. Rapid Bacterial Identification by Direct PCR Amplification of 16S rRNA Genes Using the MinIONTM Nanopore Sequencer. FEBS Open Bio 2019, 9, 548–557. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef]
- Deissová, T.; Zapletalová, M.; Kunovský, L.; Kroupa, R.; Grolich, T.; Kala, Z.; Bořilová Linhartová, P.; Lochman, J. 16S rRNA Gene Primer Choice Impacts Off-Target Amplification in Human Gastrointestinal Tract Biopsies and Microbiome Profiling. Sci. Rep. 2023, 13, 12577. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Martin, J.C.; Scott, P.; Parkhill, J.; Flint, H.J.; Scott, K.P. 16S rRNA Gene-Based Profiling of the Human Infant Gut Microbiota Is Strongly Influenced by Sample Processing and PCR Primer Choice. Microbiome 2015, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Hrovat, K.; Dutilh, B.E.; Medema, M.H.; Melkonian, C. Taxonomic Resolution of Different 16S rRNA Variable Regions Varies Strongly across Plant-Associated Bacteria. ISME Commun. 2024, 4, ycae034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, X.; Chen, A.; Li, S.; Tao, R.; Chen, K.; Huang, P.; Li, L.; Huang, J.; Li, C.; et al. Comparison of the Full-Length Sequence and Sub-Regions of 16S rRNA Gene for Skin Microbiome Profiling. mSystems 2024, 9, e00399-24. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of Complex Microbial Populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Bano, N.; Ruffin, S.; Ransom, B.; Hollibaugh, J.T. Phylogenetic Composition of Arctic Ocean Archaeal Assemblages and Comparison with Antarctic Assemblages. Appl. Environ. Microbiol. 2004, 70, 781–789. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU rRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An Integrated Catalog of Reference Genes in the Human Gut Microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Na, H.S.; Song, Y.; Yu, Y.; Chung, J. Comparative Analysis of Primers Used for 16S rRNA Gene Sequencing in Oral Microbiome Studies. Methods Protoc. 2023, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of Two 16S rRNA Primers (V3–V4 and V4–V5) for Studies of Arctic Microbial Communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the Microbiome: Advantages of Whole Genome Shotgun versus 16S Amplicon Sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Grieb, A.; Bowers, R.M.; Oggerin, M.; Goudeau, D.; Lee, J.; Malmstrom, R.R.; Woyke, T.; Fuchs, B.M. A Pipeline for Targeted Metagenomics of Environmental Bacteria. Microbiome 2020, 8, 21. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular Biological Access to the Chemistry of Unknown Soil Microbes: A New Frontier for Natural Products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Breitbart, M.; Hewson, I.; Felts, B.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Metagenomic Analyses of an Uncultured Viral Community from Human Feces. J. Bacteriol. 2003, 185, 6220–6223. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef]
- Garcia-Garcerà, M.; Garcia-Etxebarria, K.; Coscollà, M.; Latorre, A.; Calafell, F. A New Method for Extracting Skin Microbes Allows Metagenomic Analysis of Whole-Deep Skin. PLoS ONE 2013, 8, e74914. [Google Scholar] [CrossRef]
- Kunin, V.; Copeland, A.; Lapidus, A.; Mavromatis, K.; Hugenholtz, P. A Bioinformatician’s Guide to Metagenomics. Microbiol. Mol. Biol. Rev. 2008, 72, 557–578. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Hillmann, B.; Al-Ghalith, G.A.; Shields-Cutler, R.R.; Zhu, Q.; Gohl, D.M.; Beckman, K.B.; Knight, R.; Knights, D. Evaluating the Information Content of Shallow Shotgun Metagenomics. Msystems 2018, 3, e00069-18. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best Practices for Analysing Microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- La Reau, A.J.; Strom, N.B.; Filvaroff, E.; Mavrommatis, K.; Ward, T.L.; Knights, D. Shallow Shotgun Sequencing Reduces Technical Variation in Microbiome Analysis. Sci. Rep. 2023, 13, 7668. [Google Scholar] [CrossRef]
- Carter, M.M.; Olm, M.R.; Merrill, B.D.; Dahan, D.; Tripathi, S.; Spencer, S.P.; Yu, F.B.; Jain, S.; Neff, N.; Jha, A.R.; et al. Ultra-Deep Sequencing of Hadza Hunter-Gatherers Recovers Vanishing Gut Microbes. Cell 2023, 186, 3111–3124. [Google Scholar] [CrossRef]
- Rajan, S.K.; Lindqvist, M.; Brummer, R.J.; Schoultz, I.; Repsilber, D. Phylogenetic Microbiota Profiling in Fecal Samples Depends on Combination of Sequencing Depth and Choice of NGS Analysis Method. PLoS ONE 2019, 14, e0222171. [Google Scholar] [CrossRef]
- Weinroth, M.D.; Belk, A.D.; Dean, C.; Noyes, N.; Dittoe, D.K.; Rothrock, M.J.; Ricke, S.C.; Myer, P.R.; Henniger, M.T.; Ramírez, G.A.; et al. Considerations and Best Practices in Animal Science 16S Ribosomal RNA Gene Sequencing Microbiome Studies. J. Anim. Sci. 2022, 100, skab346. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. The Effect of 16S rRNA Region Choice on Bacterial Community Metabarcoding Results. Sci. Data 2019, 6, 190007. [Google Scholar] [CrossRef]
- Pacific Biosciences. Application Brief: Microbiome and Metagenome Sequencing with HiFi Reads. Available online: https://www.pacb.com/products-and-services/applications/complex-populations/microbial/ (accessed on 7 June 2025).
- Oberle, A.; Urban, L.; Falch-Leis, S.; Ennemoser, C.; Nagai, Y.; Ashikawa, K.; Ulm, P.A.; Hengstschläger, M.; Feichtinger, M. 16S rRNA Long-Read Nanopore Sequencing Is Feasible and Reliable for Endometrial Microbiome Analysis. Reprod. Biomed. Online 2021, 42, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, V.; Gobet, A.; Joyce, A. Ecosystem-Specific Microbiota and Microbiome Databases in the Era of Big Data. Environ. Microbiome 2022, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Nierychlo, M.; Andersen, K.S.; Xu, Y.; Green, N.; Jiang, C.; Albertsen, M.; Dueholm, M.S.; Nielsen, P.H. MiDAS 3: An Ecosystem-Specific Reference Database, Taxonomy and Knowledge Platform for Activated Sludge and Anaerobic Digesters Reveals Species-Level Microbiome Composition of Activated Sludge. Water Res. 2020, 182, 115955. [Google Scholar] [CrossRef] [PubMed]
| Sequencing Platform | Maximum 16S Read Length | Taxonomic Resolution (16S) | Metagenomics Read Length | Metagenomic Applications |
|---|---|---|---|---|
| Illumina | 2 × 300 bp (overlap ~50 bp) | Mainly genus level | 2 × 300 bp (overlap ~50 bp) | Assembly (fragmented), taxonomic and functional profiling |
| PacBio | ~1500 bp | Species and strain levels | ~10 kb | High-quality assembly, taxonomic and functional profiling |
| ONT | ~1500 bp | Species and strain levels | ~10 kb | High-quality assembly, taxonomic and functional profiling |
| Sequencing Platform | Initial Error Source | Initial Error Type (%) | Accuracy Improvement Strategies | Current Accuracy |
|---|---|---|---|---|
| Illumina | Library construction Sequencing process DNA damage [141] | Substitutions (after homopolymer, G/C > A/T; ~0.01–0.5%) [142] | XLEAP-SBS chemistry [143] | ~99.9% (≥85% of bases) [144] |
| PacBio | Fluorescence signals’ misinterpretation Polymerase errors [145] | Substitutions (A↔C, G↔T; ~1.7%) Deletions (~3.2%) Insertions (~8%) [145] | HiFi reads (CCS) [146] | ~99.9% (0.5–5 kb; 95% of bases; 10–15 kb; 90% of bases) [147] |
| ONT | Nanopore design leads to bias in homopolymers (A/T) [145] | Substitutions (A↔G, C↔T; ~4%) Deletions (~4%) Insertions (~4%) [145] | Kit 14 chemistry R10.4.1 flow cell Basecaller updates Duplex reads [68] | ~99.9% (duplex reads) ~99% (simplex reads) [68,69] |
| Sequencer | Manufacturer | Portability | Size |
|---|---|---|---|
| MiSeq i100 | Illumina | No | Benchtop |
| Next Seq 1000/2000 | Illumina | No | Benchtop |
| NovaSeq 6000 | Illumina | No | Production-scale |
| Vega | PacBio | No | Compact benchtop |
| Revio | PacBio | No | Benchtop |
| Flongle | ONT | Yes | Palm-sized |
| MinION Mk1D | ONT | Yes | Palm-sized |
| GridION | ONT | No | Compact benchtop |
| PromethION | ONT | No | Compact benchtop |
| Sequencing Platform | Bioinformatic Expertise |
|---|---|
| Illumina | Required (intermediate/advanced) |
| PacBio | Required (intermediate/advanced) |
| ONT | User-friendly tools (beginner to advanced) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, A.; Fullaondo, A.; Odriozola, A. Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis? Microorganisms 2025, 13, 1861. https://doi.org/10.3390/microorganisms13081861
González A, Fullaondo A, Odriozola A. Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis? Microorganisms. 2025; 13(8):1861. https://doi.org/10.3390/microorganisms13081861
Chicago/Turabian StyleGonzález, Adriana, Asier Fullaondo, and Adrian Odriozola. 2025. "Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis?" Microorganisms 13, no. 8: 1861. https://doi.org/10.3390/microorganisms13081861
APA StyleGonzález, A., Fullaondo, A., & Odriozola, A. (2025). Why Are Long-Read Sequencing Methods Revolutionizing Microbiome Analysis? Microorganisms, 13(8), 1861. https://doi.org/10.3390/microorganisms13081861







