Screening and Phenotyping of Lactic Acid Bacteria in Boza
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Instruments and Equipment
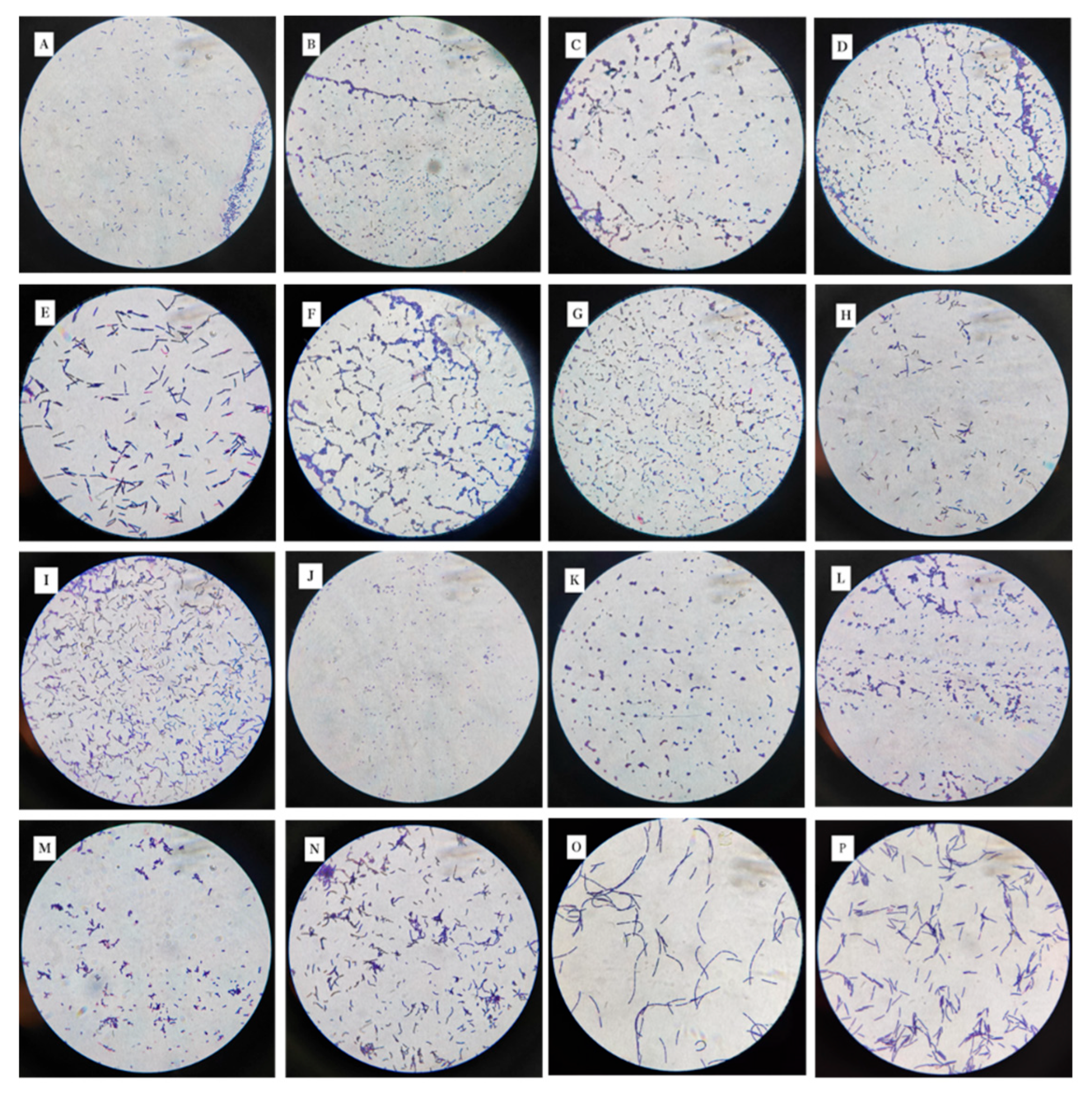
2.3. Isolation and Identification of Lactic Acid Bacteria
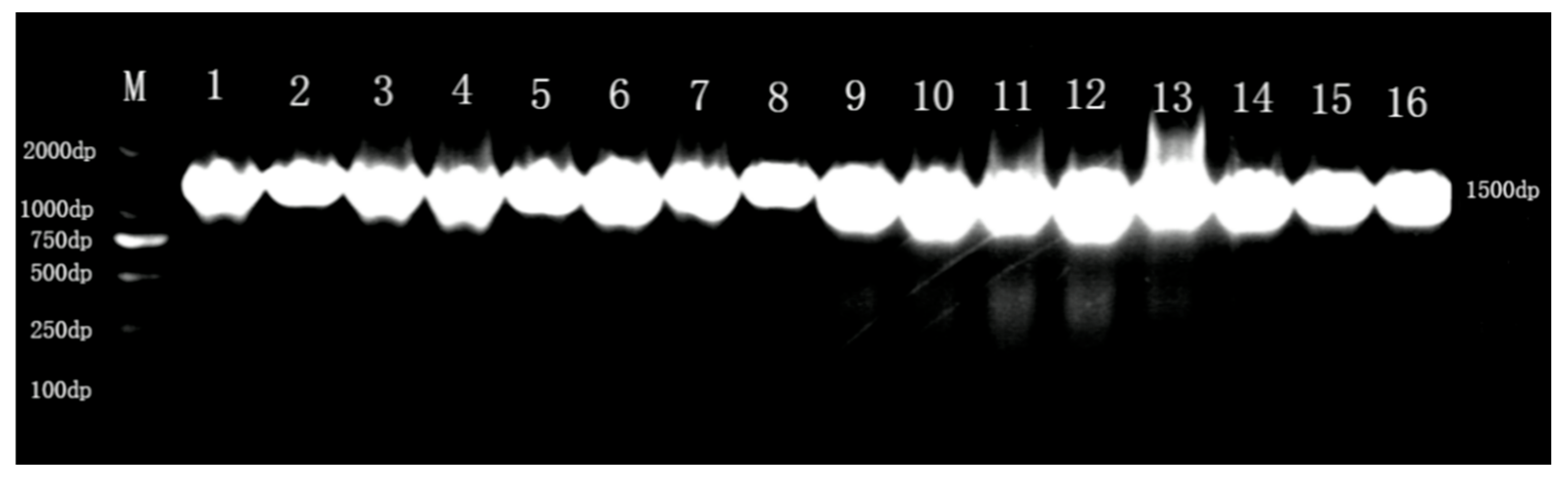
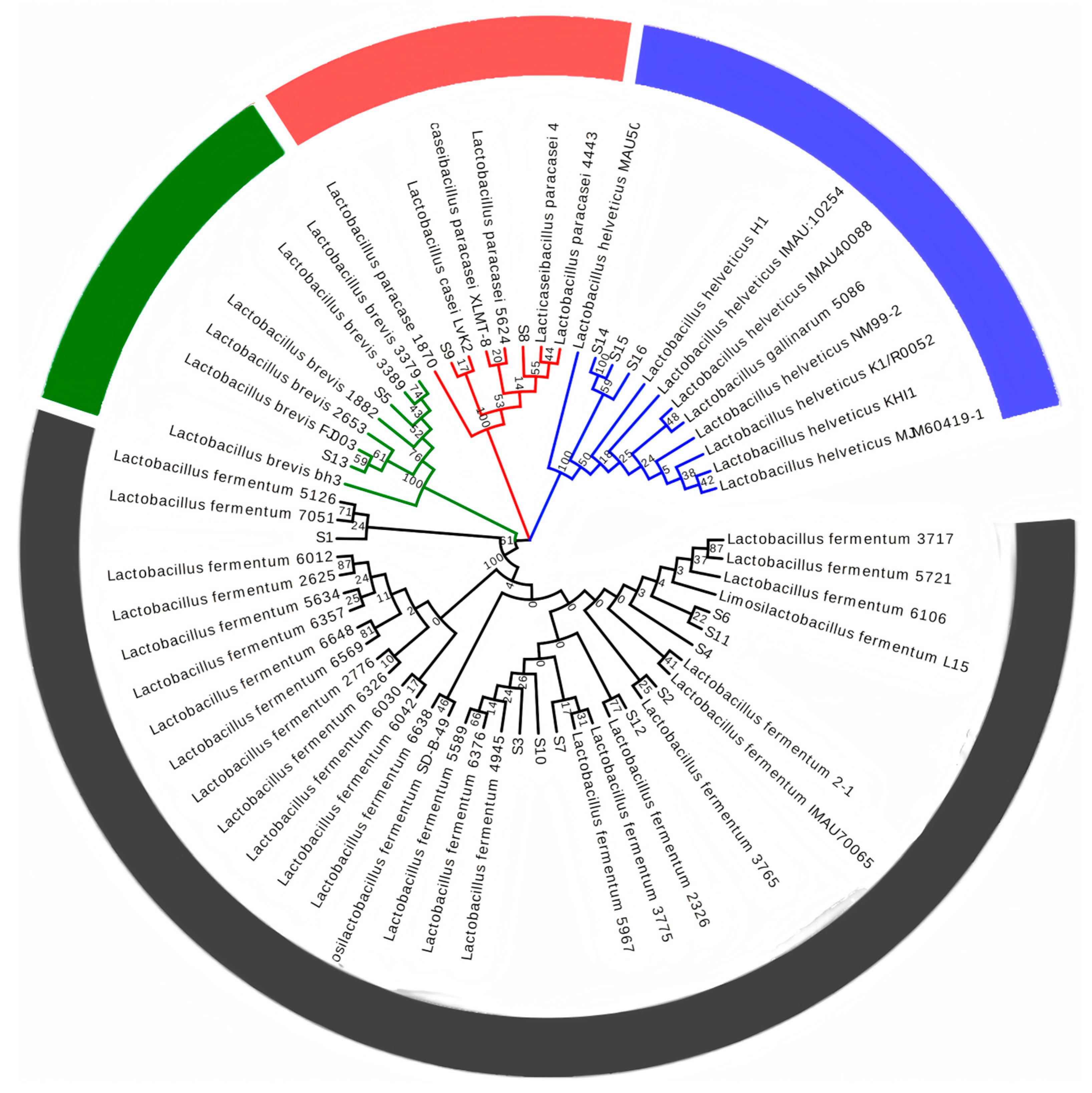
2.4. Genetic Identification of Lactic Acid Bacteria
2.5. Fermentation Characterization of Lactic Acid Bacteria
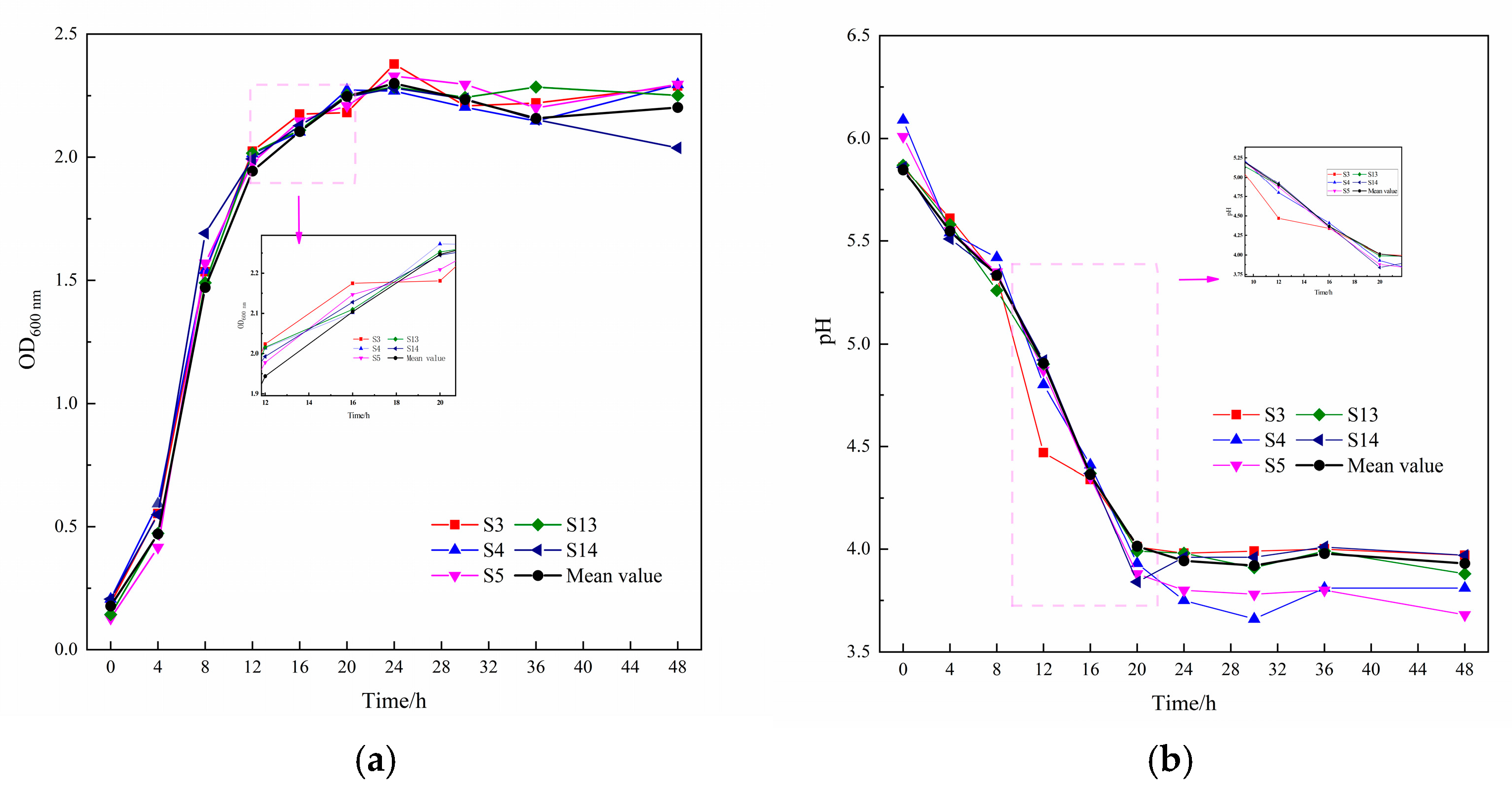
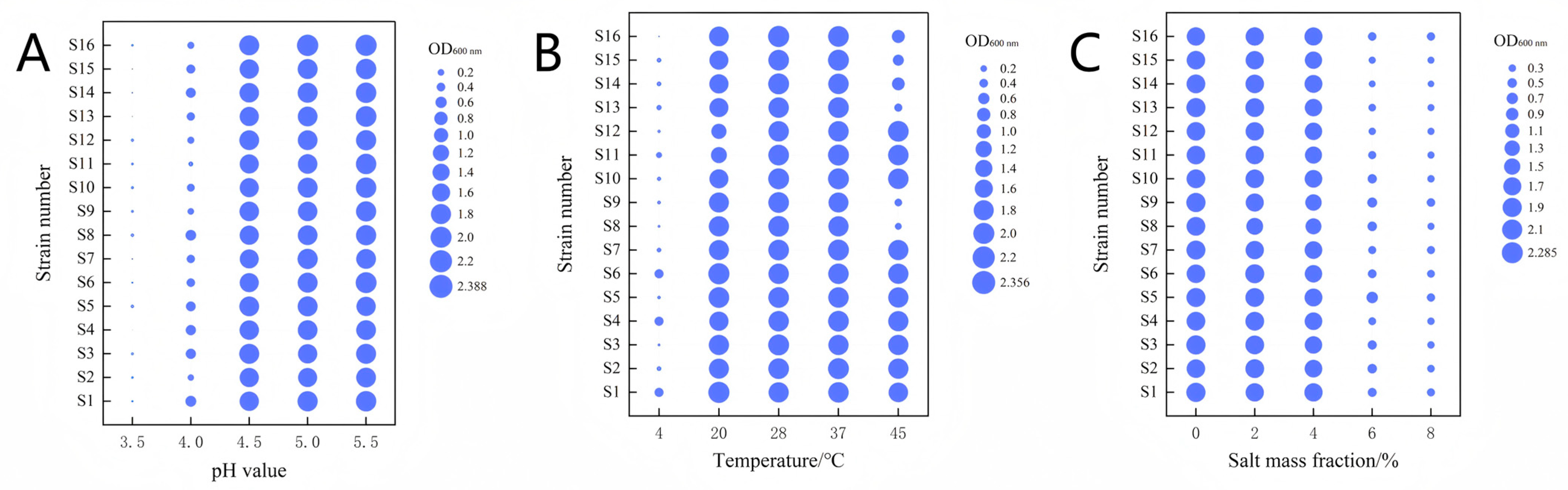
2.5.1. Growth and Acid Production Rates
2.5.2. Acid and Alkali Resistance, Salt Resistance, and Temperature Resistance
2.6. Determination of Environmental Tolerance in the Simulated Human Digestive Tract
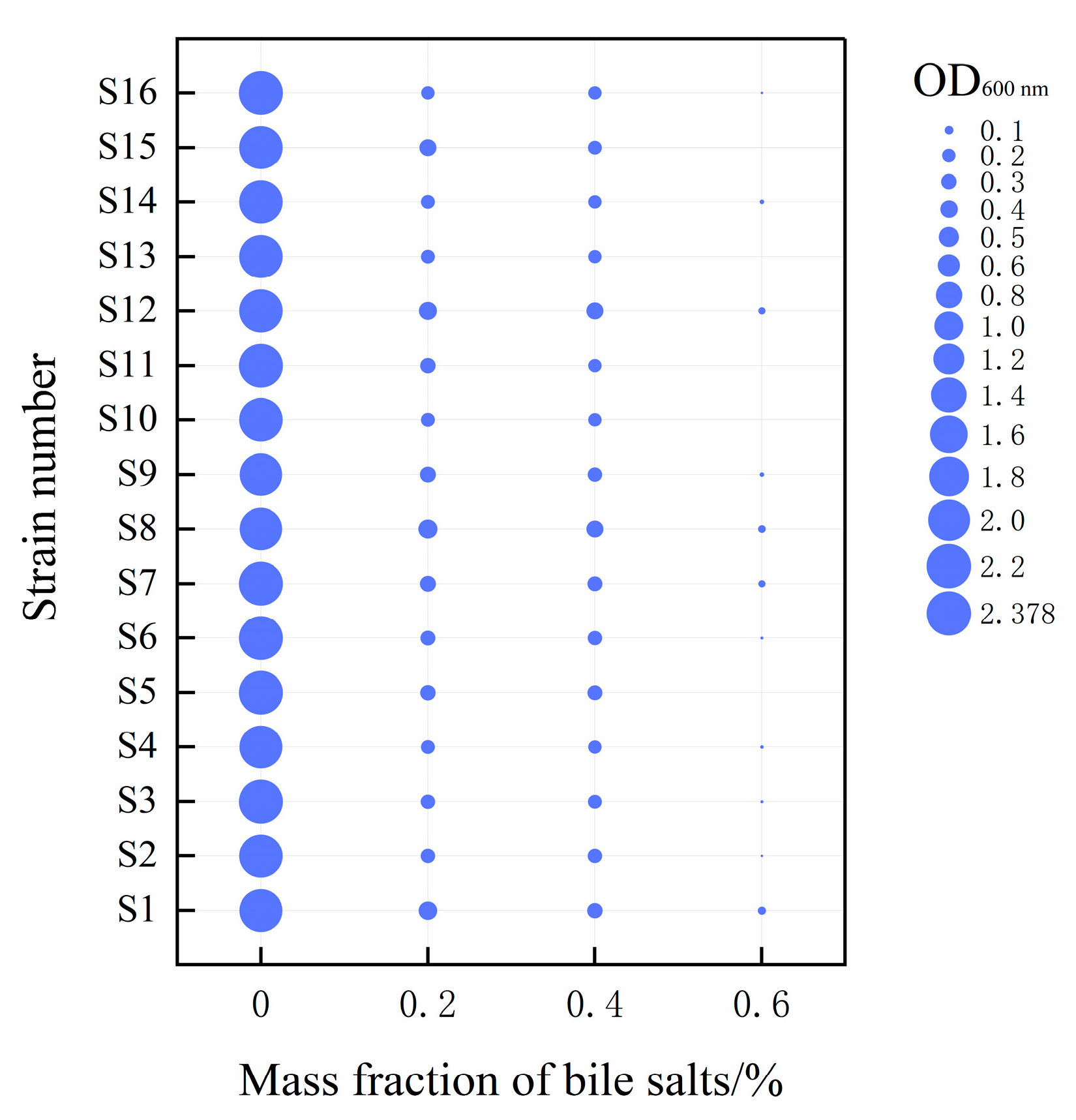
2.6.1. Bile Salt Resistance
2.6.2. Modeling of Gastrointestinal Digestion
2.7. In Vitro Characterization and Evaluation of Probiotic Properties
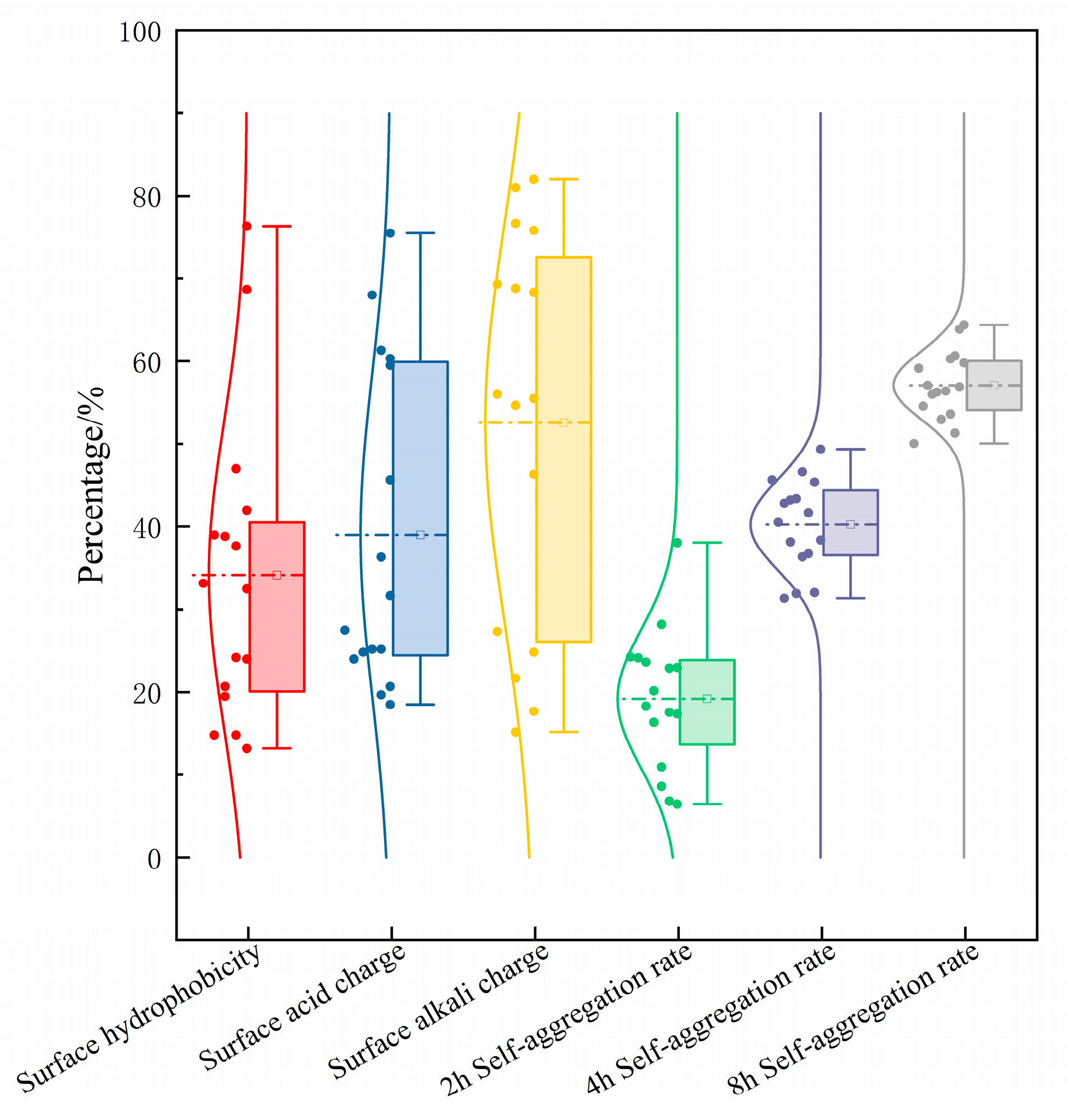
2.7.1. Surface Hydrophobicity
2.7.2. Surface Acid–Base Charge
2.7.3. Capacity for Self-Aggregation
2.8. Screening of Lactic Acid Bacteria with Antioxidant Properties
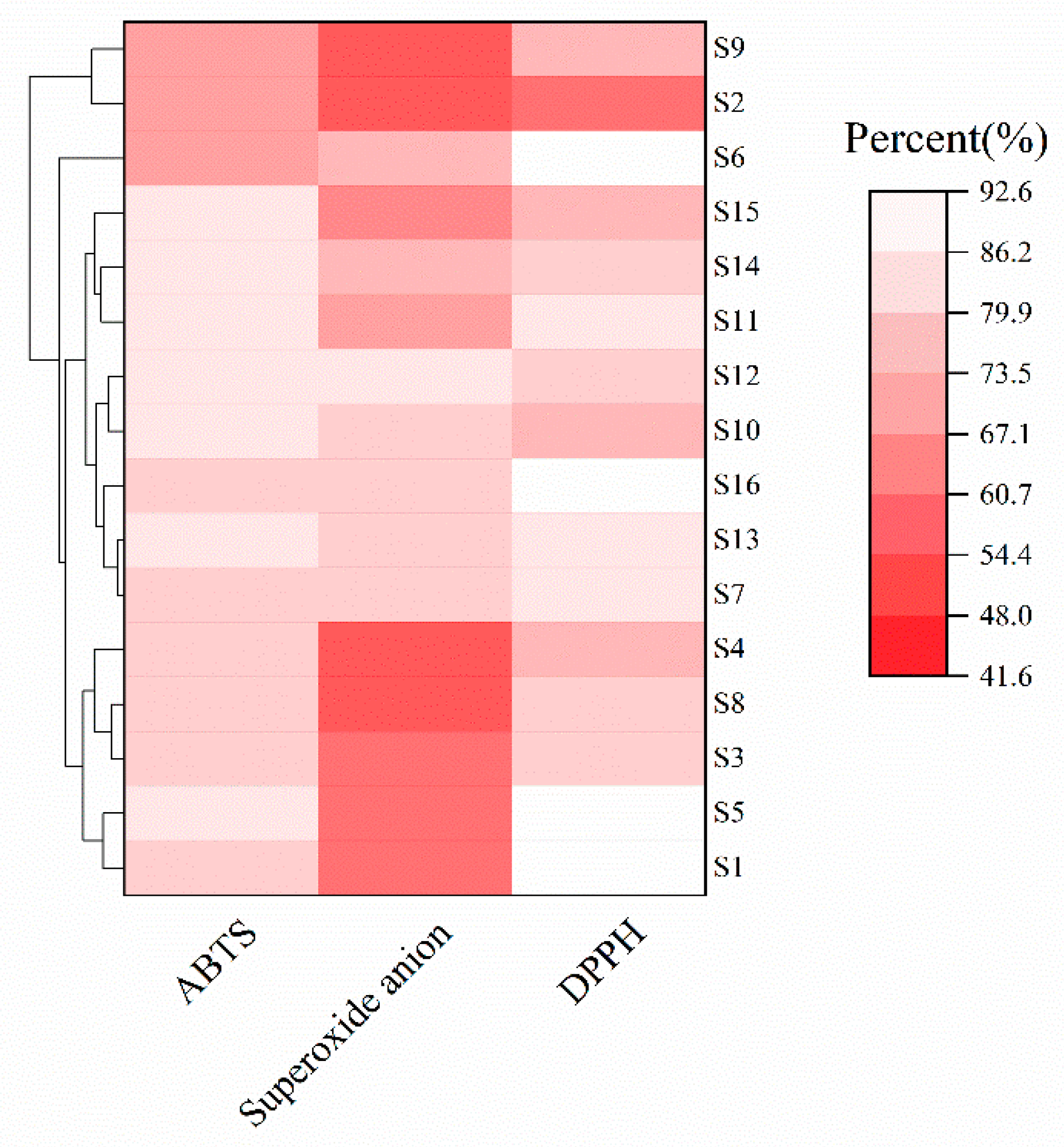
2.8.1. Measurement of DPPH Free Radical Scavenging Capacity
2.8.2. Measurement of ABTS Free Radical Scavenging Capacity
2.8.3. Determination of the Scavenging Capacity of Superoxide Anion Radicals
2.9. Safety Evaluation of Lactic Acid Bacteria
2.9.1. Amino Acid Decarboxylase Activity
2.9.2. Hemolysis
2.9.3. Antibiotic Resistance
2.9.4. Bacteriostatic Properties
2.10. Data Processing
3. Results and Discussion
3.1. Results of Isolation and Purification of Strains
3.2. Fermentation Characterization of Lactic Acid Bacteria
3.3. Simulated Human Gastrointestinal Environmental Tolerance Analysis
3.4. Analysis of In Vitro Characterization and Evaluation of Probiotic Properties
3.5. Study on Antioxidant Properties of Lactic Acid Bacteria
3.6. Evaluation of Potential Safety Risks of Lactic Acid Bacteria
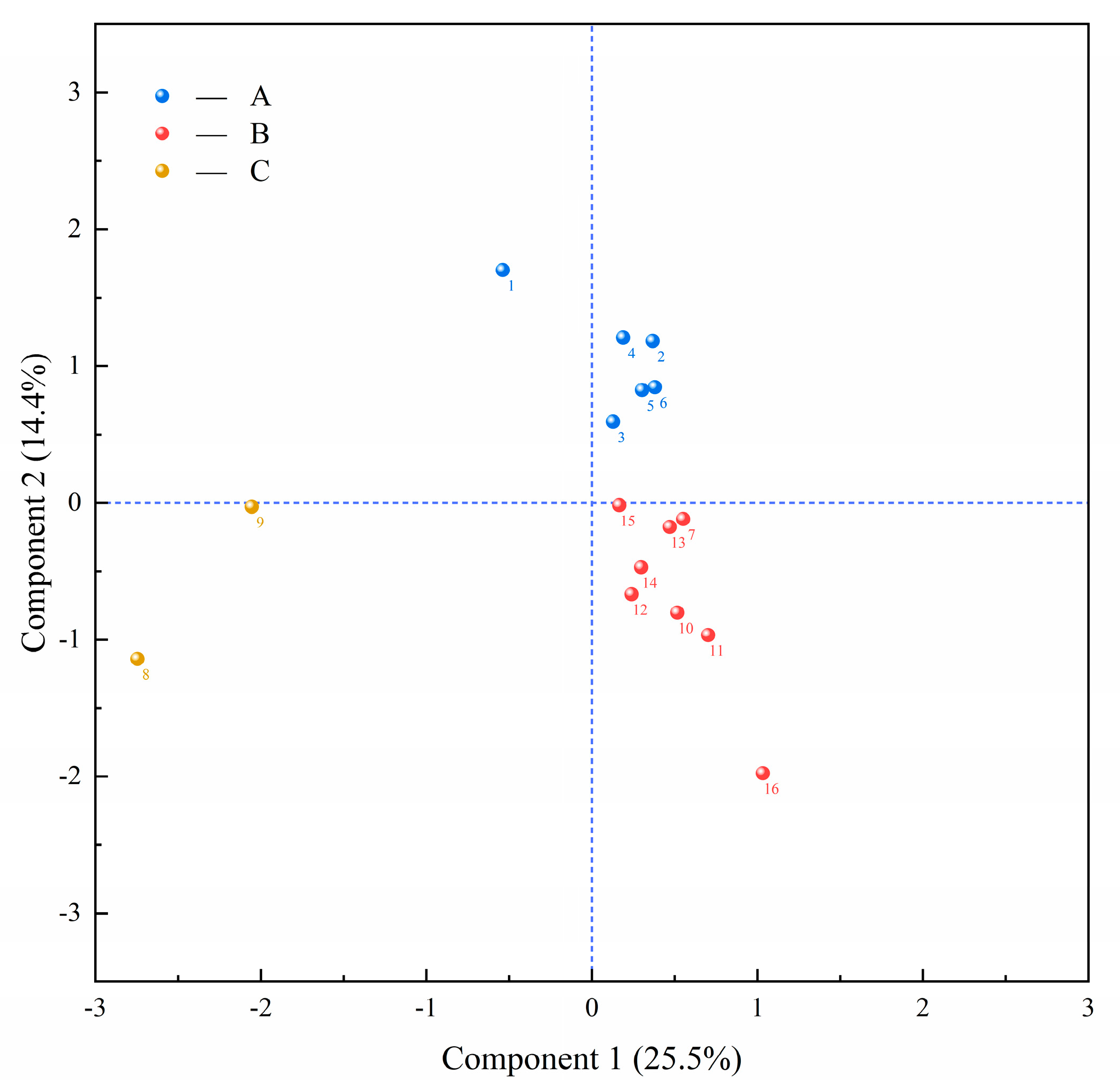
3.7. PCA-Based Phenotypic Synthesis of Strains
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jamila, S.; Janyl, I.; Mukarama, M. Milk- and cereal-based kyrgyz ethnic foods. Int. J. Astron. Food Sci. 2022, 29, 100507. [Google Scholar]
- Todorov, S.D.; Botes, M.; Guigas, C.; Schillinger, U.; Wiid, I.; Wachsman, M.B.; Holzapfel, W.H.; Dicks, L.M. Boza, a natural source of probiotic lactic acid bacteria. Appl. Microbiol. 2008, 104, 465–477. [Google Scholar] [CrossRef]
- Das, A.; Raychaudhuri, U.; Chakraborty, R. Cereal based functional food of Indian subcontinent: A review. Food Sci. Technol. 2012, 49, 665–672. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-Waterhouse, D.X.; Waterhouse, G.I.N.; Cui, C.; Ruan, Z. Fermentation-enabled wellness foods: A fresh perspective. Food Sci. 2019, 8, 203–243. [Google Scholar] [CrossRef]
- Turpin, W.; Humblot, C.; Guyot, J.P. Genetic screening of functional properties of lactic acid bacteria in a fermented pearl millet slurry and in the metagenome of fermented starchy foods. AEM 2011, 77, 8722–8734. [Google Scholar] [CrossRef]
- Dordevic, T.M.; Siler-Marinkovic, S.S.; Dimitrijevic-Brankovic, S.I. Effect of fermentation on antioxidant properties of some cereals and pseudo cereals. Food Chem. 2010, 119, 957–963. [Google Scholar] [CrossRef]
- Melda, Y.T.; Tuncay, G.; Deniz, D.A.K. Isolation and identification of lactic acid bacteria from boza produced traditionally in the thrace region of Turkey. Food Saf. Food Qual. 2021, 72, 158–163. [Google Scholar]
- Ucak, S.; Yurt, M.N.Z.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Soyucok, A.; Aydin, A.; Ozalp, V.C.; Sudagidan, M. Identification of bacterial communities of fermented cereal beverage Boza by metagenomic analysis. LWT 2022, 153, 112465. [Google Scholar] [CrossRef]
- Queiroz, L.L.; Hoffmann, C.; Lacorte, G.A.; Franco, B.D.G.d.M.; Todorov, S.D. Genomic and functional characterization of bacteriocinogenic lactic acid bacteria isolated from Boza, a traditional cereal-based beverage. Sci. Rep. 2022, 12, 1460. [Google Scholar] [CrossRef]
- Andrea, O.; Cristiana, G.; Lucia, A.; Vesna, M.; Francesca, C. Unpasteurised commercial Boza as a source of microbial diversity. Int. J. Food Microbiol. 2015, 194, 62–70. [Google Scholar]
- Yigit, M.B.; Cebeci, A. Highly potent new probiotic strains from traditional turkish fermented foods. Curr. Microbiol. 2025, 82, 97. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Li, L.; Xie, G. Impact of glutinous rice varieties from different regions on microbial community structure, metabolic profiles, and flavor characteristics of Chinese rice wine (Huangjiu). Foods 2025, 14, 1261. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.F.M.; Alonso, B.V.; Almeida, P.A.A.; Barbosa, I.V.; Braga de Paula, O.A.; Barbosa, L.R.; Bruno, L.M.; Menezes, L.D.M.; Silva, M.R.; Costa, G.M.de.; et al. Searching for antibiotic-susceptible bioprotective lactic acid bacteria to control dangerous biological agents in artisanal cheese. Food Microbiol. 2025, 130, 104762. [Google Scholar] [CrossRef]
- Tuerhong, N.; Wang, L.; Cui, J.; Shataer, D.; Yan, H.; Dong, X.; Gao, Z.; Zhang, M.; Qin, Y.; Lu, J. Targeted screening of Lactiplantibacillus plantarum strains isolated from tomatoes and its application in tomato fermented juice. Foods 2024, 13, 3569. [Google Scholar] [CrossRef]
- Aktepe, Y.; Aydın, F.; Bozoğlu, T.; Özer, G.; Çakır, İ. Molecular characterization and multifunctional evaluation of lactic acid bacteria isolated from traditional sourdough. Int. J. Food Microbiol. 2024, 423, 110845. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yang, G.; Chen, K.; Du, M.; Zalán, Z.; Hegyi, F.; Kan, J. Anti-fungal effects of lactic acid bacteria from pickles on the growth and sterigmatocystin production of Aspergillus versicolor. Int. J. Food Microbiol. 2024, 422, 110809. [Google Scholar] [CrossRef]
- Liu, J.G.; Lin, C.X.; Zhang, W.; Yang, Q.; Meng, J.; He, L.; Deng, L.; Zeng, X.F. Exploring the bacterial community for starters in traditional high-salt fermented Chinese fish (Suanyu). Food Chem. 2021, 358, 129863. [Google Scholar] [CrossRef]
- Yang, Z.; Ni, L.; Tian, W.; Chi, H. Screening and identification of goat-milk-derived lactic acid bacteria with bacteriocin-like activity and probiotic potentials. Microorganisms 2023, 11, 849. [Google Scholar] [CrossRef]
- Nagpal, R.; Wang, S.; Ahmadi, S.; Hayes, J.; Gagliano, J.; Subashchandrabose, S.; Kitzman, D.W.; Becton, T.; Read, R.; Yadav, H. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep. 2018, 8, 12649. [Google Scholar] [CrossRef]
- Bernet, M.F.; Brassart, D.; Neeser, J.R.; Servin, A.L. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl. Environ. Microbiol. 1993, 59, 4121–4128. [Google Scholar] [CrossRef]
- Susana, C.; Ribeiro, P.M.; O’Connor, R.P.; Ross, C.S.; Silva, C.C.G. An anti-listerial Lactococcus lactis strain isolated from azorean pico cheese produces lacticin 481. Int. Dairy J. 2016, 63, 18–28. [Google Scholar]
- Ruiz-Moyano, S.; dos Santos, M.T.P.G.; Galván, A.I.; Merchán, A.V.; González, E.; de Guía Córdoba, M.; Benito, M.J. Screening of autochthonous lactic acid bacteria strains from artisanal soft cheese: Probiotic characteristics and prebiotic metabolism. LWT 2019, 114, 108388. [Google Scholar] [CrossRef]
- Jung, J.; Jang, H.J.; Eom, S.J.; Choi, N.S.; Lee, N.K.; Paik, H.D. Fermentation of red ginseng extract by the probiotic Lactobacillus plantarum KCCM 11613P: Ginsenoside conversion and antioxidant effects. J. Ginseng Res. 2019, 43, 20–26. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Liu, W.; Wang, H.; Pang, X.; Yao, W.; Gao, X. Characterization and antioxidant activity of two low-molecular-weight polysaccharides purified from the fruiting bodies of Ganoderma lucidum. Int. J. Biol. Macromol. 2010, 46, 451–457. [Google Scholar] [CrossRef]
- Moreno, I.; Marasca, E.T.G.; de Sá, P.B.Z.R.; de Souza Moitinho, J.; Marquezini, M.G.; Alves, M.R.C.; Bromberg, R. Evaluation of probiotic potential of bacteriocinogenic lactic acid bacteria strains isolated from meat products. Probiotics Antimicrob. Proteins 2018, 10, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Hossein, S.; Rasoul, S.; Yousef, N.; Jalil, K.; Bahman, P. Potential probiotic characterization of lactic acid bacteria isolated from Duimaj, an Iranian traditional snack food, using biochemical, molecular and computational approaches. LWT 2023, 184, 115091. [Google Scholar]
- Parlindungan, E.; Lugli, G.A.; Ventura, M.; Van, S.D.; Mahony, J. Lactic acid bacteria diversity and characterization of probiotic candidates in fermented meats. Foods 2021, 10, 1519. [Google Scholar] [CrossRef]
- Zeng, H.; Liu, Y.; Huang, K.; Chen, H.; Yang, B.; Wang, J. Lactiplantibacillus plantarum A1, C1 and C10 are potential probiotics isolated from pineapple residual silage. Microorganisms 2022, 11, 29. [Google Scholar] [CrossRef]
- Bonger, Z.T.; Abera, M.K.; Habitu, T.A.; Abera, A.A.; Tenagashaw, M.W.; Tiruneh, A.T.; Awol, S.J.; Wudineh, T.A.; Asres, D.T.; Yehuala, T.F. Isolation and identification of dominant lactic acid bacteria and yeast species from teff (Eragrostis teff) injera dough fermentation. J. Food 2023, 21, 718–734. [Google Scholar]
- Yang, Q.; Markwick, R.K.; Mutukumira, A.N. Identification of dominant lactic acid bacteria and yeast in rice sourdough produced in New Zealand. CRFS 2021, 4, 729–736. [Google Scholar] [CrossRef] [PubMed]
- NgongolaManani, T.A.; Wicklund, T.; Mwangwela, A.M.; Hilde, M. Identification and characterisation of lactic acid bacteria involved in natural and lactic acid. Food Biotechnol. 2015, 29, 20–50. [Google Scholar]
- Qureshi, N.; Gu, Q.; Li, P. Whole genome sequence analysis and in vitro probiotic characteristics of a lactobacillus strain Lactobacillus paracasei ZFM54. J. Appl. Microbiol. 2020, 129, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Evivie, S.E.; Muhammad, Z.; Luo, G.W.; Liang, H.Z.; Wang, N.N.; Huo, G.C. In vitro assessment of the antimicrobial potentials of Lactobacillus helveticus strains isolated from traditional cheese in Sinkiang China against food-borne pathogens. Food Funct. 2016, 7, 789–797. [Google Scholar] [CrossRef]
- Paik, H.D.; Lee, J.Y. Investigation of reduction and tolerance capability of lactic acid bacteria isolated from kimchi against nitrate and nitrite in fermented sausage condition. Meat Sci. 2014, 97, 609–614. [Google Scholar] [CrossRef]
- Kun, W.; Mengmeng, N.; Dawei, S.; Xuejian, S.; Jing, Z.; Yue, W.; Baoxin, L.; Guangcai, N. Preparation, partial characterization and biological activity of exopolysaccharides produced from Lactobacillus fermentum S1. J. Biosic. Bioeng. 2020, 129, 206–214. [Google Scholar]
- Wang, Y.; He, L.; Xing, Y.; Zhou, W.; Pian, R.; Yang, F.; Chen, X.; Zhang, Q. Bacterial diversity and fermentation quality of Moringa oleifera leaves silage prepared with lactic acid bacteria inoculants and stored at different temperatures. Bioresour. Technol. 2019, 284, 349–358. [Google Scholar] [CrossRef]
- Melo, J.; Andrew, P.W.; Faleiro, M.L. Different assembly of acid and salt tolerance response in two dairy Listeria monocytogenes wild strains. Arch. Microbiol. 2013, 195, 339–348. [Google Scholar] [CrossRef]
- Fan, Y.; Yang, X.; Hu, C.; Wei, B.; Xu, F.; Guo, Q. Fermentation performance evaluation of lactic acid bacteria strains for sichuan radish paocai production. Foods 2024, 13, 1813. [Google Scholar] [CrossRef]
- Kübra, C.T.; Mükerrem, K.; Güzin, K. Probiotic properties of lactic acid bacteria strains isolated from pastırma. LWT 2020, 134, 110216. [Google Scholar]
- Das, S.; Mishra, B.K.; Hati, S. Techno-functional characterization of indigenous lactobacillus isolates from the traditional fermented foods of Meghalaya, India. Curr. Res. Food Sci. 2020, 3, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tsigkrimani, M.; Panagiotarea, K.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial ecology of sheep milk, artisanal feta, and kefalograviera cheeses. Part II: Technological, safety, and probiotic attributes of lactic acid bacteria isolates. Foods 2022, 11, 459. [Google Scholar] [CrossRef]
- Han, Q.; Kong, B.H.; Chen, Q.; Sun, F.D.; Zhang, H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J. Funct. Foods 2017, 32, 391–400. [Google Scholar] [CrossRef]
- Sensoy, I. A review on the food digestion in the digestive tract and the used in vitro models. Curr. Res. Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Leeuwendaal, N.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. The potential of non-starter lactic acid bacteria from Cheddar cheese to colonise the gut. J. Funct. Foods 2021, 83, 104425. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Faseleh, J.M.; Ho, Y.W. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed. Res. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef] [PubMed]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; de Cadiñanos, L.P.G.; Requena, T.; Peláez, C.; Martínez-Cuesta, M.C. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Hernández-Alcántara, A.M.; Wacher, C.; Llamas, M.G.; López, P.; Pérez-Chabela, M.L. Probiotic properties and stress response of thermotolerant lactic acid bacteria isolated from cooked meat products. LWT 2018, 91, 249–257. [Google Scholar] [CrossRef]
- Menezes, A.G.T.; Ramos, C.L.; Cenzi, G.; Melo, D.S.; Dias, D.R.; Schwan, R.F. Probiotic potential, antioxidant activity, and phytase production of indigenous yeasts isolated from indigenous fermented foods. Probiotics Antimicrob. Proteins 2020, 12, 280–288. [Google Scholar] [CrossRef]
- Jang, H.J.; Song, M.W.; Lee, N.K.; Paik, H.D. Antioxidant effects of live and heat-killed probiotic Lactobacillus plantarum Ln1 isolated from kimchi. Food Sci. Technol. 2018, 55, 3174–3180. [Google Scholar] [CrossRef]
- Fekri, A.; Torbati, M.; Yari, K.A.; Bagherpour Shamloo, H.; Azadmard-Damirchi, S. Functional effects of phytate-degrading, probiotic lactic acid bacteria and yeast strains isolated from Iranian traditional sourdough on the technological and nutritional properties of whole wheat bread. Food Chem. 2020, 306, 125620. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Yang, Q.; Wan, W.; Zhu, Q.; Zeng, X. Physicochemical properties and adaptability of amine-producing enterobacteriaceae isolated from traditional Chinese fermented fish (Suan yu). Food Chem. 2022, 369, 130885. [Google Scholar] [CrossRef] [PubMed]
- Ali-Vehmas, T.; Vikerpuur, M.; Pyörälä, S.; Atroshi, F. Characterization of hemolytic activity of Staphylococcus aureus strains isolated from bovine mastitic milk. Microbiol. Res. 2001, 155, 339–344. [Google Scholar] [CrossRef]
- Shubham, G.; Upasana, M.; Ranendra, K.M. Isolation and characterisation of lactic acid bacteria from traditional fermented fish product shidal of India with reference to their probiotic potential. LWT 2021, 146, 111641. [Google Scholar]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic lactobacillus species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef]
- Moradi, M.; Kousheh, S.A.; Almasi, H.; Alizadeh, A.; Guimarães, J.T.; Yılmaz, N.; Lotfi, A. Postbiotics produced by lactic acid bacteria: The next frontier in food safety. CRFSFS 2020, 19, 3390–3415. [Google Scholar] [CrossRef]
- Nikheel, B.R.; Girija, G.P.; Giulia, T.; Anuya, M.; Rahul, C.R.; Asif, P.; Fatih, O. Recent advances in bio-preservatives impacts of lactic acid bacteria and their metabolites on aquatic food products. Food Biosci. 2021, 44, 101440. [Google Scholar]
- Yang, S.; Liu, L.; Wang, J.; Guo, S.; Liu, G.; Chen, X.; Deng, X.; Tu, M.; Tao, Y.; Rao, Y. Antimicrobial activity against staphylococcus aureus and genome features of Lactiplantibacillus plantarum LR-14 from Sichuan pickles. Arch. Microbiol. 2022, 204, 637. [Google Scholar] [CrossRef]
- Petkova, M.; Stefanova, P.; Gotcheva, V.; Angelov, A. Isolation and characterization of lactic acid bacteria and yeasts from typical bulgarian sourdoughs. Microorganisms 2021, 9, 1346. [Google Scholar] [CrossRef]
| Strain Number | Gastric Survival | Intestinal Survival |
|---|---|---|
| (VI%) | (VI%) | |
| S1 | 52.0 ± 4.2 gh | 28.6 ± 0.4 bc |
| S2 | 66.4 ± 2.0 ef | 24.6 ± 0.7 bcde |
| S3 | 58.7 ± 1.2 fg | 20.5 ± 1.0 de |
| S4 | 97.6 ± 7.9 a | 24.4 ± 0.4 bcde |
| S5 | 45.9 ± 5.6 hi | 21.4 ± 3.5 de |
| S6 | 64.7 ± 5.3 f | 28.9 ± 6.8 b |
| S7 | 69.0 ± 1.2 ef | 19.2 ± 3.8 e |
| S8 | 96.2 ± 11.8 ab | 24.8 ± 4.4 bcde |
| S9 | 85.6 ± 2.4 bcd | 36.4 ± 3.5 a |
| S10 | 76.8 ± 6.4 de | 21.8 ± 3.4 cde |
| S11 | 84.4 ± 6.1 cd | 20.4 ± 1.2 de |
| S12 | 66.8 ± 3.0 ef | 19.6 ± 0.4 e |
| S13 | 40.4 ± 2.7 i | 25.9 ± 5.5 bcde |
| S14 | 85.2 ± 6.9 bcd | 20.8 ± 4.0 de |
| S15 | 86.5 ± 1.1 abcd | 28.8 ± 4.0 b |
| S16 | 91.9 ± 8.6 abc | 26.8 ± 1.6 bcd |
| Strain Number | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | S16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tetracycline (TET) | S | S | S | S | S | S | S | S | S | S | S | S | I | S | S | S |
| Kanamycin (KAN) | I | R | R | R | R | R | R | R | R | R | R | R | R | I | I | R |
| Ampicillin (AMP) | S | S | S | S | S | S | S | S | S | S | S | S | I | I | S | S |
| Gentamycin (GEN) | I | I | I | I | I | R | I | I | I | I | I | I | R | R | R | I |
| Erythromycin (E) | S | S | S | S | S | R | S | S | S | S | S | S | S | S | S | S |
| Chloramphenicol (C) | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S | S |
| Clindamycin (CC) | S | S | S | S | S | I | S | S | S | S | S | S | S | I | I | S |
| Vancomycin (VAN) | S | S | I | S | S | I | S | S | S | S | S | S | R | I | R | S |
| Streptomycin (S) | I | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| Staphylococcus aureus | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Colibacillus | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Bacillus cereous | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Pei, L.; Wang, X.; Luo, M.; Hou, S.; Ye, X.; Liu, W.; Zhou, Y. Screening and Phenotyping of Lactic Acid Bacteria in Boza. Microorganisms 2025, 13, 1767. https://doi.org/10.3390/microorganisms13081767
Zhao X, Pei L, Wang X, Luo M, Hou S, Ye X, Liu W, Zhou Y. Screening and Phenotyping of Lactic Acid Bacteria in Boza. Microorganisms. 2025; 13(8):1767. https://doi.org/10.3390/microorganisms13081767
Chicago/Turabian StyleZhao, Xudong, Longying Pei, Xinqi Wang, Mingming Luo, Sihan Hou, Xingqian Ye, Wei Liu, and Yuting Zhou. 2025. "Screening and Phenotyping of Lactic Acid Bacteria in Boza" Microorganisms 13, no. 8: 1767. https://doi.org/10.3390/microorganisms13081767
APA StyleZhao, X., Pei, L., Wang, X., Luo, M., Hou, S., Ye, X., Liu, W., & Zhou, Y. (2025). Screening and Phenotyping of Lactic Acid Bacteria in Boza. Microorganisms, 13(8), 1767. https://doi.org/10.3390/microorganisms13081767






