Dysbiosis in the Nasal Mycobiome of Infants Born in the Aftermath of Hurricane Maria
Abstract
1. Introduction
2. Materials and Methods
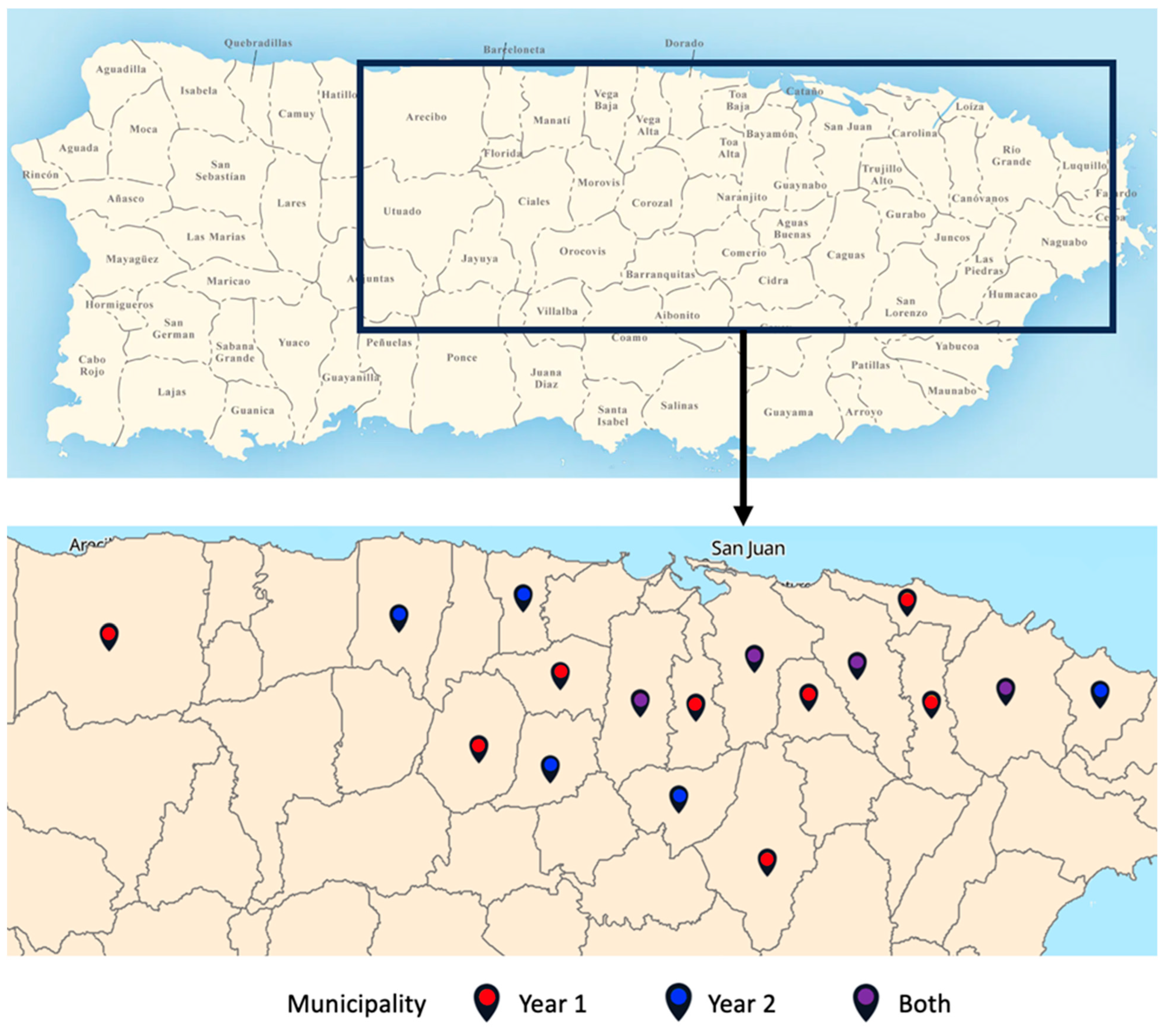
2.1. Cohort Recruitment
2.2. Sample Collection, Processing, and Sequencing
2.3. Sequencing Data Analysis
3. Results
3.1. The HOLA Cohort Study and Profiling of the Infant Nasal Mycobiome
3.2. Mycobiome Comparisons at the Community Level
3.3. Mycobiome Comparisons at the Genus Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HOLA | Hurricane as the Origin of Later Alterations in Microbiome |
| ITS | Internally Transcribed Spacer |
| ASV | Amplicon Sequence Variance |
| ANOSIM | Analysis of Similarity |
| PCoA | Principal Coordinate Analysis |
| COPD | Chronic Obstructive Pulmonary Disease |
References
- Seidel, D.; Wurster, S.; Jenks, J.D.; Sati, H.; Gangneux, J.P.; Egger, M.; Alastruey-Izquierdo, A.; Ford, N.P.; Chowdhary, P.A.; Sprute, R.; et al. Impact of climate change and natural disasters on fungal infections. Lancet Microbe 2024, 5, e594–e605. [Google Scholar] [CrossRef]
- Poole, J.A.; Barnes, C.S.; Demain, J.G.; Bernstein, J.A.; Padukudru, M.A.; Sheehan, W.J.; Fogelbach, G.G.; Wedner, J.; Codina, R.; Levetin, E.; et al. Impact of weather and climate change with indoor and outdoor air quality in asthma: A Work Group Report of the AAAAI Environmental Exposure and Respiratory Health Committee. J. Allergy Clin. Immunol. 2019, 143, 1702–1710. [Google Scholar] [CrossRef]
- D’Amato, G.; Chong-Neto, H.J.; Monge Ortega, O.P.; Vitale, C.; Ansotegui, I.; Rosario, N.; Haahtela, T.; Galan, C.; Pawankar, R.; Murrieta-Aguttes, M.; et al. The effects of climate change on respiratory allergy and asthma induced by pollen and mold allergens. Allergy 2020, 75, 2219–2228. [Google Scholar] [CrossRef]
- Solomon, G.M.; Hjelmroos-Koski, M.; Rotkin-Ellman, M.; Hammond, S.K. Airborne Mold and Endotoxin Concentrations in New Orleans, Louisiana, after Flooding, October through November 2005. Environ. Health Perspect. 2006, 114, 1381–1386. [Google Scholar] [CrossRef]
- Vélez-Torres, L.N.; Bolaños-Rosero, B.; Godoy-Vitorino, F.; Rivera-Mariani, F.E.; Maestre, J.P.; Kinney, K.; Cavallin, H. Hurricane María drives increased indoor proliferation of filamentous fungi in San Juan, Puerto Rico: A two-year culture-based approach. PeerJ 2022, 10, e12730. [Google Scholar] [CrossRef]
- Toda, M.; Williams, S.; Jackson, B.R.; Wurster, S.; Serpa, J.A.; Nigo, M.; Grimes, C.Z.; Atmar, R.L.; Chiller, T.M.; Ostrosky-Zeichner, L.; et al. Invasive Mold Infections Following Hurricane Harvey—Houston, Texas. Open Forum Infect. Dis. 2023, 10, ofad093. [Google Scholar] [CrossRef]
- Riggs, M.A.; Rao, C.Y.; Brown, C.M.; Van Sickle, D.; Cummings, K.J.; Dunn, K.H.; Deddens, J.A.; Ferdinands, J.; Callahan, D.; Moolenaar, R.L.; et al. Resident cleanup activities, characteristics of flood-damaged homes and airborne microbial concentrations in New Orleans, Louisiana, October 2005. Environ. Res. 2008, 106, 401–409. [Google Scholar] [CrossRef]
- Rao, C.Y.; Riggs, M.A.; Chew, G.L.; Muilenberg, M.L.; Thorne, P.S.; Van Sickle, D.; Dunn, K.H.; Brown, C. Characterization of Airborne Molds, Endotoxins, and Glucans in Homes in New Orleans after Hurricanes Katrina and Rita. Appl. Environ. Microbiol. 2007, 73, 1630–1634. [Google Scholar] [CrossRef]
- Magyar, D.; Tischner, Z.; Szabó, B.; Freiler-Nagy, Á.; Papp, T.; Allaga, H.; Kredics, L. Characterization of Indoor Molds after Ajka Red Mud Spill, Hungary. Pathogens 2023, 13, 22. [Google Scholar] [CrossRef]
- Emerson, J.B.; Keady, P.B.; Brewer, T.E.; Clements, N.; Morgan, E.E.; Awerbuch, J.; Miller, S.L.; Fierer, N. Impacts of Flood Damage on Airborne Bacteria and Fungi in Homes after the 2013 Colorado Front Range Flood. Environ. Sci. Technol. 2015, 49, 2675–2684. [Google Scholar] [CrossRef]
- Oluyomi, A.O.; Panthagani, K.; Sotelo, J.; Gu, X.; Armstrong, G.; Luo, D.N.; Hoffman, K.L.; Rohlman, D.; Tidwell, L.; Hamilton, W.J.; et al. Houston hurricane Harvey health (Houston-3H) study: Assessment of allergic symptoms and stress after hurricane Harvey flooding. Environ. Health 2021, 20, 9. [Google Scholar] [CrossRef]
- Stark, P.C.; Burge, H.A.; Ryan, L.M.; Milton, D.K.; Gold, D.R. Fungal Levels in the Home and Lower Respiratory Tract Illnesses in the First Year of Life. Am. J. Respir. Crit. Care Med. 2003, 168, 232–237. [Google Scholar] [CrossRef]
- Hossain, M.A.; Ahmed, M.S.; Ghannoum, M.A. Attributes of Stachybotrys chartarum and its association with human disease. J. Allergy Clin. Immunol. 2004, 113, 200–208. [Google Scholar] [CrossRef]
- O’Driscoll, B.R.; Hopkinson, L.C.; Denning, D.W. Mold sensitization is common amongst patients with severe asthma requiring multiple hospital admissions. BMC Pulm. Med. 2005, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Varga, M.K.; Moshammer, H.; Atanyazova, O. Childhood asthma and mould in homes—A meta-analysis. Wien. Klin. Wochenschr. 2025, 137, 79–88. [Google Scholar] [CrossRef]
- Howard, E.; Orhurhu, V.; Huang, L.; Guthrie, B.; Phipatanakul, W. The Impact of Ambient Environmental Exposures to Microbial Products on Asthma Outcomes from Birth to Childhood. Curr. Allergy Asthma Rep. 2019, 19, 59. [Google Scholar] [CrossRef]
- Zhang, A.; De Ángel Solá, D.; Acevedo Flores, M.; Cao, L.; Wang, L.; Kim, J.G.; Tarr, P.I.; Warner, B.B.; Matos, N.R.; Wang, L. Infants exposed in utero to Hurricane Maria have gut microbiomes with reduced diversity and altered metabolic capacity. mSphere 2023, 8, e00134-23. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Zhang, A.; Flores, M.A.; De Ángel Solá, D.; Cao, L.; Bolanos-Rosero, B.; Wang, L.; Godoy-Vitorino, F.; Matos, N.R.; Wang, L. Prenatal exposure to Hurricane Maria is associated with an altered infant nasal microbiome. J. Allergy Clin. Immunol. Glob. 2022, 1, 128–137. [Google Scholar] [CrossRef]
- Wang, L.; De Ángel Solá, D.; Acevedo Flores, M.; Schriefer, A.; Wang, L.; Gerónimo López, K.; Chang, A.; Warner, B.; Shan, L.; Holtz, L.R.; et al. Prenatal food insecurity post Hurricane Maria is associated with decreased Veillonella in the infant gut. Pediatr. Res. 2020, 88, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; May, T.W.; Frøslev, T.G.; Pawlowska, J.; Lindahl, B.; Põldmaa, K.; Truong, C.; et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: Sequences, taxa and classifications reconsidered. Nucleic Acids Res. 2024, 52, D791–D797. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Agache, I.; Hernandez, M.L.; Radbel, J.M.; Renz, H.; Akdis, C.A. An Overview of Climate Changes and Its Effects on Health: From Mechanisms to One Health. J. Allergy Clin. Immunol. Pract. 2025, 13, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Bignier, C.; Havet, L.; Brisoux, M.; Omeiche, C.; Misra, S.; Gonsard, A.; Drummond, D. Climate change and children’s respiratory health. Paediatr. Respir. Rev. 2025, 53, 64–73. [Google Scholar] [CrossRef]
- Salo, P.M.; Yin, M.; Arbes Jr, S.J.; Cohn, R.D.; Sever, M.; Muilenberg, M.; Burge, H.A.; London, S.J.; Zeldin, D.C. Dustborne Alternaria alternata antigens in US homes: Results from the National Survey of Lead and Allergens in Housing. J. Allergy Clin. Immunol. 2005, 116, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Grimsley, L.F.; Chulada, P.C.; Kennedy, S.; White, L.; Wildfire, J.; Cohn, R.D.; Mitchell, H.; Thornton, E.; El-Dahr, J.; Mvula, M.M.; et al. Indoor Environmental Exposures for Children with Asthma Enrolled in the HEAL Study, Post-Katrina New Orleans. Environ. Health Perspect. 2012, 120, 1600–1606. [Google Scholar] [CrossRef]
- Soffer, N.; Green, B.J.; Acosta, L.; Divjan, A.; Sobek, E.; Lemons, A.R.; Rundle, A.G.; Jacobson, J.S.; Goldstein, I.F.; Miller, R.L.; et al. Alternaria is associated with asthma symptoms and exhaled NO among NYC children. J. Allergy Clin. Immunol. 2018, 142, 1366–1368.e10. [Google Scholar] [CrossRef]
- Bush, R.K.; Prochnau, J.J. Alternaria-induced asthma. J. Allergy Clin. Immunol. 2004, 113, 227–234. [Google Scholar] [CrossRef]
- Lemanske, R.F. Issues in understanding pediatric asthma: Epidemiology and genetics. J. Allergy Clin. Immunol. 2002, 109, S521–S524. [Google Scholar] [CrossRef]
- Rivas, C.M.; Schiff, H.V.; Moutal, A.; Khanna, R.; Kiela, P.R.; Dussor, G.; Price, T.J.; Vagner, J.; DeFea, K.A.; Boitano, S. Alternaria alternata-induced airway epithelial signaling and inflammatory responses via protease-activated receptor-2 expression. Biochem. Biophys. Res. Commun. 2022, 591, 13–19. [Google Scholar] [CrossRef]
- Ogawa, H.; Fujimura, M.; Takeuchi, Y.; Makimura, K. The influence of Schizophyllum commune on asthma severity. Lung 2011, 189, 485–492. [Google Scholar] [CrossRef][Green Version]
- Oguma, T.; Ishiguro, T.; Kamei, K.; Tanaka, J.; Suzuki, J.; Hebisawa, A.; Obase, Y.; Mukae, H.; Tanosaki, T.; Furusho, S.; et al. Clinical characteristics of allergic bronchopulmonary mycosis caused by Schizophyllum commune. Clin. Transl. Allergy 2024, 14, e12327. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Liu, H.Y.; Tan, X.L.; Ji, Y.; Jiang, Y.X.; Prabhakar, M.; Rong, Z.-H.; Zhou, H.-W.; Zhang, G.-X. Sputum Bacterial and Fungal Dynamics during Exacerbations of Severe COPD. PLoS ONE 2015, 10, e0130736. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Pagán, K.; Bolaños-Rosero, B.; Pérez, C.; AP, O.; Godoy-Vitorino, F. Ecological competition in the oral mycobiome of Hispanic adults living in Puerto Rico associates with periodontitis. J. Oral Microbiol. 2024, 16, 2316485. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liao, H.; Liu, Z.; Yang, W.; Liu, Q.; Li, Q. Association between early life exposure to indoor environmental factors and childhood asthma. Build. Environ. 2022, 226, 109740. [Google Scholar] [CrossRef]



| Variables. | Year 1 Group (n = 26) | Year 2 Group (n = 32) | p-Value |
|---|---|---|---|
| Male sex: no. (%) | 9 (34.62) | 18 (56.25) | 0.17 |
| Age (wk), mean (SD) | 16.10 (5.51) | 16.17 (4.44) | 0.81 |
| Gestational age (wk), mean (SD) | 38.84 (0.94) | 38.85 (1.40) | 0.96 |
| Sibling presence at home, no. (%) | 19 (73.08) | 22 (68.75) | 0.94 |
| Exclusive breastfeeding, no. (%) | 7 (26.92) | 8 (25.00) | 0.99 |
| Maternal exposure to mold, no. (%) | 8 (30.77) | 0 (0) | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; de Ángel Solá, D.; Rivera-Mariani, F.E.; Bolaños Rosero, B.; Rosario Matos, N.; Wang, L. Dysbiosis in the Nasal Mycobiome of Infants Born in the Aftermath of Hurricane Maria. Microorganisms 2025, 13, 1784. https://doi.org/10.3390/microorganisms13081784
Wang R, de Ángel Solá D, Rivera-Mariani FE, Bolaños Rosero B, Rosario Matos N, Wang L. Dysbiosis in the Nasal Mycobiome of Infants Born in the Aftermath of Hurricane Maria. Microorganisms. 2025; 13(8):1784. https://doi.org/10.3390/microorganisms13081784
Chicago/Turabian StyleWang, Ruochen, David de Ángel Solá, Félix E. Rivera-Mariani, Benjamín Bolaños Rosero, Nicolás Rosario Matos, and Leyao Wang. 2025. "Dysbiosis in the Nasal Mycobiome of Infants Born in the Aftermath of Hurricane Maria" Microorganisms 13, no. 8: 1784. https://doi.org/10.3390/microorganisms13081784
APA StyleWang, R., de Ángel Solá, D., Rivera-Mariani, F. E., Bolaños Rosero, B., Rosario Matos, N., & Wang, L. (2025). Dysbiosis in the Nasal Mycobiome of Infants Born in the Aftermath of Hurricane Maria. Microorganisms, 13(8), 1784. https://doi.org/10.3390/microorganisms13081784






