Molecular Epidemiology of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Guizhou Angus Calves: Dominance of Angus Cattle-Adapted Genotypes and Zoonotic Potential of E. bieneusi
Abstract
1. Introduction
2. Materials and Methods
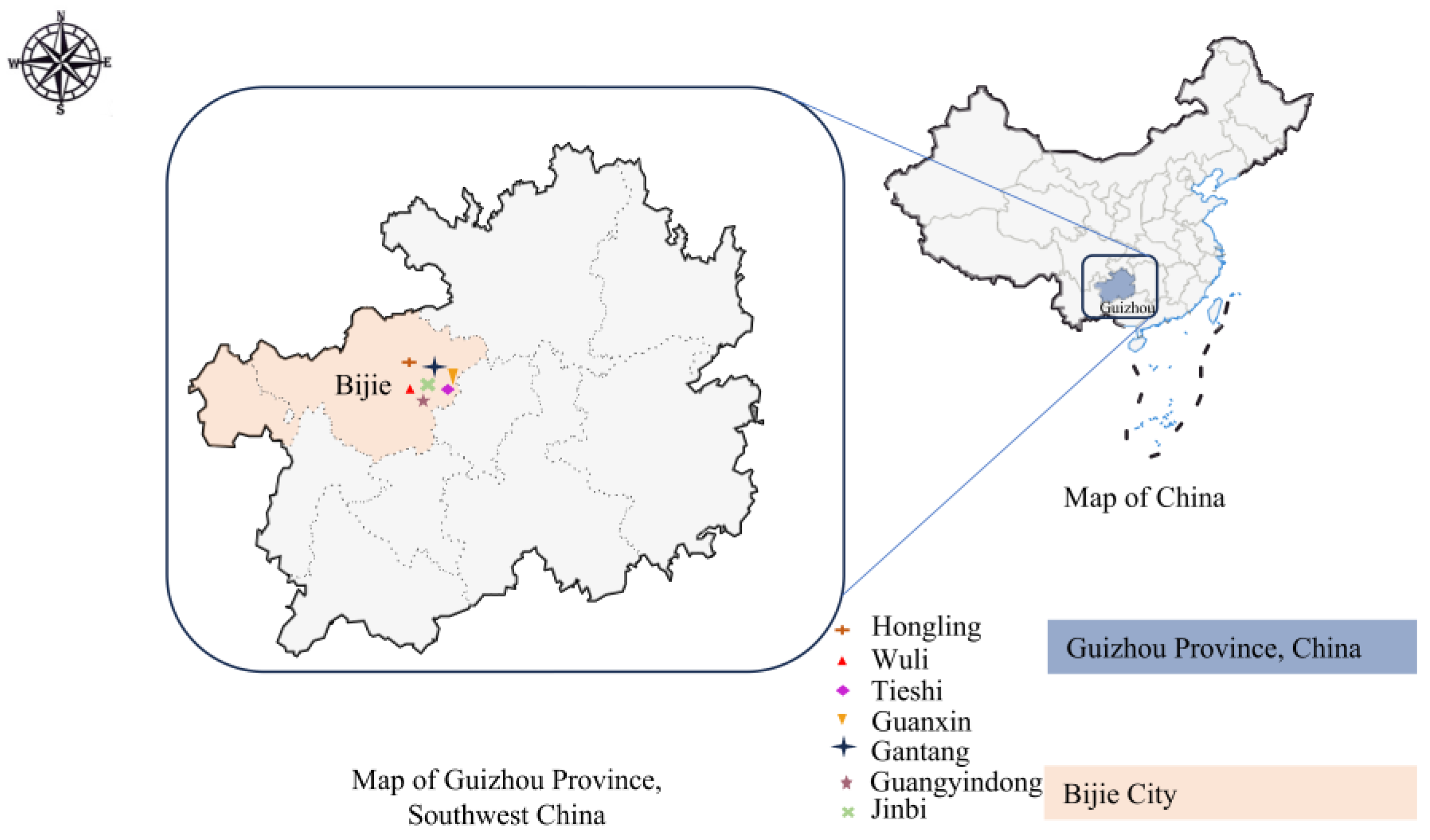
2.1. Sample Collection
2.2. DNA Extraction and PCR Analysis
2.3. Sequence Analysis
2.4. Statistical Analysis
3. Results
3.1. Cryptosporidium bovis Dominance: 89.6% Prevalence with Geographic Clustering
3.2. Giardia duodenalis Assemblage E Prevails: Low Zoonotic Risk but High Farm-Level Variability
3.3. Enterocytozoon bieneusi Zoonotypes: BEB4/I Co-Circulation and Diarrhea Association
4. Discussion
4.1. Why Guizhou’s Infection Rates Surpass Sichuan and Yunnan: A Climate Lens?
4.2. From Animal-Adapted to Zoonotic: Evolutionary Risks of G. duodenalis Assemblage E
4.3. BEB4/I Co-Prevalence: Waterborne Transmission in Karst Hydrology
4.4. Implications for Sustainable Disease Control and Prevention
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guérin, A.; Striepen, B. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host Microbe 2020, 28, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Rojas-López, L.; Marques, R.C.; Svärd, S.G. Giardia duodenalis. Trends Parasitol. 2022, 38, 605–606. [Google Scholar] [CrossRef] [PubMed]
- Nourrisson, C.; Lavergne, R.A.; Moniot, M.; Morio, F.; Poirier, P. Enterocytozoon bieneusi, a human pathogen. Emerg. Microbes Infect. 2024, 13, 2406276. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Fayer, R.; Ryan, U.; Upton, S.J. Cryptosporidium taxonomy: Recent advances and implications for public health. Clin. Microbiol. Rev. 2004, 17, 72–97. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ryan, U.; Feng, Y.; Xiao, L. Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol. 2022, 38, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.; Wieckowski, P.; Matechou, E.; Katzer, F.; Tsaousis, A.D.; Farré, M. Global prevalence of Cryptosporidium infections in cattle: A meta-analysis. Curr. Res. Parasitol. Vector Borne Dis. 2025, 7, 100264. [Google Scholar] [CrossRef] [PubMed]
- Einarsson, E.; Ma’ayeh, S.; Svärd, S.G. An up-date on Giardia and giardiasis. Curr. Opin. Microbiol. 2016, 34, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Hijjawi, N.; Feng, Y.; Xiao, L. Giardia: An under-reported foodborne parasite. Int. J. Parasitol. 2019, 49, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, J.; Qin, H.; Wang, L.; Li, J.; Zhang, L. Cryptosporidium parvum and gp60 genotype prevalence in dairy calves worldwide: A systematic review and meta-analysis. Acta Trop. 2023, 240, 106843. [Google Scholar] [CrossRef] [PubMed]
- de Aquino, M.C.C.; Inácio, S.V.; Rodrigues, F.S.; de Barros, L.D.; Garcia, J.L.; Headley, S.A.; Gomes, J.F.; Bresciani, K.D.S. Cryptosporidiosis and giardiasis in buffaloes (Bubalus bubalis). Front. Vet. Sci. 2020, 7, 557967. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.M.; Feng, Y.; Fayer, R.; Xiao, L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia—A 50 year perspective (1971–2021). Int. J. Parasitol. 2021, 51, 1099–1119. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, H.; Sun, Y.; Zhang, L.; Jian, F.; Qi, M.; Ning, C.; Xiao, L. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J. Clin. Microbiol. 2011, 49, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Wang, R.J.; Ren, G.J.; Yu, Z.Q.; Zhang, L.X.; Zhang, S.Y.; Lu, H.; Peng, X.Q.; Zhao, G.H. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitol. Res. 2016, 115, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, H.; Zhang, Z.; Li, J.; Wang, C.; Zhao, J.; Hu, S.; Wang, R.; Zhang, L.; Wang, M. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 2016, 219, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cai, M.; Jiang, W.; Wang, Y.; Jin, Y.; Li, N.; Guo, Y.; Feng, Y.; Xiao, L. High genetic diversity of Giardia duodenalis assemblage E in pre-weaned dairy calves in Shanghai, China, revealed by multilocus genotyping. Parasitol. Res. 2017, 116, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, T.; Koehler, A.V.; Hu, M.; Gasser, R.B. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasit. Vectors 2017, 10, 519. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, Z.; Yan, F.; Zhang, Z.; Zhang, G.; Zhang, L.; Jian, F.; Zhang, S.; Ning, C.; Wang, R. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet. Parasitol. 2017, 248, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, G.; Li, X.; Zhang, X.; Karanis, G.; Jian, Y.; Ma, L.; Karanis, P. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in 1–2-month-old highland yaks in Qinghai Province, China. Parasitol. Res. 2018, 117, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Dan, J.; Yan, G.; Tu, R.; Tian, Y.; Cao, S.; Shen, L.; Deng, J.; Yu, S.; Geng, Y.; et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 2018, 25, 45. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gong, X.; Zhu, K.; Li, N.; Yu, Z.; Guo, Y.; Weng, Y.; Kváč, M.; Feng, Y.; Xiao, L. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit. Vectors 2019, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, N.; Jiang, W.; Guo, Y.; Wang, X.; Jin, Y.; Feng, Y.; Xiao, L. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol. Res. 2019, 118, 3053–3060. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chang, Y.; Zhang, X.; Chen, Y.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Jian, F.; et al. Molecular characterization and distribution of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi from yaks in Tibet, China. BMC Vet. Res. 2019, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 62. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, W.; Dong, Z.; Cheng, F.; Zhou, X.; Li, B.; Zhang, J. Detection of Infectious agents causing neonatal calf diarrhea on two large dairy farms in Yangxin County, Shandong Province, China. Front. Vet. Sci. 2020, 7, 589126. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.X.; Zou, Y.; Li, T.S.; Chen, H.; Wang, S.S.; Cao, F.Q.; Yang, J.F.; Sun, X.L.; Zhu, X.Q.; Zou, F.C. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in Yunling cattle in Yunnan Province, southwestern China. Microb. Pathog. 2021, 158, 105025. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, L.; Li, W.; Li, C.; Gu, Y. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J. Vet. Med. Sci. 2022, 84, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, Z.S.; Han, W.X.; Yang, B.; Chai, H.L.; Wang, M.Y.; Wang, Y.; Zhang, S.; Zhao, W.H.; Ma, Y.M.; et al. Prevalence and molecular characterization of Giardia duodenalis in dairy cattle in Central Inner Mongolia, Northern China. Sci. Rep. 2023, 13, 13960. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Yang, B.; Huang, M.; Qi, M.; Xu, C.; Jing, B.; Zhang, Z. Molecular detection and genetic characteristics of Giardia duodenalis in dairy cattle from large-scale breeding farms in Xinjiang, China. Parasitol. Res. 2024, 123, 106. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Y.; Wang, M.; Zhang, S.; Wang, L.; Zhang, Z.; Chai, H.; Yi, C.; Fan, W.; Liu, Y. First report of Giardia duodenalis in dairy cattle and beef cattle in Shanxi, China. Mol. Biol. Rep. 2024, 51, 403. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Liu, A.; Li, H.; Zhao, Y.; Yao, L.; Yang, B.; Zhang, W.; Yang, F. Molecular characterization and zoonotic potential of Cryptosporidium spp. and Giardia duodenalis in humans and domestic animals in Heilongjiang Province, China. Parasit. Vectors 2024, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhang, S.; Zhang, Z.S.; Qian, X.Y.; Chai, H.L.; Wang, Y.; Fan, W.J.; Yi, C.; Ding, Y.L.; Han, W.X.; et al. Prevalence and molecular characterization of Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. Vet. Res. Commun. 2024, 48, 2629–2643. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Alderisio, K.; Limor, J.; Royer, M.; Lal, A.A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 2000, 66, 5492–5498. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wan, M.; Yang, F.; Li, N.; Xiao, L.; Feng, Y.; Guo, Y. Development and application of a gp60-based subtyping tool for Cryptosporidium bovis. Microorganisms 2021, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Beck, R.; Lalle, M.; Marinculic, A.; Pozio, E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int. J. Parasitol. 2008, 38, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.M.; Fayer, R.; Lal, A.A.; Trout, J.M.; Schaefer, F.W., 3rd; Xiao, L. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003, 69, 4495–4501. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhao, G.; Gong, Y.; Zhang, L. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 Years. Front. Microbiol. 2017, 8, 1823. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, N.Z.; Gong, Q.L.; Zhao, Q.; Zhang, X.X. Prevalence of Cryptosporidium in dairy cattle in China during 2008-2018: A systematic review and meta-analysis. Microb. Pathog. 2019, 132, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Cao, X.F.; Deng, L.; Li, W.; Huang, X.M.; Lan, J.C.; Xiao, Q.C.; Zhong, Z.J.; Feng, F.; Zhang, Y.; et al. Epidemiology of Cryptosporidium infection in cattle in China: A review. Parasite 2017, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.W.; Shu, F.F.; Pu, L.H.; Zou, Y.; Yang, J.F.; Zou, F.C.; Zhu, X.Q.; Li, Z.; He, J.J. Occurrence and molecular characterization of Cryptosporidium spp. in dairy cattle and dairy buffalo in Yunnan Province, Southwest China. Animals 2022, 12, 1031. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yue, D.; Qi, M.; Wang, R.; Zhao, J.; Li, J.; Shi, K.; Wang, M.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 2014, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Zhang, X.; Li, X.; Karanis, G.; Ma, L.; Karanis, P. Prevalence and molecular characterization of Giardia duodenalis in cattle and sheep from the Qinghai-Tibetan Plateau Area (QTPA), northwestern China. Vet. Parasitol. 2018, 250, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Chen, C.; Qin, Y.F.; Yang, X.B.; Li, M.H.; Meng, X.Z.; Zhao, Z.Y.; Ma, N.; Cai, Y.; Zhang, Y.; et al. Prevalence and related factors of Enterocytozoon bieneusi in cattle: A global systematic review and meta-analysis. Prev. Vet. Med. 2022, 208, 105775. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Wang, K.S.; Yang, J.F.; Mao, H.M.; Pu, L.H.; Zou, Y.; Ma, J.; Zhu, X.Q.; Zou, F.C.; He, J.J. Prevalence and novel genotypes identification of Enterocytozoon bieneusi in dairy cattle in Yunnan Province, China. Animals 2021, 11, 3014. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, W. Characteristics of an underground stope channel supplied by atmospheric precipitation and its water disaster prevention in the karst mining areas of Guizhou. Sci. Rep. 2023, 13, 15892. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Liu, G.; Xiang, Q.; Liu, Y. Spatio-temporal variation characteristics of stable isotopes of tap water and its potential as a proxy for surface water in Sichuan, China. Sci. Total Environ. 2024, 912, 168755. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Shang, C.; Yang, K.; Luo, Y. Dynamic response of surface water temperature in urban lakes under different climate scenarios-a case study in Dianchi lake, China. Int. J. Environ. Res. Public Health 2022, 19, 12142. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, H.; Jing, B.; Wang, R.; Jian, F.; Ning, C.; Zhang, L. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, Northwestern China. Parasit. Vectors 2016, 9, 546. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, M.; Zhang, K.; Li, J.; Huang, J.; Ning, C.; Zhang, L. Prevalence and genotyping of Giardia duodenalis isolated from sheep in Henan Province, central China. Infect. Genet. Evol. 2016, 39, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Dan, J.; Zhang, X.; Ren, Z.; Wang, L.; Cao, S.; Shen, L.; Deng, J.; Zuo, Z.; Yu, S.; Wang, Y.; et al. Occurrence and multilocus genotyping of Giardia duodenalis from post-weaned dairy calves in Sichuan province, China. PLoS ONE 2019, 14, e0224627. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Fei, J.; Cai, J.; Wang, X.; Li, N.; Guo, Y.; Feng, Y.; Xiao, L. Multilocus genotyping of Giardia duodenalis in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 2017, 247, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zou, Y.; Zhang, X.L.; Wang, P.; Chen, X.Q.; Zhu, X.Q. Prevalence and multilocus genotyping of Giardia lamblia in cattle in Jiangxi Province, China: Novel assemblage E subtypes identified. Korean J. Parasitol. 2020, 58, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Heng, Z.J.; Yang, J.F.; Xie, X.Y.; Xu, C.R.; Chen, J.R.; Ma, J.; He, J.J.; Mao, H.M. Prevalence and multilocus genotyping of Giardia duodenalis in Holstein cattle in Yunnan, China. Front. Vet. Sci. 2022, 9, 949462. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011, 24, 110–140. [Google Scholar] [CrossRef] [PubMed]
- Adam, R.D. Biology of Giardia lamblia. Clin. Microbiol. Rev. 2001, 14, 447–475. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Qin, R.L.; Mei, J.J.; Zou, Y.; Zhang, Z.H.; Zheng, W.B.; Liu, Q.; Zhu, X.Q.; Gao, W.W.; Xie, S.C. Molecular detection and genotyping of Enterocytozoon bieneusi in beef cattle in Shanxi Province, North China. Animals 2022, 12, 2961. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Jing, B.; Jian, F.; Wang, R.; Zhang, S.; Wang, H.; Ning, C.; Zhang, L. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, Northwest China. Parasitol. Int. 2017, 66, 960–963. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Qi, M.; Sun, M.F.; Li, D.F.; Wang, R.J.; Zhang, S.M.; Zhao, J.F.; Li, J.Q.; Cui, Z.H.; Chen, Y.C.; et al. Prevalence and population genetics analysis of Enterocytozoon bieneusi in dairy cattle in China. Front. Microbiol. 2019, 10, 1399. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhao, Z.; Zhao, J.; Fu, Y.; Lang, J.; Zhang, J.; Liang, G.; Zhang, L.; Li, J.; Zhao, G. Molecular characterization and zoonotic potential of Enterocytozoon bieneusi in ruminants in northwest China. Acta Trop. 2022, 234, 106622. [Google Scholar] [CrossRef] [PubMed]
- Speich, B.; Marti, H.; Ame, S.M.; Ali, S.M.; Bogoch, I.I.; Utzinger, J.; Albonico, M.; Keiser, J. Prevalence of intestinal protozoa infection among school-aged children on Pemba Island, Tanzania, and effect of single-dose albendazole, nitazoxanide and albendazole-nitazoxanide. Parasit. Vectors 2013, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Wanke, C.A.; DeGirolami, P.; Federman, M. Enterocytozoon bieneusi infection and diarrheal disease in patients who were not infected with human immunodeficiency virus: Case report and review. Clin. Infect. Dis. 1996, 23, 816–818. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Chen, Y.; Ndegwa, P. Anaerobic digestion process deactivates major pathogens in biowaste: A meta-analysis. Renew. Sust. Energy Rev. 2022, 153, 111752. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, Q.; Gao, S.; Zhang, X.; Wang, Z.; Wu, P.; Zeng, J. Coupled controls of the infiltration of rivers, urban activities and carbonate on trace elements in a karst groundwater system from Guiyang, Southwest China. Ecotoxicol. Environ. Saf. 2023, 249, 114424. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Toze, S.; Veal, C.; Fisher, P.; Zhang, Q.; Zhu, Z.; Staley, C.; Sadowsky, M.J. Comparative decay of culturable faecal indicator bacteria, microbial source tracking marker genes, and enteric pathogens in laboratory microcosms that mimic a sub-tropical environment. Sci. Total Environ. 2021, 751, 141475. [Google Scholar] [CrossRef] [PubMed]
- Betancourt, W.Q.; Rose, J.B. Drinking water treatment processes for removal of Cryptosporidium and Giardia. Vet. Parasitol. 2004, 126, 219–234. [Google Scholar] [CrossRef] [PubMed]
| Targets | Primer Names | Sequence (5′-3′) | Annealing Temperature |
|---|---|---|---|
| 18rRNA [33] | 18S-F1 | TTCTAGAGCTAATACATGCG | 55 °C |
| 18S-R1 | CCCATTTCCTTCGAAACAGGA | ||
| 18S-F2 | GGAAGGGTTGTATTTATTAGATAAAG | ||
| 18S-R2 | CTCATAAGGTGCTGAAGGAGTA | ||
| gp60 [34] | Bovis-gp60-F1 | ATGCGACTTACGCTCTACATTACTCT | |
| Bovis-gp60-R1 | GACAAAATGAAGGCTGAGATAGATGGGA | ||
| Bovis-gp60-F2 | CCTCTCGGCATTTATTGCCCT | ||
| Bovis-gp60-R2 | ATACCTAAGGCCAAATGCTGATGAA | ||
| bg [35] | bg-F1 | AAGCCCGACGACCTCACCCGCAGTGC | 65 °C |
| bg-R1 | GAGGCCGCCCTGGATCTTCGAGACGAC | ||
| bg-F2 | GAACGAGATCGAGGTCCG | 55 °C | |
| bg-R2 | CTCGACGAGCTTCGTGTT | ||
| gdh [35] | gdh-F1 | TTCCGTGTCCAGTACAACTC | 50 °C |
| gdh-R1 | GCCAGCTTCTCCTCGTTGAA | ||
| gdh-F2 | CGCTTCCACCCCTCTGTCAAT | ||
| gdh-R2 | TGTTGTCCTTGCACATCTC | ||
| tpi [35] | tpi-F1 | AATAAATIATGCCTGCTCGTCG | 54 °C |
| tpi-R1 | ATGGACITCCTCTGCCTGCTC | ||
| tpi-F2 | CCCTTCATCGGIGGTAACTTCAA | 58 °C | |
| tpi-R2 | GTGGCCACCACICCCGTGCC | ||
| ITS [36] | Eb-ITS-F1 | GATGGTCATAGGGATGAAGAGCTT | 55 °C |
| Eb-ITS-R1 | TATGCTTAAGTCCAGGGAG | ||
| Eb-ITS-F2 | AGGGATGAAGAGCTTCGGCTCTG | ||
| Eb-ITS-R2 | AGTGATCCTGTATTAGGGATATT |
| Factors | No. Tested | No. Positive | p Value | Species (n) |
|---|---|---|---|---|
| (%, 95% CI) | ||||
| Location | ||||
| Hongling | 174 | 21 (12.1, 7.7–17.9) | <0.001 | C. bovis (19), C. ryanae (1), C. bovis & C. ryanae (1) |
| Wuli | 168 | 47 (27.9, 21.3–35.3) | C. bovis (42), C. ryanae (2), C. andersoni (3) | |
| Tieshi | 102 | 34 (33.3, 24.3–43.3) | C. bovis (28), C. ryanae (6) | |
| Guanxin | 93 | 18 (19.4, 11.9–28.9) | C. bovis (18) | |
| Gantang | 100 | 26 (26.0, 17.7–35.7) | C. bovis (23), C. ryanae (2), C. andersoni (1) | |
| Guangyindong | 97 | 28 (28.9, 20.1–39.0) | C. bovis (24), C. ryanae (4) | |
| Jinbi | 83 | 18 (21.7, 13.4–32.1) | C. bovis (18) | |
| Season | ||||
| spring | 663 | 155 (23.4, 20.2–26.8) | 0.963 | C. bovis (141), C. ryanae (12), C. andersoni (1), C. bovis & C. ryanae (1) |
| summer | 154 | 37 (24.0, 17.5–31.6) | C. bovis (31), C. ryanae (3), C. andersoni (3) | |
| Age (week) | ||||
| 0–2 | 107 | 2 (4.7, 1.5–10.6) | <0.01 | C. bovis (2) |
| 2–4 | 200 | 43 (21.5, 16.0–27.8) | C. bovis (41), C. ryanae (2) | |
| 4–8 | 479 | 143 (29.9, 25.8–34.1) | C. bovis (126), C. ryanae (13), C. andersoni (3), C. bovis & C. ryanae (1) | |
| 8–12 | 31 | 4 (12.9, 3.6–29.8) | C. bovis (3), C. andersoni (1) | |
| Diarrhea | ||||
| Yes | 93 | 21 (22.6, 14.6–32.4) | 0.817 | C. bovis (18), C. andersoni (3) |
| No | 724 | 171 (23.6, 20.6–26.9) | C. bovis (154), C. ryanae (12), C. andersoni (4), C. bovis & C. ryanae (1) | |
| Gender | ||||
| Male | 178 | 32 (17.9, 12.6–24.4) | 0.032 | C. bovis (26), C. ryanae (4), C. andersoni (2) |
| Female | 639 | 160 (25.0, 21.7–28.6) | C. bovis (146), C. ryanae (11), C. andersoni (2), C. bovis & C. ryanae (1) | |
| Total | 817 | 192 (23.5, 20.6–26.6) | C. bovis (172), C. ryanae (15), C. andersoni (4), C. bovis & C. ryanae (1) | |
| Farm | Subtypes (n) |
|---|---|
| Hongling | XXVIf (18)\XXVIe (1) |
| Wuli | XXVIb (13)\XXVIc (5)\XXVId (11)\XXVIe (12)\mixed infection(XXVIb and XXVIc) (1) |
| Tieshi | XXVIe (23)\XXVIf (5) |
| Guanxin | XXVIe (13)\XXVIf (5) |
| Gantang | XXVIa (1)\XXVId (21)\XXVIf (2) |
| Guangyindong | XXVIa (12)\XXVId (10)\XXVIe (1)\XXVIf (1) |
| Jinbi | XXVIb (1)\XXVIc (17) |
| Factors | No. Tested | No. Positive | p Value | Assemblages (n) | ||
|---|---|---|---|---|---|---|
| (%, 95% CI) | ||||||
| Location | bg gene (n) | gdh gene (n) | tpi gene (n) | |||
| Hongling | 174 | 33 (18.9, 13.4–25.6) | <0.001 | A (2), E (22) | E (18) | A (2), E (15) |
| Wuli | 168 | 44 (26.2, 19.7–33.5) | A (2), E (34) | A (3), E (23) | A (1), E (18) | |
| Tieshi | 102 | 5 (4.9, 1.6–11.1) | E (5) | E (4) | E (3) | |
| Guanxin | 93 | 37 (39.8, 29.8–50.5) | A (1), E (30) | A (1), E (23) | E (21) | |
| Gantang | 100 | 55 (55.0, 44.7–65.0) | E (42) | E (34) | E (18) | |
| Guangyindong | 97 | 50 (51.6, 41.2–61.8) | E (37) | E (26) | E (16) | |
| Jinbi | 83 | 32 (38.5, 28.1–49.9) | E (23) | E (12) | E (11) | |
| Season | ||||||
| spring | 663 | 224 (33.8, 30.2–37.5) | 0.569 | E (219), A (5) | ||
| summer | 154 | 32 (20.8, 14.7–28.0) | E (31), A (1) | |||
| Age (week) | Assemblage | |||||
| 0–2 | 107 | 19 (17.7, 11.0–26.3) | <0.001 | E (19) | ||
| 2–4 | 200 | 49 (24.5, 18.7–31.1) | E (48), A (1) | |||
| 4–8 | 479 | 178 (37.2, 32.8–41.7) | E (173), A (5) | |||
| 8–12 | 31 | 10 (32.2, 16.7–51.4) | E (10) | |||
| Diarrhea | Assemblage | |||||
| Yes | 93 | 23 (24.7, 16.3–34.8) | 0.166 | E (23) | ||
| No | 724 | 233 (32.2, 28.8–35.7) | E (227), A (6) | |||
| Gender | Assemblage | |||||
| Male | 178 | 53 (29.8, 23.2–37.1) | 0.629 | E (52), A (1) | ||
| Female | 639 | 203 (31.8, 28.2–35.5) | E (198), A (5) | |||
| Total | 817 | 256 (31.3, 28.1–34.6) | E (250), A (6) | |||
| Factors | No. Tested | No. Positive | p Value | Genotypes (n) |
|---|---|---|---|---|
| (%, 95% CI) | ||||
| Location | ||||
| Hongling | 174 | 0 (0.0, 0.0–2.1) | <0.001 | - |
| Wuli | 168 | 31 (18.5, 12.9–25.2) | I (11), BEB4 (7), J (13) | |
| Tieshi | 102 | 24 (23.5, 15.7–32.9) | BEB4 (24) | |
| Guanxin | 93 | 0 (0.0, 0.0–3.9) | - | |
| Gantang | 100 | 39 (39.0, 29.4–49.3) | I (27), BEB4 (9), CHPM1 (3) | |
| Guangyindong | 97 | 38 (39.2, 29.4–49.6) | I (13), BEB4 (25) | |
| Jinbi | 83 | 29 (34.9, 24.8–46.2) | I (14), BEB4 (15) | |
| Season | ||||
| spring | 663 | 142 (21.4, 18.4–24.7) | 0.843 | I (59), BEB4 (80), CHPM1 (3) |
| summer | 154 | 19 (12.0, 7.6–18.6) | I (6), J (13) | |
| Age (week) | ||||
| 0–2 | 107 | 7 (6.5, 2.7–13.0) | <0.001 | I (5), BEB4 (2) |
| 2–4 | 200 | 25 (12.5, 8.3–17.9) | I (11), BEB4 (12), CHPM1 (1), J (1) | |
| 4–8 | 479 | 125 (26.1, 22.2–30.3) | I (48), BEB4 (64), CHPM1 (2), J (11) | |
| 8–12 | 31 | 4 (12.9, 3.6–29.8) | I (1), BEB4 (2), J (1) | |
| Diarrhea | ||||
| Yes | 93 | 33 (35.5, 25.8–46.1) | <0.01 | I (13), BEB4 (20) |
| No | 724 | 128 (17.7, 15.0–20.7) | I (52), BEB4 (60), CHPM1 (3), J (13) | |
| Gender | ||||
| Male | 178 | 22 (12.4, 7.9–18.1) | 0.065 | I (8), BEB4 (11), J (3) |
| Female | 639 | 139 (21.8, 18.6–25.2) | I (57), BEB4 (69), CHPM1 (3), J (10) | |
| Total | 817 | 161 (19.7, 17.0–22.6) | I (65), BEB4 (80), CHPM1 (3), J (13) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, P.; Tao, Z.; Shi, K.; Zhao, J.; Huang, B.; Liu, H.; Wang, C.; Yin, J.; Zhu, G.; Cacciò, S.M.; et al. Molecular Epidemiology of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Guizhou Angus Calves: Dominance of Angus Cattle-Adapted Genotypes and Zoonotic Potential of E. bieneusi. Microorganisms 2025, 13, 1735. https://doi.org/10.3390/microorganisms13081735
Qin P, Tao Z, Shi K, Zhao J, Huang B, Liu H, Wang C, Yin J, Zhu G, Cacciò SM, et al. Molecular Epidemiology of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Guizhou Angus Calves: Dominance of Angus Cattle-Adapted Genotypes and Zoonotic Potential of E. bieneusi. Microorganisms. 2025; 13(8):1735. https://doi.org/10.3390/microorganisms13081735
Chicago/Turabian StyleQin, Peixi, Zhuolin Tao, Kaizhi Shi, Jiaxian Zhao, Bingyan Huang, Hui Liu, Chunqun Wang, Jigang Yin, Guan Zhu, Simone M. Cacciò, and et al. 2025. "Molecular Epidemiology of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Guizhou Angus Calves: Dominance of Angus Cattle-Adapted Genotypes and Zoonotic Potential of E. bieneusi" Microorganisms 13, no. 8: 1735. https://doi.org/10.3390/microorganisms13081735
APA StyleQin, P., Tao, Z., Shi, K., Zhao, J., Huang, B., Liu, H., Wang, C., Yin, J., Zhu, G., Cacciò, S. M., & Hu, M. (2025). Molecular Epidemiology of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Guizhou Angus Calves: Dominance of Angus Cattle-Adapted Genotypes and Zoonotic Potential of E. bieneusi. Microorganisms, 13(8), 1735. https://doi.org/10.3390/microorganisms13081735






