Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strains
2.2. Identification of the Microorganism
2.3. Cloning of the α-Amylase Gene
2.4. Expression and Purification of Recombinant α-Amylase
2.5. Enzyme Assay
2.6. Effects of Temperature on the Activity and Stability of Recombinant α-Amylase
2.7. Effects of pH on the Activity and Stability of Recombinant α-Amylase
2.8. Effects of CaCl2 on the Activity and Thermal Stability of Recombinant α-Amylase
2.9. Bioinformatics and Structural Analyses
3. Results
3.1. Bacterial Strain Identification and Cloning of the α-Amylase Gene
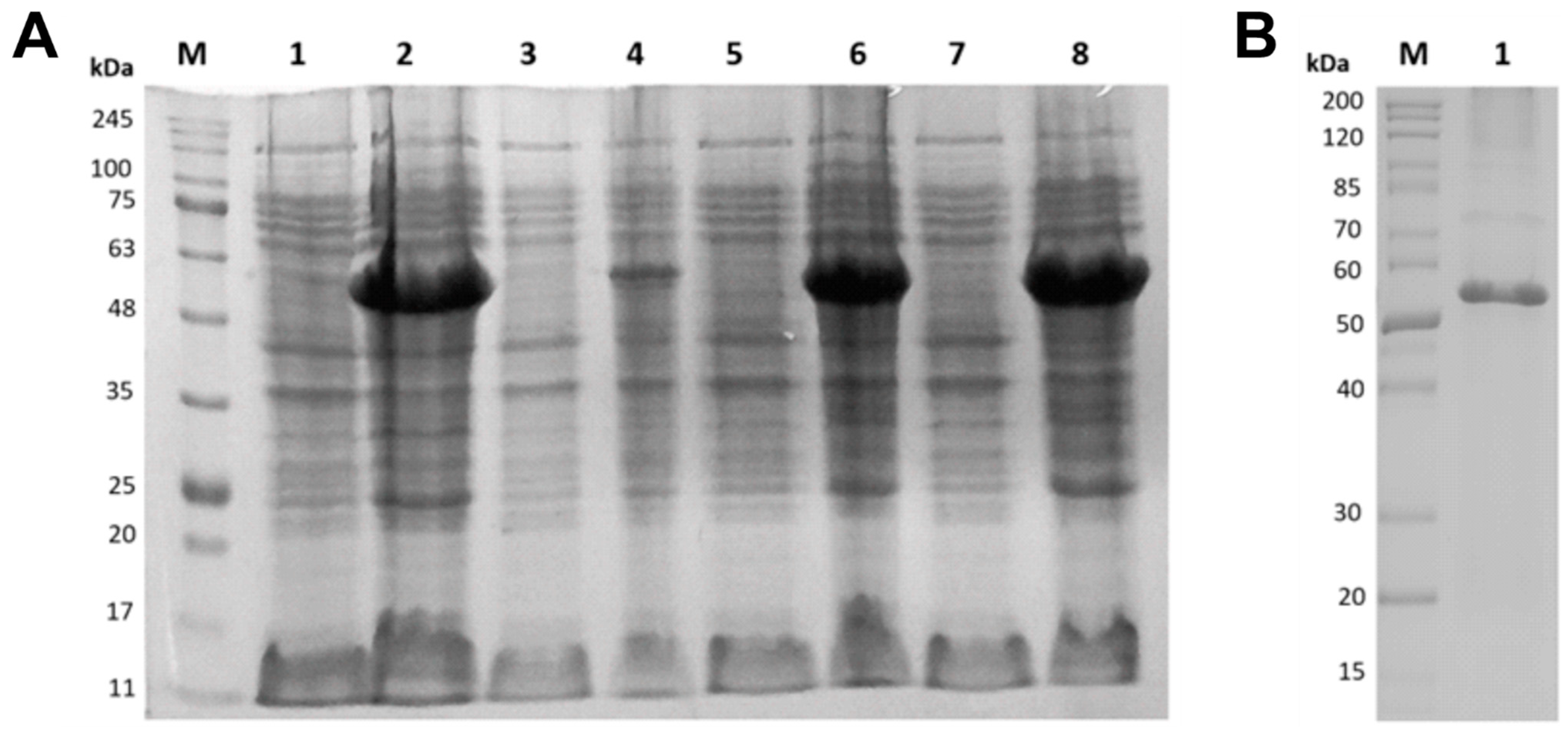
3.2. Expression and Purification of Recombinant α-Amylase
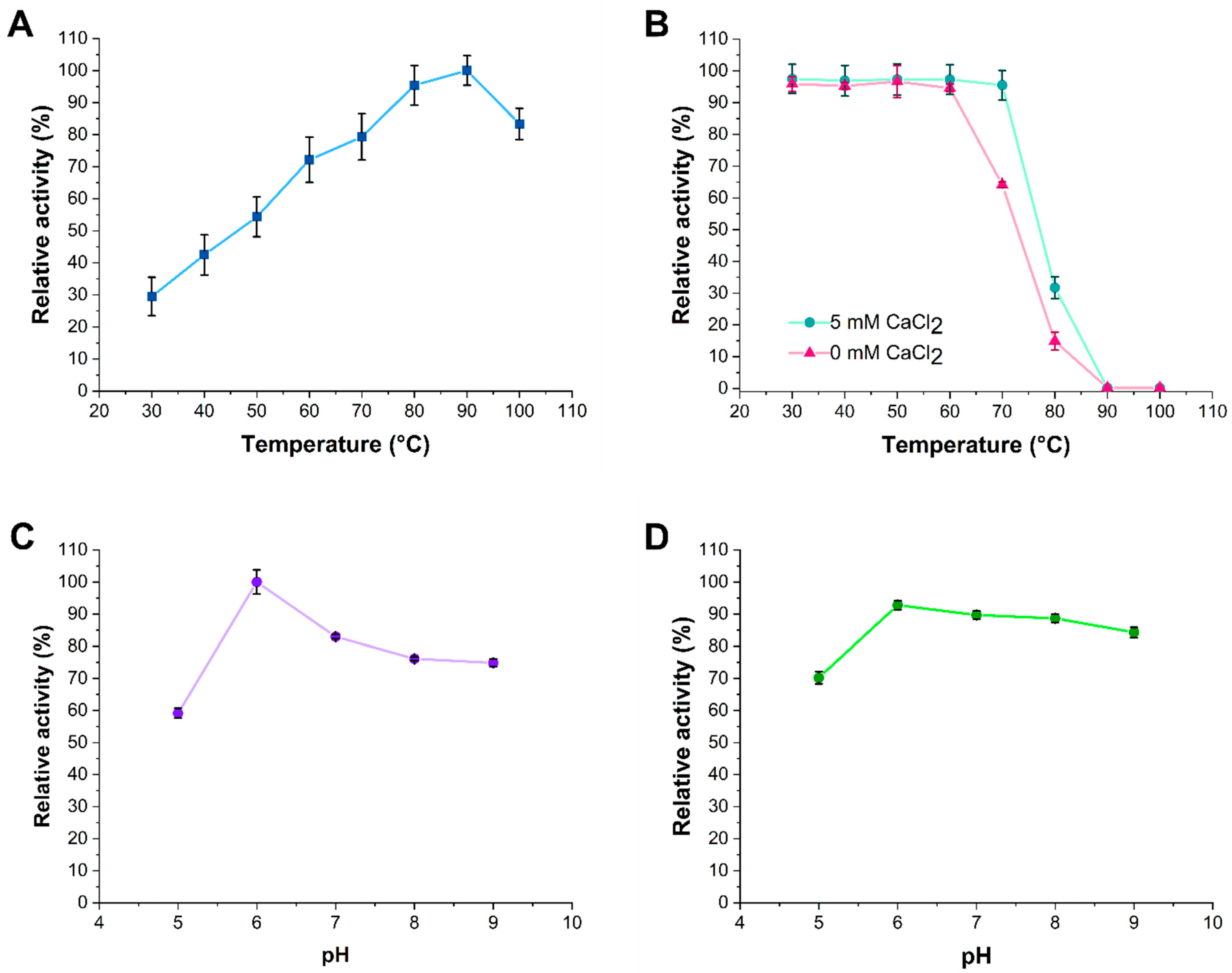
3.3. Effects of Temperature, pH, and Ca2+ Ions on α-Amylase Activity and Stability
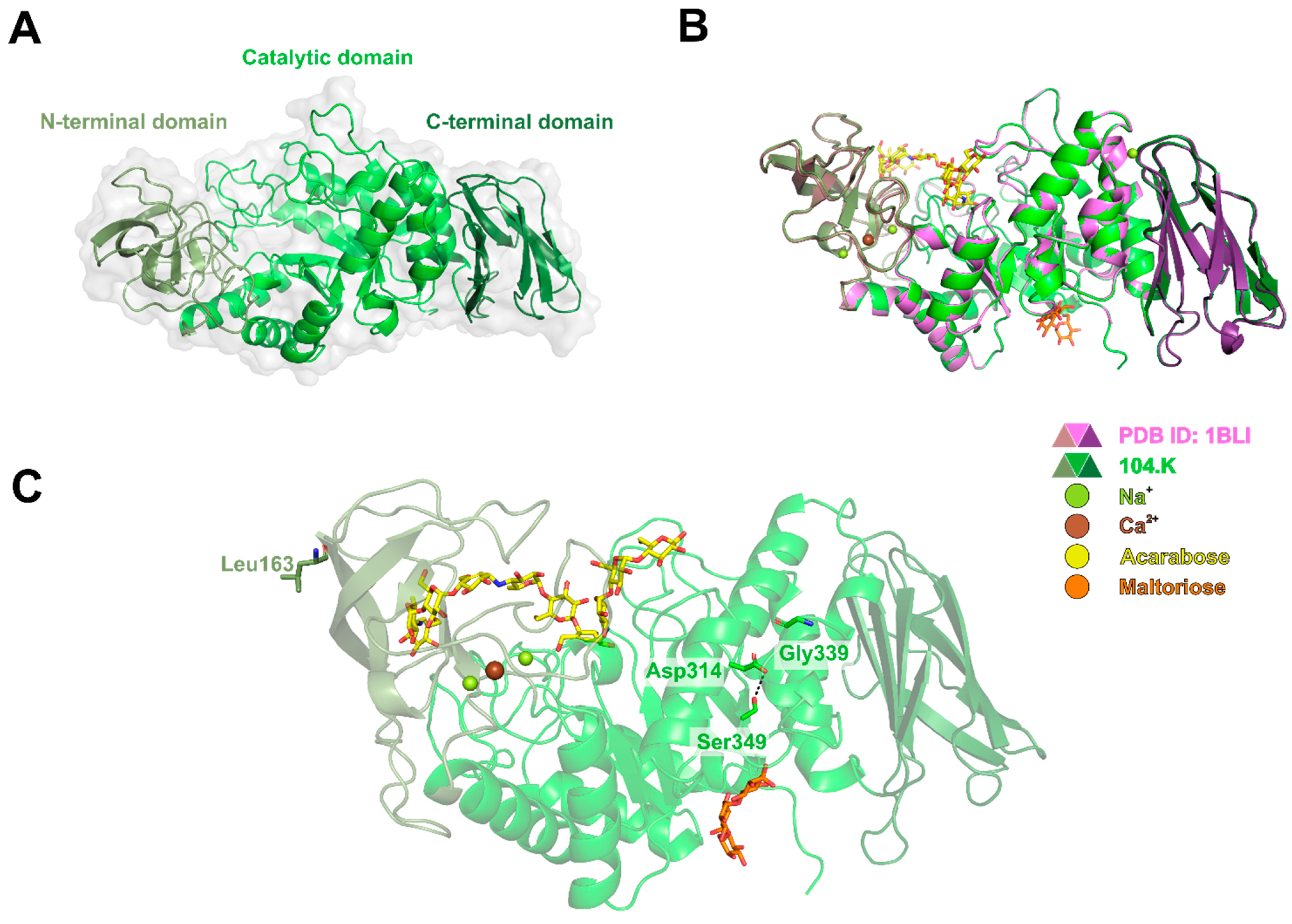
3.4. Phylogenetic and Computational Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shad, M.; Hussain, N.; Usman, M.; Akhtar, M.W.; Sajjad, M. Exploration of Computational Approaches to Predict the Structural Features and Recent Trends in α-amylase Production for Industrial Applications. Biotechnol. Bioeng. 2023, 120, 2092–2116. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abdullah, M.; Yasin, M.T.; Amanat, K.; Sultan, M.; Rahim, A.; Sarwar, F. Recent Trends in Production and Potential Applications of Microbial Amylases: A Comprehensive Review. Protein Expr. Purif. 2025, 227, 106640. [Google Scholar] [CrossRef] [PubMed]
- Miłek, J. Determination of Activation Energies and the Optimum Temperatures of Hydrolysis of Starch by α-Amylase from Porcine Pancreas. Molecules 2021, 26, 4117. [Google Scholar] [CrossRef] [PubMed]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated Carbohydrate-Active Enzyme and Substrate Annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B. How Many α-Amylase GH Families Are There in the CAZy Database? Amylase 2022, 6, 1–10. [Google Scholar] [CrossRef]
- Paul, J.S.; Gupta, N.; Beliya, E.; Tiwari, S.; Jadhav, S.K. Aspects and Recent Trends in Microbial α-Amylase: A Review. Appl. Biochem. Biotechnol. 2021, 193, 2649–2698. [Google Scholar] [CrossRef]
- Tran, T.N.; Chen, S.-C.; Doan, C.T.; Wang, S.-L. Unlocking the Potential of Pomelo Albedo: A Novel Substrate for Alpha-Amylase Production Using Bacillus licheniformis. Fermentation 2025, 11, 336. [Google Scholar] [CrossRef]
- Tesfaye, E.L.; Kumar, P.; Jutur, P.P.; Tefera, A.T.; Jiru, T.M.; Gaur, N.A. The Role of Amylase in Bioethanol Production: Advances in Amylase-Producing Strains Using CRISPR/Cas9 Technology. Fuel Commun. 2025, 23, 100142. [Google Scholar] [CrossRef]
- Zafar, M.G.; Mumtaz, A.; Akbar, A.; Liaqat, I.; Hassan, M.; Mahmood, M.; Mustafa, G.; Safi, S.Z.; Syed-Hassan, S.S.A.; Ansari, A.R.; et al. Distinctive role of enzymes in textile industry. In Enzymes in Textile Processing: A Climate Changes Mitigation Approach; Arshad, M., Ed.; SDGs and Textiles; Springer Nature: Singapore, 2025; pp. 1–17. ISBN 978-981-97-8057-0. [Google Scholar]
- Sinshaw, G.; Ayele, A.; Korsa, G.; Bekele, G.K.; Gemeda, M.T. Industrially Important Microbial Enzymes Production and Their Applications. In Microbial Enzymes; Chowdhary, P., Yadav, D., Anand, G., Gaur, R.K., Eds.; Wiley: Hoboken, NJ, USA, 2025; pp. 149–172. ISBN 978-3-527-35290-6. [Google Scholar]
- Km, D.; Tp, N.; Kt, R. Bacillus subtilis EMEA 22, potent mangrove rhizosphere amylase producer: Isolation and characterization. J. Microbiol. Biotechnol. Food Sci. 2025, 14, e10321. [Google Scholar] [CrossRef]
- Singh, J.; Kundu, D.; Das, M.; Banerjee, R. Enzymatic Processing of Juice From Fruits/Vegetables: An Emerging Trend and Cutting Edge Research in Food Biotechnology. In Enzymes in Food Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 419–432. ISBN 978-0-12-813280-7. [Google Scholar]
- Mahfudz, M.K.; Jaikhan, S.; Phirom-on, K.; Apiraksakorn, J. Cost-Effective Strategy and Feasibility for Amylase Production from Okara by Bacillus subtilis J12. Fermentation 2024, 10, 561. [Google Scholar] [CrossRef]
- Sahu, P.K.; Singh, R.; Shrivastava, M.; Darjee, S.; Mageshwaran, V.; Phurailtpam, L.; Rohatgi, B. Microbial Production of α-Amylase from Agro-Waste: An Approach towards Biorefinery and Bio-Economy. Energy Nexus 2024, 14, 100293. [Google Scholar] [CrossRef]
- Li, L.; Liu, X.; Bai, Y.; Yao, B.; Luo, H.; Tu, T. High-Throughput Screening Techniques for the Selection of Thermostable Enzymes. J. Agric. Food Chem. 2024, 72, 3833–3845. [Google Scholar] [CrossRef]
- Nazir, A.; Ijaz, M.; Rehman, H.M.; Sajjad, M. Rigidifying Flexible Sites: A Promising Strategy to Improve Thermostability of Lysophospholipase From Pyrococcus abyssi. Proteins 2025, 93, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.J.; Hazwani-Oslan, S.N.; Oslan, S.N. Purification and Characterisation of Thermostable α-Amylases from Microbial Sources. BioResources 2019, 15, 2005–2029. [Google Scholar] [CrossRef]
- Ashaolu, T.J.; Malik, T.; Soni, R.; Prieto, M.A.; Jafari, S.M. Extremophilic Microorganisms as a Source of Emerging Enzymes for the Food Industry: A Review. Food Sci. Nutr. 2025, 13, e4540. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Anbu, P.; Arshad, M.K.M.; Lakshmipriya, T.; Voon, C.H.; Hashim, U.; Chinni, S.V. Biotechnological Processes in Microbial Amylase Production. BioMed Res. Int. 2017, 2017, 1272193. [Google Scholar] [CrossRef]
- Soghomonyan, T.; Hambardzumyan, A.; Mkhitaryan, A.; Khoyetsyan, L.; Paronyan, M.; Izmailyan, M.; Kinosyan, M.; Bagiyan, V.; Ghochikyan, V.; Panosyan, H.; et al. Obtaining and Characterizing Thermostable α-Amylases Secreted by Bacillus subtilis, Originating from Bacillus amyloliquefaciens and Bacillus subtilis. Fermentation 2024, 10, 547. [Google Scholar] [CrossRef]
- Abd-Elhalim, B.T.; Gamal, R.F.; El-Sayed, S.M.; Abu-Hussien, S.H. Optimizing Alpha-Amylase from Bacillus amyloliquefaciens on Bread Waste for Effective Industrial Wastewater Treatment and Textile Desizing through Response Surface Methodology. Sci. Rep. 2023, 13, 19216. [Google Scholar] [CrossRef] [PubMed]
- Hashim, S.O. Starch-Modifying Enzymes. In Alkaliphiles in Biotechnology; Mamo, G., Mattiasson, B., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer International Publishing: Cham, Switzerland, 2019; Volume 172, pp. 221–244. ISBN 978-3-030-49735-4. [Google Scholar]
- Souza, P.M.D.; Magalhães, P.D.O.E. Application of Microbial α-Amylase in Industry—A Review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef]
- Mehta, D.; Satyanarayana, T. Bacterial and Archaeal α-Amylases: Diversity and Amelioration of the Desirable Characteristics for Industrial Applications. Front. Microbiol. 2016, 7, 1129. [Google Scholar] [CrossRef]
- Jiang, T.; Cai, M.; Huang, M.; He, H.; Lu, J.; Zhou, X.; Zhang, Y. Characterization of a Thermostable Raw-Starch Hydrolyzing α-Amylase from Deep-Sea Thermophile Geobacillus sp. Protein Expr. Purif. 2015, 114, 15–22. [Google Scholar] [CrossRef]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and Utilization of Thermophiles and Thermostable Enzymes in Biorefining. Microb. Cell Factories 2007, 6, 9. [Google Scholar] [CrossRef]
- Jaiswal, N.; Jaiswal, P. Thermostable α-Amylases and Laccases: Paving the Way for Sustainable Industrial Applications. Processes 2024, 12, 1341. [Google Scholar] [CrossRef]
- Mao, S.; Jiang, J.; Xiong, K.; Chen, Y.; Yao, Y.; Liu, L.; Liu, H.; Li, X. Enzyme Engineering: Performance Optimization, Novel Sources, and Applications in the Food Industry. Foods 2024, 13, 3846. [Google Scholar] [CrossRef]
- Whitman, W.B.; Rainey, F.A.; Kämpfer, P.; Trujillo, M.E.; DeVos, P.; Hedlund, B.; Dedysh, S. (Eds.) Bergey’s Manual of Systematics of Archaea and Bacteria, 1st ed.; Wiley: Hoboken, NJ, USA, 2015; ISBN 978-1-118-96060-8. [Google Scholar]
- Salvà-Serra, F.; Svensson-Stadler, L.; Busquets, A.; Jaén-Luchoro, D.; Karlsson, R.; Moore, E.R.B.; Gomila, M. A Protocol for Extraction and Purification of High-Quality and Quantity Bacterial DNA Applicable for Genome Sequencing: A Modified Version of the Marmur Procedure. Protoc. Exch. 2018. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Abele, M.; Soleymaniniya, A.; Bayer, F.P.; Lomp, N.; Doll, E.; Meng, C.; Neuhaus, K.; Scherer, S.; Wenning, M.; Wantia, N.; et al. Proteomic Diversity in Bacteria: Insights and Implications for Bacterial Identification. Mol. Cell. Proteom. 2025, 24, 100917. [Google Scholar] [CrossRef]
- Lasch, P.; Beyer, W.; Bosch, A.; Borriss, R.; Drevinek, M.; Dupke, S.; Ehling-Schulz, M.; Gao, X.; Grunow, R.; Jacob, D.; et al. A MALDI-ToF Mass Spectrometry Database for Identification and Classification of Highly Pathogenic Bacteria. Sci. Data 2025, 12, 187. [Google Scholar] [CrossRef]
- Tsuchida, S.; Tamura, M.; Hamaue, N.; Aoki, T. Screening of Recombinant Escherichia coli Using Activation of Green Fluorescent Protein as an Indicator. Biochem. Biophys. Res. Commun. 2014, 452, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Roche Molecular Biochemicals. Lab FAQs: Find a Quick Solution, 2nd ed.; Roche Applied Science: Mannheim, Germany, 2003; pp. 105–106. ISBN 3-88630-245-8. [Google Scholar]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The Protein Sequence Classification Resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. ISBN 978-1-58829-343-5. [Google Scholar]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, F.; Li, Y.; Yin, X.; Wang, Y.; Gao, C. Characterisation of Mutagenised Acid-Resistant Alpha-Amylase Expressed in Bacillus subtilis WB600. Appl. Microbiol. Biotechnol. 2008, 78, 85–94. [Google Scholar] [CrossRef]
- Radzlin, N.; Mohamad Ali, M.S.; Goh, K.M.; Yaakop, A.S.; Zakaria, I.I.; Kahar, U.M. Exploring a Novel GH13_5 α-Amylase from Jeotgalibacillus malaysiensis D5T for Raw Starch Hydrolysis. AMB Express 2024, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Farias, T.C.; Kawaguti, H.Y.; Bello Koblitz, M.G. Microbial Amylolytic Enzymes in Foods: Technological Importance of the Bacillus Genus. Biocatal. Agric. Biotechnol. 2021, 35, 102054. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Lawson, D.M.; Turkenburg, J.P.; Bisgaard-Frantzen, H.; Svendsen, A.; Borchert, T.V.; Dauter, Z.; Wilson, K.S.; Davies, G.J. Structural Analysis of a Chimeric Bacterial α-Amylase. High-Resolution Analysis of Native and Ligand Complexes, Biochemistry 2000, 39, 9099–9107. [Google Scholar] [CrossRef]
- Aras, S.; Altıntas, R.; Aygun, E.; Goren, G.; Sisecioglu, M. Purification of α-Amylase from Thermophilic Bacillus Licheniformis SO5 by Using a Novel Method, Alternating Current Magnetic-Field Assisted Three-Phase Partitioning: Molecular Docking and Bread Quality Improvement. Food Chem. 2025, 484, 144258. [Google Scholar] [CrossRef]
- Yang, T.; Hu, Q.; Liu, Y.; Xu, R.; Wang, D.; Chang, Z.; Jin, M.; Huang, J. Biochemical Characteristics and Potential Application of a Thermostable Starch Branching Enzyme from Bacillus licheniformis. AMB Express 2023, 13, 8. [Google Scholar] [CrossRef]
- Suthar, S.; Joshi, D.; Patel, H.; Patel, D.; Kikani, B.A. Optimization and Purification of a Novel Calcium-Independent Thermostable, α-Amylase Produced by Bacillus licheniformis UDS-5. World J. Microbiol. Biotechnol. 2024, 40, 385. [Google Scholar] [CrossRef]
- Yadav, J.K. A Differential Behavior of α-Amylase, in Terms of Catalytic Activity and Thermal Stability, in Response to Higher Concentration CaCl2. Int. J. Biol. Macromol. 2012, 51, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.K.; Sharma, A.; Soni, R. Microbial Enzyme Systems in the Production of Second Generation Bioethanol. Sustainability 2023, 15, 3590. [Google Scholar] [CrossRef]
- Zhai, W.; Zhu, M.; Lin, L.; Wei, W.; Wei, D. Computer-Aided Protein Surface Modification Strategy to Improve the Thermostability of α-Amylase. Starch Stärke 2025, 77, 2300288. [Google Scholar] [CrossRef]
- Liu, J.; Han, L.; Li, J.; Du, G.; Zhang, G. Modification of Flexible Regions for Enhanced Thermal Stability of Alkaline Amylase. J. Agric. Food Chem. 2025, 73, 9973–9982. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kholikov, A.; Vokhidov, K.; Murtozoyev, A.; Tóth, Z.S.; Nagy, G.N.; Vértessy, B.G.; Makhsumkhanov, A. Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications. Microorganisms 2025, 13, 1757. https://doi.org/10.3390/microorganisms13081757
Kholikov A, Vokhidov K, Murtozoyev A, Tóth ZS, Nagy GN, Vértessy BG, Makhsumkhanov A. Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications. Microorganisms. 2025; 13(8):1757. https://doi.org/10.3390/microorganisms13081757
Chicago/Turabian StyleKholikov, Askar, Khushnut Vokhidov, Azizjon Murtozoyev, Zoé S. Tóth, Gergely N. Nagy, Beáta G. Vértessy, and Akhmadzhan Makhsumkhanov. 2025. "Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications" Microorganisms 13, no. 8: 1757. https://doi.org/10.3390/microorganisms13081757
APA StyleKholikov, A., Vokhidov, K., Murtozoyev, A., Tóth, Z. S., Nagy, G. N., Vértessy, B. G., & Makhsumkhanov, A. (2025). Characterization of a Thermostable α-Amylase from Bacillus licheniformis 104.K for Industrial Applications. Microorganisms, 13(8), 1757. https://doi.org/10.3390/microorganisms13081757






