Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
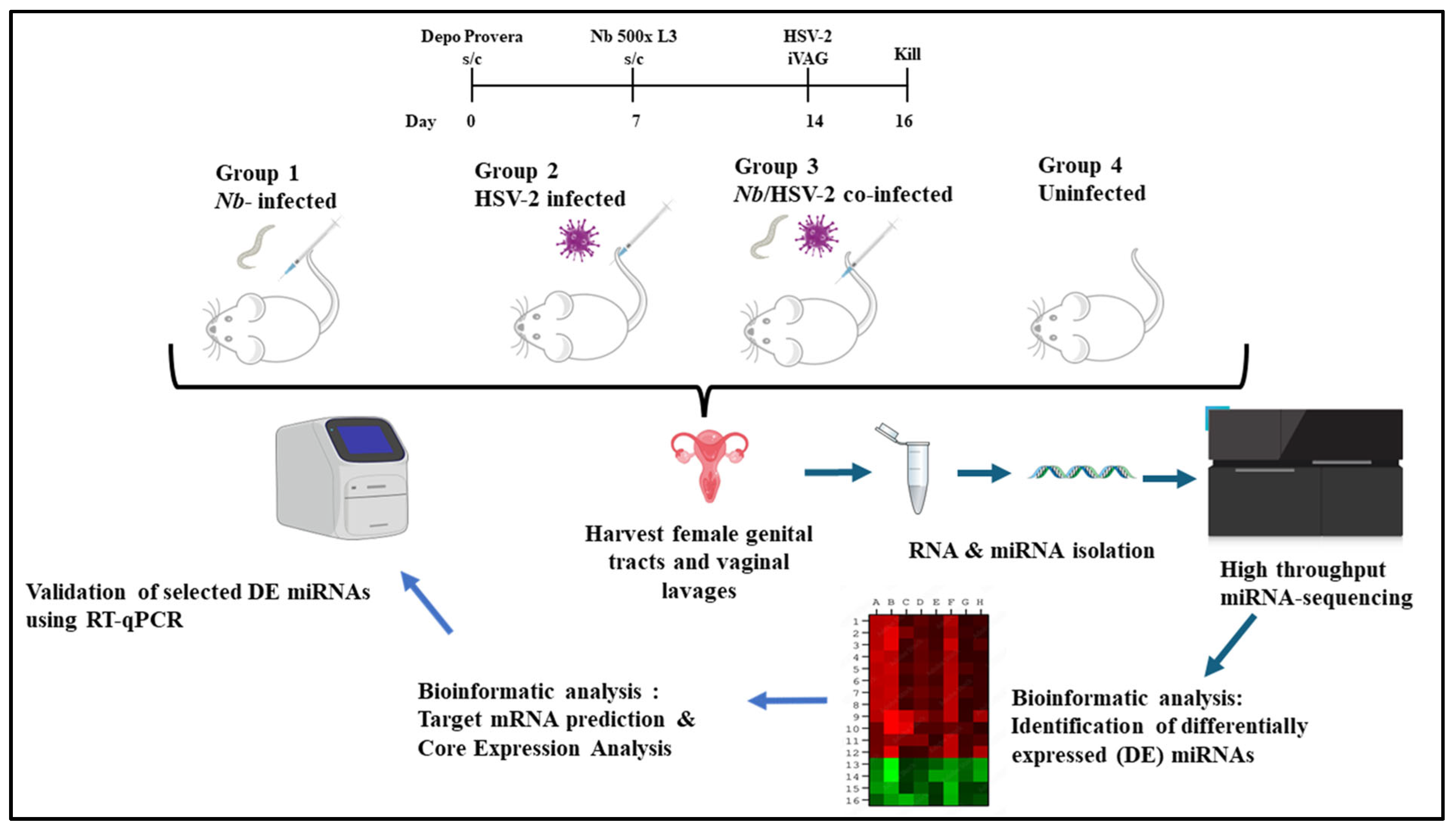
2.2. Experimental Models
2.2.1. Animals
2.2.2. Nb Maintenance and Infection
2.2.3. Virus
2.3. Total RNA Extraction and Quality Control
2.4. Library Preparation and Sequencing
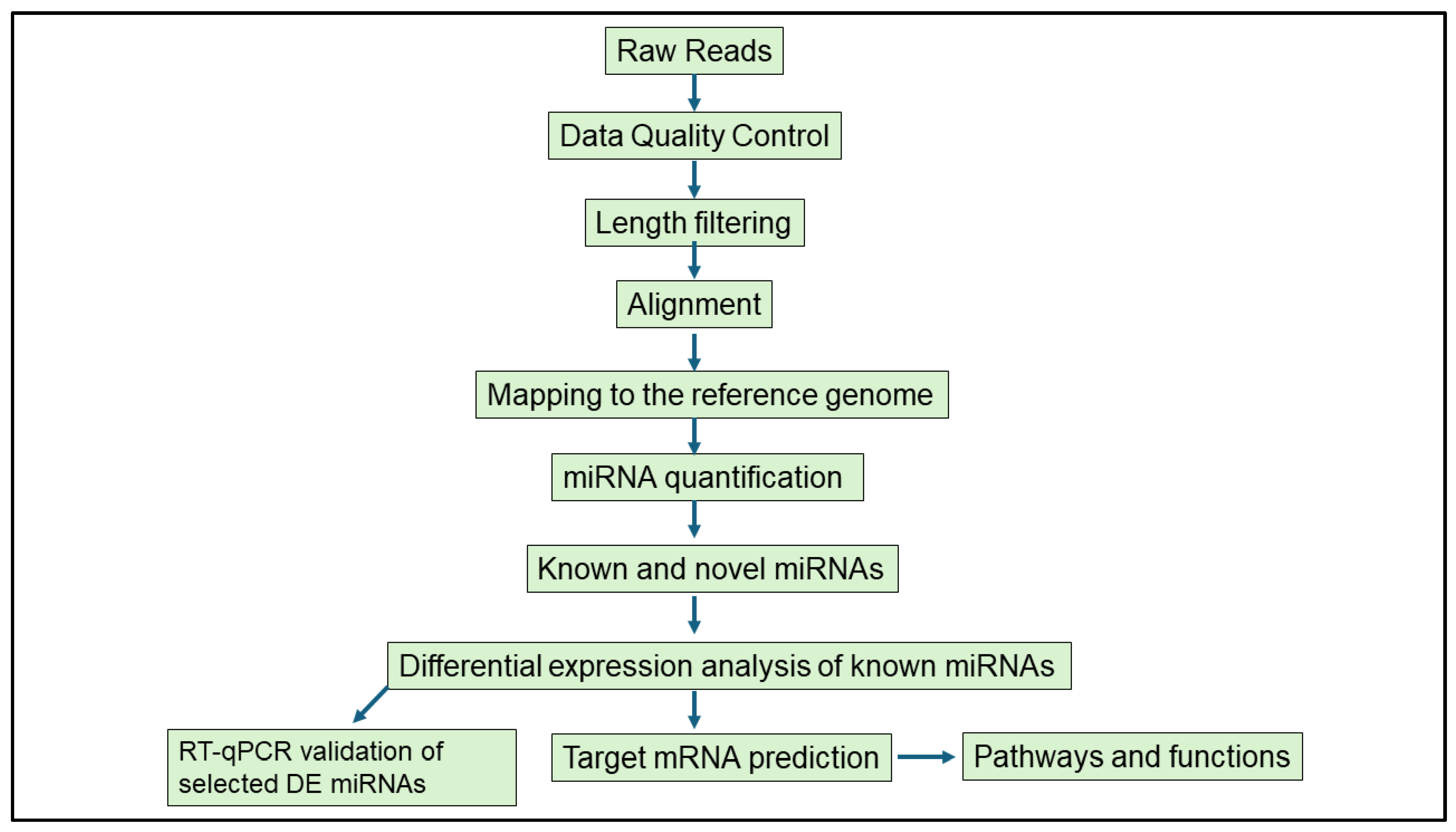
2.5. Bioinformatics Analysis
2.5.1. Detecting Host-Derived miRNAs and DE miRNAs
2.5.2. MiRNA Target Gene Prediction; Network and Core Expression Analysis
2.6. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.7. Statistical Analysis
3. Results
3.1. Animal Infection
3.2. Basic Characteristics of Libraries Obtained from miRNA-Sequencing
3.3. Differential Expression of miRNAs
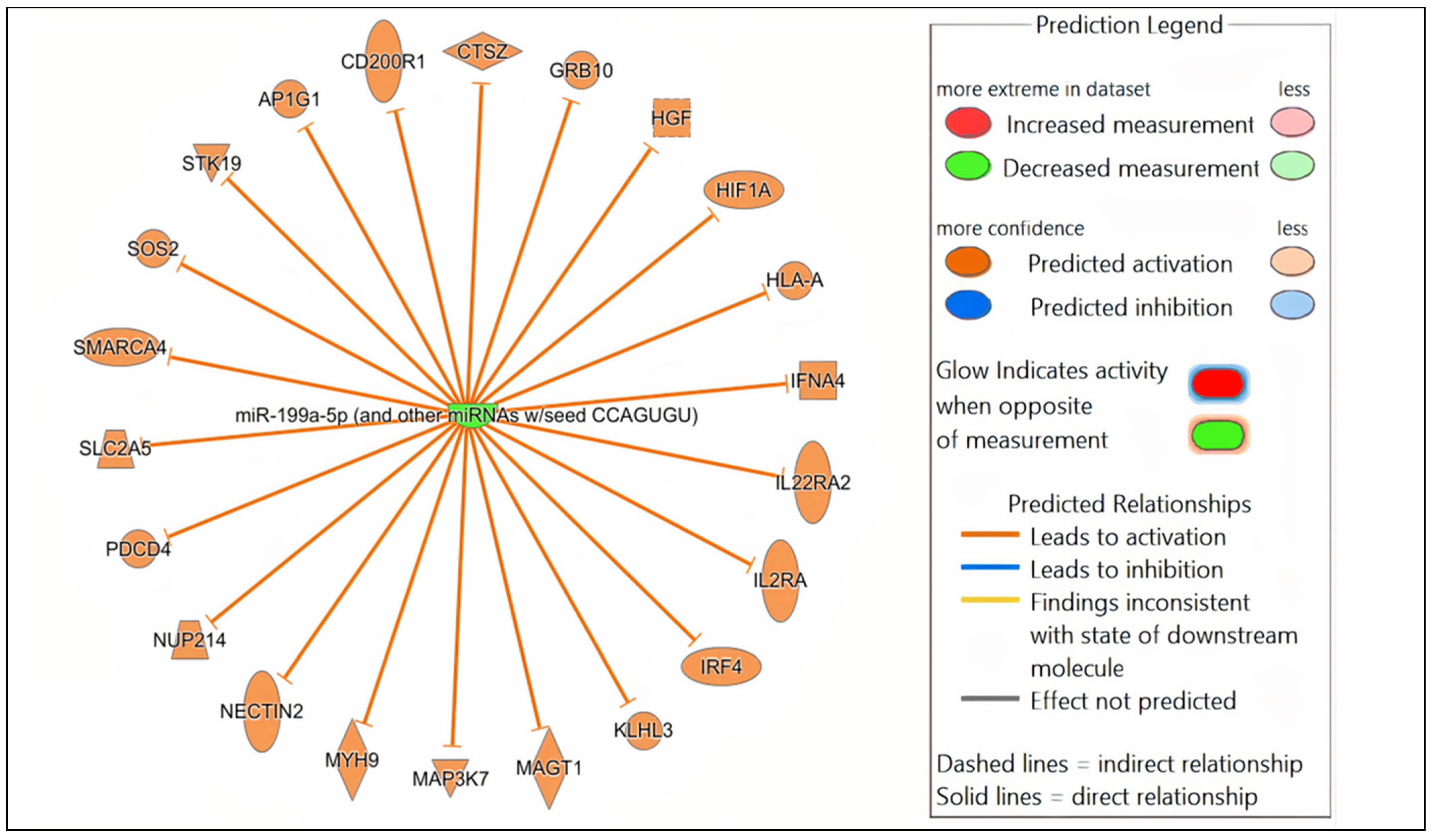
3.4. MiRNA Target Gene Prediction, Network and Core Expression Analysis
3.5. Confirmation of DE miRNAs Using Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4. Discussion
4.1. Immune-Related DE miRNAs and Pathways
4.2. Pathology-Related DE miRNAs and Pathways
5. Limitations of the Study and Future Work
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- James, C.; Harfouche, M.; Welton, N.J.; Turner, K.M.; Abu-Raddad, L.J.; Gottlieb, S.L.; Looker, K.J. Herpes simplex virus: Global infection prevalence and incidence estimates, 2016. Bull. World Health Organ. 2020, 98, 315–329. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Soil-Transmitted Helminth Infections. Available online: https://www.who.int/news-room/fact-sheets/detail/soil-transmitted-helminth-infections (accessed on 7 June 2025).
- Mathew, J., Jr.; Sapra, A. Herpes Simplex Type 2. Available online: https://www.ncbi.nlm.nih.gov/books/NBK554427/ (accessed on 7 June 2025).
- Sausen, D.G.; Shechter, O.; Gallo, E.S.; Dahari, H.; Borenstein, R. Herpes Simplex Virus, Human Papillomavirus, and Cervical Cancer: Overview, Relationship, and Treatment Implications. Cancers 2023, 15, 3692. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.; Barra, N.G.; Lee, A.J.; Ashkar, A.A. Innate and adaptive immunity against herpes simplex virus type 2 in the genital mucosa. J. Reprod. Immunol. 2011, 88, 210–218. [Google Scholar] [CrossRef] [PubMed]
- McSorley, H.J.; Maizels, R.M. Helminth infections and host immune regulation. Clin. Microbiol. Rev. 2012, 25, 585–608. [Google Scholar] [CrossRef] [PubMed]
- Schlosser-Brandenburg, J.; Midha, A.; Mugo, R.M.; Ndombi, E.M.; Gachara, G.; Njomo, D.; Rausch, S.; Hartmann, S. Infection with soil-transmitted helminths and their impact on coinfections. Front. Parasitol. 2023, 2, 1197956. [Google Scholar] [CrossRef] [PubMed]
- Mkhize-Kwitshana, Z.L.; Taylor, M.; Jooste, P.; Mabaso, M.L.H.; Walzl, G. The influence of different helminth infection phenotypes on immune responses against HIV in co-infected adults in South Africa. BMC Infect. Dis. 2011, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, O.A.; Yogeswaran, P.; Wright, G. Intestinal helminth infections amongst HIV-infected adults in Mthatha General Hospital, South Africa. Afr. J. Prim. Health Care Fam. Med. 2015, 7, 910. [Google Scholar] [CrossRef] [PubMed]
- Mpaka-Mbatha, M.N.; Naidoo, P.; Bhengu, K.N.; Islam, M.M.; Singh, R.; Nembe-Mafa, N.; Mkhize-Kwitshana, Z.L. Cytokine Gene Expression Profiles during HIV and Helminth Coinfection in Underprivileged Peri-Urban South African Adults. Diagnostics 2023, 13, 2475. [Google Scholar] [CrossRef] [PubMed]
- Gravitt, P.E.; Marks, M.; Kosek, M.; Huang, C.; Cabrera, L.; Olortegui, M.P.; Medrano, A.M.; Trigoso, D.R.; Qureshi, S.; Bardales, G.S.; et al. Soil-Transmitted Helminth Infections Are Associated with an Increase in Human Papillomavirus Prevalence and a T-Helper Type 2 Cytokine Signature in Cervical Fluids. J. Infect. Dis. 2016, 213, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Omondi, M.A.; Kamassa, E.H.; Katawa, G.; Tchopba, C.N.; Vogelbusch, C.; Parcina, M.; Tchadié, E.P.; Amessoudji, O.M.; Arndts, K.; Karou, S.D.; et al. Hookworm infection associates with a vaginal Type 1/Type 2 immune signature and increased HPV load. Front. Immunol. 2022, 13, 1009968. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, A.D.; Tamayo, M.A.; Beheim, B.; Trumble, B.C.; Stieglitz, J.; Hooper, P.L.; Martin, M.; Kaplan, H.; Gurven, M. Helminth infection, fecundity, and age of first pregnancy in women. Science 2015, 350, 970–972. [Google Scholar] [CrossRef] [PubMed]
- Chetty, A.; Omondi, M.A.; Butters, C.; Smith, K.A.; Katawa, G.; Ritter, M.; Layland, L.; Horsnell, W. Impact of Helminth Infections on Female Reproductive Health and Associated Diseases. Front. Immunol. 2020, 11, 577516. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.H.; Gilman, R.H.; Chiao, E.Y.; Gravitt, P.E. Gut Helminth Infection-Induced Immunotolerance and Consequences for Human Papillomavirus Persistence. Am. J. Trop. Med. Hyg. 2021, 105, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Damane, B.; Mulaudzi, T.; Kader, S.; Naidoo, P.; Dlamini, Z.; Mkhize-Kwitshana, Z. HIV-Helminth Co-Infections and Immune Checkpoints: Implications for Cancer Risk in South Africa. Viruses 2025, 17, 451. [Google Scholar] [CrossRef] [PubMed]
- Chetty, A.; Darby, M.G.; Vornewald, P.M.; Martín-Alonso, M.; Filz, A.; Ritter, M.; McSorley, H.J.; Masson, L.; Smith, K.; Brombacher, F.; et al. Il4ra-independent vaginal eosinophil accumulation following helminth infection exacerbates epithelial ulcerative pathology of HSV-2 infection. Cell Host Microbe 2021, 29, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, B.C.; Charchar, F.J.; Lin, R.C.; McMullen, J.R. A microRNA guide for clinicians and basic scientists: Background and experimental techniques. Heart Lung Circ. 2012, 21, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Chandan, K.; Gupta, M.; Sarwat, M. Role of Host and Pathogen-Derived MicroRNAs in Immune Regulation During Infectious and Inflammatory Diseases. Front. Immunol. 2020, 10, 3081. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Iizasa, H.; Kanehiro, Y.; Fekadu, S.; Yoshiyama, H. Herpesviral microRNAs in Cellular Metabolism and Immune Responses. Front. Microbiol. 2017, 8, 1318. [Google Scholar] [CrossRef] [PubMed]
- Anwesha, B.; Anupam, M. Herpesviridae and microRNAs. In Current Perspectives on Viral Disease Outbreaks; David, C., Ed.; IntechOpen: Rijeka, Croatia, 2021; p. 81. [Google Scholar]
- Entwistle, L.J.; Wilson, M.S. MicroRNA-mediated regulation of immune responses to intestinal helminth infections. Parasite Immunol. 2017, 39, e12406. [Google Scholar] [CrossRef] [PubMed]
- Miśkiewicz, J.; Mielczarek-Palacz, A.; Gola, J.M. MicroRNAs as Potential Biomarkers in Gynecological Cancers. Biomedicines 2023, 11, 1704. [Google Scholar] [CrossRef] [PubMed]
- Frey, K.G.; Bishop-Lilly, K.A. Chapter 15—Next-Generation Sequencing for Pathogen Detection and Identification. In Methods in Microbiology; Sails, A., Tang, Y.-W., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 42, pp. 525–554. [Google Scholar]
- Tang, S.; Patel, A.; Krause, P.R. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 2009, 83, 1433–1442. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Wang, K.; Tang, S.; Krause, P.R.; Mont, E.K.; Cohen, J.I.; Cullen, B.R. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 2010, 84, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Kuang, L.; Deng, Y.; Liu, X.; Zou, Z.; Mi, L. Differential expression of mRNA and miRNA in guinea pigs following infection with HSV2v. Exp. Ther. Med. 2017, 14, 2577–2583. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Deng, Y.; Chen, H.; Lin, Y.; Yang, X.; Sun, B.; Pan, D. Neuronal miR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1. J. Virol. 2022, 96, e0034922. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.-C.; Xu, M.-J.; Alasaad, S.; Song, H.-Q.; Peng, L.; Tao, J.-P.; Zhu, X.-Q. Comparative analysis of microRNA profiles between adult Ascaris lumbricoides and Ascaris suum. BMC Vet. Res. 2014, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Tritten, L.; Tam, M.; Vargas, M.; Jardim, A.; Stevenson, M.M.; Keiser, J.; Geary, T.G. Excretory/secretory products from the gastrointestinal nematode Trichuris muris. Exp. Parasitol. 2017, 178, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Eichenberger, R.M.; Ryan, S.; Jones, L.; Buitrago, G.; Polster, R.; Montes de Oca, M.; Zuvelek, J.; Giacomin, P.R.; Dent, L.A.; Engwerda, C.R.; et al. Hookworm Secreted Extracellular Vesicles Interact with Host Cells and Prevent Inducible Colitis in Mice. Front. Immunol. 2018, 9, 850. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.P.; Fromm, B.; Andersen, S.D.; Marcilla, A.; Andersen, K.L.; Borup, A.; Williams, A.R.; Jex, A.R.; Gasser, R.B.; Young, N.D.; et al. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite–host cross talk. J. Extracell. Vesicles 2019, 8, 1578116. [Google Scholar] [CrossRef] [PubMed]
- Marshak, J.O.; Dong, L.; Koelle, D.M. The murine intravaginal HSV-2 challenge model for investigation of DNA vaccines. Methods Mol. Biol. 2014, 1144, 305–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Evans, J.; Bhagwate, A.; Middha, S.; Bockol, M.; Yan, H.; Kocher, J.-P. CAP-miRSeq: A comprehensive analysis pipeline for microRNA sequencing data. BMC Genom. 2014, 15, 423. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Xi, D.; Zhang, X.; Huang, Z.; Tang, N.; Liu, Y.; Wang, L.; Tang, Y.; Zhong, H.; He, F. Screening and validation of differentially expressed microRNAs and target genes in hypertensive mice induced by cytomegalovirus infection. Biosci. Rep. 2020, 40, BSR20202387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, C.; Zhang, B.; Li, Z.; Zeng, W.; Luo, R.; Cao, J.; Cheng, G.; Fan, S.; He, Q. Differential expression and correlation analysis of miRNA–mRNA profiles in swine testicular cells infected with porcine epidemic diarrhea virus. Sci. Rep. 2021, 11, 1868. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-M.; Zhao, S.-S.; Tao, D.-L.; Li, J.-Y.; Yang, X.; Fan, Y.-Y.; Song, J.-K.; Liu, Q.; Zhao, G.-H. Temporal transcriptomic changes in microRNAs involved in the host immune response and metabolism during Neospora caninum infection. Parasites Vectors 2023, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.L.; Yu, M.; Wang, X.; Chen, H.J.; Ji, X.S.; Zhao, Y. Global changes in gene expression of mRNA and miRNA in liver tissues of Micropterus salmoides after infection with Aeromonas hydrophila. Aquac. Rep. 2025, 42, 102756. [Google Scholar] [CrossRef]
- Camberis, M.; Le Gros, G.; Urban, J., Jr. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr. Protoc. Immunol. 2003, 55, 19-12. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Wu, F.; Liu, T.; Wang, W.; Liu, T.; Jin, X.; Xu, L.; Ma, Y.; Huang, G.; Chen, Z. JieZe-1 Alleviates HSV-2 Infection-Induced Genital Herpes in Balb/c Mice by Inhibiting Cell Apoptosis via Inducing Autophagy. Front. Pharmacol. 2021, 12, 775521. [Google Scholar] [CrossRef] [PubMed]
- Biton, M.; Levin, A.; Slyper, M.; Alkalay, I.; Horwitz, E.; Mor, H.; Kredo-Russo, S.; Avnit-Sagi, T.; Cojocaru, G.; Zreik, F.; et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium–T cell crosstalk. Nat. Immunol. 2011, 12, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Okoye, I.S.; Czieso, S.; Ktistaki, E.; Roderick, K.; Coomes, S.M.; Pelly, V.S.; Kannan, Y.; Perez-Lloret, J.; Zhao, J.L.; Baltimore, D.; et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc. Natl. Acad. Sci. USA 2014, 111, E3081–E3090. [Google Scholar] [CrossRef] [PubMed]
- Knolle, M.D.; Chin, S.B.; Rana, B.M.J.; Englezakis, A.; Nakagawa, R.; Fallon, P.G.; Git, A.; McKenzie, A.N.J. MicroRNA-155 Protects Group 2 Innate Lymphoid Cells from Apoptosis to Promote Type-2 Immunity. Front. Immunol. 2018, 9, 2232. [Google Scholar] [CrossRef] [PubMed]
- Entwistle, L.; Aegerter, H.; Czieso, S.; Amaniti, E.; Guidi, R.; Sesay, A.; Nikolov, N.; Chakravaty, P.; Huynh, A.; Mills, J.; et al. Inhibition of miR-99a-5p prevents allergen-driven airway exacerbations without compromising type-2 memory responses in the intestine following helminth infection. Mucosal Immunol. 2021, 14, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, B.; Sørensen Rossen, L.; Zippor, M.; Boysen, A.T.; Indira Chandran, V.; Skallerup, P.; Thamsborg, S.M.; Nejsum, P. Micro RNA profiles of host extracellular vesicles are modulated by Ascaris suum infection but parasite extracellular vesicle miRNAs are systemically undetectable using in-depth miRNA sequencing. Int. J. Parasitol. 2024, 54, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.M.; Akula, S.M. miRNA-36 inhibits KSHV, EBV, HSV-2 infection of cells via stifling expression of interferon induced transmembrane protein 1 (IFITM1). Sci. Rep. 2017, 7, 17972. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Dass, D.; Dhotre, K.; Wakchoure, P.; More, A.; Rana, S.; Khan, A.A.; Mukherjee, A. Combinatorial Effects of miRNAs in HSV-2 Infection of Macrophages: An In Silico and In Vitro Integration Approach. Vaccines 2023, 11, 1488. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhou, L.; Gao, Q.; Zhang, A.; Wei, J.; Hong, D.; Chu, Y.; Duan, X.; Zhang, Y.; Xu, G. MicroRNA-144-3p inhibits autophagy activation and enhances Bacillus Calmette-Guérin infection by targeting ATG4a in RAW264.7 macrophage cells. PLoS ONE 2017, 12, e0179772. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, C.M.; Podyminogin, R.L.; Diercks, A.H.; Treuting, P.M.; Peschon, J.J.; Rodriguez, D.; Gundapuneni, M.; Weiss, M.J.; Aderem, A. miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis. PLoS Pathog. 2017, 13, e1006305. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, I.S.; Lee, S.-G.; Kim, Y.J.; Silwal, P.; Kim, J.Y.; Kim, J.K.; Seo, W.; Chung, C.; Cho, H.K.; et al. MiR-144-3p is associated with pathological inflammation in patients infected with Mycobacteroides abscessus. Exp. Mol. Med. 2021, 53, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Tang, H.; Ye, B.C. MicroRNA-144-3p Inhibits Host Lipid Catabolism and Autophagy by Targeting PPARα and ABCA1 During Mycobacterium tuberculosis Infection. ACS Infect. Dis. 2024, 10, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Igietseme, J.U.; Omosun, Y.; Partin, J.; Goldstein, J.; He, Q.; Joseph, K.; Ellerson, D.; Ansari, U.; Eko, F.O.; Bandea, C. Prevention of Chlamydia-induced infertility by inhibition of local caspase activity. J. Infect. Dis. 2013, 207, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Benyeogor, I.; Simoneaux, T.; Wu, Y.; Lundy, S.; George, Z.; Ryans, K.; McKeithen, D.; Pais, R.; Ellerson, D.; Lorenz, W.W.; et al. A unique insight into the MiRNA profile during genital chlamydial infection. BMC Genom. 2019, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Bai, X.; Xie, X.; Chen, G.; Jia, X.; Lei, M.; Li, C.; Lai, S. Negative effects of heat stress on ovarian tissue in female rabbit. Front. Vet. Sci. 2022, 9, 1009182. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, F.; Hou, Y.; Lin, X.; Liang, R.; Hu, X.; Zhao, J.; Wang, J.; Olsen, N.; Zheng, S.G. TGF-β-induced CD4+ FoxP3+ regulatory T cell-derived extracellular vesicles modulate Notch1 signaling through miR-449a and prevent collagen-induced arthritis in a murine model. Cell. Mol. Immunol. 2021, 18, 2516–2529. [Google Scholar] [CrossRef] [PubMed]
- Reinkens, T.; Stalke, A.; Huge, N.; Vajen, B.; Eilers, M.; Schäffer, V.; Dittrich-Breiholz, O.; Schlegelberger, B.; Illig, T.; Skawran, B. Ago-RIP sequencing identifies new MicroRNA-449a-5p target genes increasing sorafenib efficacy in hepatocellular carcinoma. J. Cancer 2022, 13, 62. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Yoshitomi, H.; Furu, M.; Ishikawa, M.; Shibuya, H.; Ito, H.; Matsuda, S. MicroRNA-451 down-regulates neutrophil chemotaxis via p38 MAPK. Arthritis Rheumatol. 2014, 66, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xing, F.; Fu, Q.; He, B.; Jia, Z.; Du, J.; Li, Y.; Zhang, X.; Chen, X. hUC-MSCs exosomal miR-451 alleviated acute lung injury by modulating macrophage M2 polarization via regulating MIF-PI3K-AKT signaling pathway. Environ. Toxicol. 2022, 37, 2819–2831. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Rau, C.-S.; Jeng, J.C.; Chen, Y.-C.; Lu, T.-H.; Wu, C.-J.; Wu, Y.-C.; Tzeng, S.-L.; Yang, J.C.-S. Whole blood-derived microRNA signatures in mice exposed to lipopolysaccharides. J. Biomed. Sci. 2012, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.-H.; Yang, J.C.-S.; Jeng, J.C.; Chen, Y.-C.; Lu, T.-H.; Tzeng, S.-L.; Wu, Y.-C.; Wu, C.-J.; Rau, C.-S. Circulating microRNA signatures in mice exposed to lipoteichoic acid. J. Biomed. Sci. 2013, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-C.; Yang, J.C.-S.; Rau, C.-S.; Chen, Y.-C.; Lu, T.-H.; Lin, M.-W.; Tzeng, S.-L.; Wu, Y.-C.; Wu, C.-J.; Hsieh, C.-H. Profiling circulating microRNA expression in experimental sepsis using cecal ligation and puncture. PLoS ONE 2013, 8, e77936. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Tian, M.; Chang, J.; Li, F.; Zhang, G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed. Pharmacother. 2020, 122, 109692. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Pan, Q.; Cao, H.X.; Xin, F.Z.; Zhao, Z.H.; Yang, R.X.; Zeng, J.; Zhou, H.; Fan, J.G. Lipotoxic hepatocyte-derived exosomal microRNA 192-5p activates macrophages through rictor/Akt/forkhead box transcription factor O1 signaling in nonalcoholic fatty liver disease. Hepatology 2020, 72, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Nagura, Y.; Matsuura, K.; Iio, E.; Fujita, K.; Inoue, T.; Matsumoto, A.; Tanaka, E.; Nishiguchi, S.; Kang, J.-H.; Matsui, T.; et al. Serum miR-192-5p levels predict the efficacy of pegylated interferon therapy for chronic hepatitis B. PLoS ONE 2022, 17, e0263844. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Giri, B.R.; Liu, J.; He, X.; Cai, P.; Jing, Z.; Cheng, G. Characterization of MicroRNA Cargo of Extracellular Vesicles Isolated from the Plasma of Schistosoma japonicum-Infected Mice. Front. Cell Infect. Microbiol. 2022, 12, 803242. [Google Scholar] [CrossRef] [PubMed]
- Smita, S.; Ahad, A.; Ghosh, A.; Biswas, V.K.; Koga, M.M.; Gupta, B.; Acha-Orbea, H.; Raghav, S.K. Importance of EMT Factor ZEB1 in cDC1 “MutuDC Line” Mediated Induction of Th1 Immune Response. Front. Immunol. 2018, 9, 2604. [Google Scholar] [CrossRef] [PubMed]
- Lutz, G.; Jurak, I.; Kim, E.T.; Kim, J.Y.; Hackenberg, M.; Leader, A.; Stoller, M.; Fekete, D.; Weitzman, M.; Coen, D.; et al. Viral Ubiquitin Ligase Stimulates Selective Host MicroRNA Expression by Targeting ZEB Transcriptional Repressors. Viruses 2017, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, Z.; Mertz, J.E. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog. 2007, 3, e194. [Google Scholar] [CrossRef] [PubMed]
- Bao, C.; Li, Y.; Huan, L.; Zhang, Y.; Zhao, F.; Wang, Q.; Liang, L.; Ding, J.; Liu, L.; Chen, T. NF-κB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1α. Sci. Signal. 2015, 8, ra75. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Liu, C.; Zou, X.; Wu, W.; Zhang, C.; Yuan, D. MiRNA-194 regulates palmitic acid-induced toll-like receptor 4 inflammatory responses in THP-1 cells. Nutrients 2015, 7, 3483–3496. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Uribe, J.; Zaldívar-López, S.; Aguilar, C.; Entrenas-García, C.; Bautista, R.; Claros, M.G.; Garrido, J.J. Study of microRNA expression in Salmonella Typhimurium-infected porcine ileum reveals miR-194a-5p as an important regulator of the TLR4-mediated inflammatory response. Vet. Res. 2022, 53, 35. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, H.; Humphreys, N.; Renauld, J.C.; Van Snick, J.; Grencis, R. Interleukin-9 is involved in host protective immunity to intestinal nematode infection. Eur. J. Immunol. 1997, 27, 2536–2540. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, H.; Renauld, J.-C.; Van Snick, J.; Grencis, R. Interleukin-9 enhances resistance to the intestinal nematode Trichuris muris. Infect. Immun. 1998, 66, 3832–3840. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Grencis, R.K.; Humphreys, N.E.; Renauld, J.-C.; Van Snick, J. Anti-IL-9 vaccination prevents worm expulsion and blood eosinophilia in Trichuris muris-infected mice. Proc. Natl. Acad. Sci. USA 2000, 97, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Licona-Limón, P.; Henao-Mejia, J.; Temann, A.U.; Gagliani, N.; Licona-Limón, I.; Ishigame, H.; Hao, L.; De’Broski, R.H.; Flavell, R.A. Th9 cells drive host immunity against gastrointestinal worm infection. Immunity 2013, 39, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Conickx, G.; Mestdagh, P.; Avila Cobos, F.; Verhamme, F.M.; Maes, T.; Vanaudenaerde, B.M.; Seys, L.J.M.; Lahousse, L.; Kim, R.Y.; Hsu, A.C.; et al. MicroRNA Profiling Reveals a Role for MicroRNA-218-5p in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2017, 195, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Pan, Y.; Gao, J.; Xu, Y.; Li, X.; Tian, Z.; Chen, H.; Wang, Y. Downregulation of miR-218 by porcine reproductive and respiratory syndrome virus facilitates viral replication via inhibition of type I interferon responses. J. Biol. Chem. 2021, 296, 100683. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhang, Q.; Yang, N.; Wang, Q.; Zhang, Y.; Xu, X. PEDV inhibits HNRNPA3 expression by miR-218-5p to enhance cellular lipid accumulation and promote viral replication. mBio 2024, 15, e0319723. [Google Scholar] [CrossRef] [PubMed]
- Fang, A.; Yuan, Y.; Sui, B.; Wang, Z.; Zhang, Y.; Zhou, M.; Chen, H.; Fu, Z.F.; Zhao, L. Inhibition of miR-200b-3p confers broad-spectrum resistance to viral infection by targeting TBK1. mBio 2023, 14, e0086723. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Connell, A.L.; Iempridee, T.; Xu, I.; Mertz, J.E. Cellular MicroRNAs 200b and 429 Regulate the Epstein-Barr Virus Switch between Latency and Lytic Replication. J. Virol. 2010, 84, 10329–10343. [Google Scholar] [CrossRef] [PubMed]
- Lee Kyoung, H.; Lim Beom, J.; Ferreira Victor, H.; Min Seo, Y.; Hong, Y.-M.; Jo, J.-H.; Han Sang, H. Expression of human miR-200b-3p and -200c-3p in cytomegalovirus-infected tissues. Biosci. Rep. 2018, 38, BSR20180961. [Google Scholar] [CrossRef]
- Wang, H.; Gao, H.; Duan, S.; Song, X. Inhibition of microRNA-199a-5p reduces the replication of HCV via regulating the pro-survival pathway. Virus Res. 2015, 208, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Suemasa, F.; Sagara, H.; Nakamura, S.; Ino, Y.; Kobayashi, K.; Hiramatsu, H.; Haraguchi, T.; Kurokawa, K.; Todo, T.; et al. MiR-199a Inhibits Secondary Envelopment of Herpes Simplex Virus-1 Through the Downregulation of Cdc42-specific GTPase Activating Protein Localized in Golgi Apparatus. Sci. Rep. 2017, 7, 6650. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yan, Y.; Li, W.; Li, Y.; Yang, H. Expression Profile of miR-199a and Its Role in the Regulation of Intestinal Inflammation. Animals 2023, 13, 1979. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.H.; Ren, H.J.; Peng, R.Y.; Li, Y.; Ming, L. Comparative expression profiles of host circulating miRNAs in response to Trichinella spiralis infection. Vet. Res. 2020, 51, 39. [Google Scholar] [CrossRef] [PubMed]
- Deruaz, M.; Luster, A.D. Chemokine-mediated immune responses in the female genital tract mucosa. Immunol. Cell Biol. 2015, 93, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Bonne-Année, S.; Kerepesi, L.A.; Hess, J.A.; O’Connell, A.E.; Lok, J.B.; Nolan, T.J.; Abraham, D. Human and mouse macrophages collaborate with neutrophils to kill larval Strongyloides stercoralis. Infect. Immun. 2013, 81, 3346–3355. [Google Scholar] [CrossRef] [PubMed]
- Bonne-Année, S.; Kerepesi, L.A.; Hess, J.A.; Wesolowski, J.; Paumet, F.; Lok, J.B.; Nolan, T.J.; Abraham, D. Extracellular traps are associated with human and mouse neutrophil and macrophage mediated killing of larval Strongyloides stercoralis. Microbes Infect. 2014, 16, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wu, W.; Millman, A.; Craft, J.F.; Chen, E.; Patel, N.; Boucher, J.L.; Urban, J.F.; Kim, C.C.; Gause, W.C. Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat. Immunol. 2014, 15, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.E.; Logan, N.; Rückerl, D.; Humbles, A.A.; Allan, S.M.; Papayannopoulos, V.; Stockinger, B.; Maizels, R.M.; Allen, J.E. Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat. Immunol. 2014, 15, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Heeb, L.E.M.; Egholm, C.; Boyman, O. Evolution and function of interleukin-4 receptor signaling in adaptive immunity and neutrophils. Genes Immun. 2020, 21, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Oeser, K.; Schwartz, C.; Voehringer, D. Conditional IL-4/IL-13-deficient mice reveal a critical role of innate immune cells for protective immunity against gastrointestinal helminths. Mucosal Immunol. 2015, 8, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Thery, F.; Eggermont, D.; Impens, F. Proteomics Mapping of the ISGylation Landscape in Innate Immunity. Front. Immunol. 2021, 12, 720765. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.-F.; Zhuang, Y.-J.; Wang, Y.; Zhang, Z.-Y.; Xu, X.-Z.; Mao, Y.-R.; Yu, J.-J. Tumor suppressor miR-192-5p targets TRPM7 and inhibits proliferation and invasion in cervical cancer. Kaohsiung J. Med. Sci. 2021, 37, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ma, H.; Li, Y.; Su, R. MiR-192-5p-Modified Tumor-Associated Macrophages-Derived Exosome Suppressed Endometrial Cancer Progression Through Targeting IRAK1/NF-κB Signaling. Reprod. Sci. 2022, 29, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Dou, K.; Wu, Y.; Ma, Y.; Sun, J. The NF-κB modulated miR-194-5p/IGF1R/PPFIBP axis is crucial for the tumorigenesis of ovarian cancer. J. Cancer 2020, 11, 3433–3445. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, K.; Zhao, Z.; Pang, Y.; Liu, F.; Wang, P.; Wang, Z.; Yang, X. miR-451a suppresses the proliferation and migration of high-grade serous ovarian cancer by targeting RAB5A through the Ras/Raf/MEK/ERK pathway. J. Gene Med. 2024, 26, e3649. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; He, Q.; Lu, Y.; Tao, F.; Zhao, L.; Ou, R. MicroRNA-218-5p inhibits cell growth and metastasis in cervical cancer via LYN/NF-κB signaling pathway. Cancer Cell Int. 2018, 18, 198. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhao, Y.; Li, F.; Qiao, B. MiR-144-3p: A novel tumor suppressor targeting MAPK6 in cervical cancer. J. Physiol. Biochem. 2019, 75, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yao, B.; Wu, Z. miRNA-199a-5p suppresses proliferation and invasion by directly targeting NF-κB1 in human ovarian cancer cells. Oncol. Lett. 2018, 16, 4543–4550. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhao, Y.; Chen, L.; Sun, Y.; Fan, S. Effects of miRNA-199a-5p on cell proliferation and apoptosis of uterine leiomyoma by targeting MED12. Open Med. 2022, 17, 151–159. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jiang, Z.; Wang, J.; Han, Z. Mechanism of miR-200b-3p-induced FOSL2 inhibition of endometrial cancer cell proliferation and metastasis. Sci. Rep. 2025, 15, 15742. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Wang, Q.; Zhao, Y.; Wu, J. miR-205-3p Functions as a Tumor Suppressor in Ovarian Carcinoma. Reprod. Sci. 2020, 27, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Xue, J.; Zhang, Q.; Li, F.; Zhang, W.; Chen, H.; Huang, Y.; Zheng, F. MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol. Rep. 2014, 32, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, Y.; Xiong, W.; Ye, W.; Zhao, W.; Hua, Y. MicroRNA-449a Is Downregulated in Cervical Cancer and Inhibits Proliferation, Migration, and Invasion. Oncol. Res. Treat. 2019, 42, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, Q.-M.; Zhang, X.-W.; Sheng, Q.; Yan, X.-T. MiR-376a promotion of proliferation and metastases in ovarian cancer: Potential role as a biomarker. Life Sci. 2017, 173, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Yang, J.; Meng, X.; Zhang, J.; Zhang, Y. The promotional effect of microRNA-103a-3p in cervical cancer cells by regulating the ubiquitin ligase FBXW7 function. Hum. Cell 2022, 35, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Zhang, D.; Zhu, L.; Xue, J. Animal models of human herpesvirus infection. Anim. Models Exp. Med. 2025, 8, 615–628. [Google Scholar] [CrossRef] [PubMed]
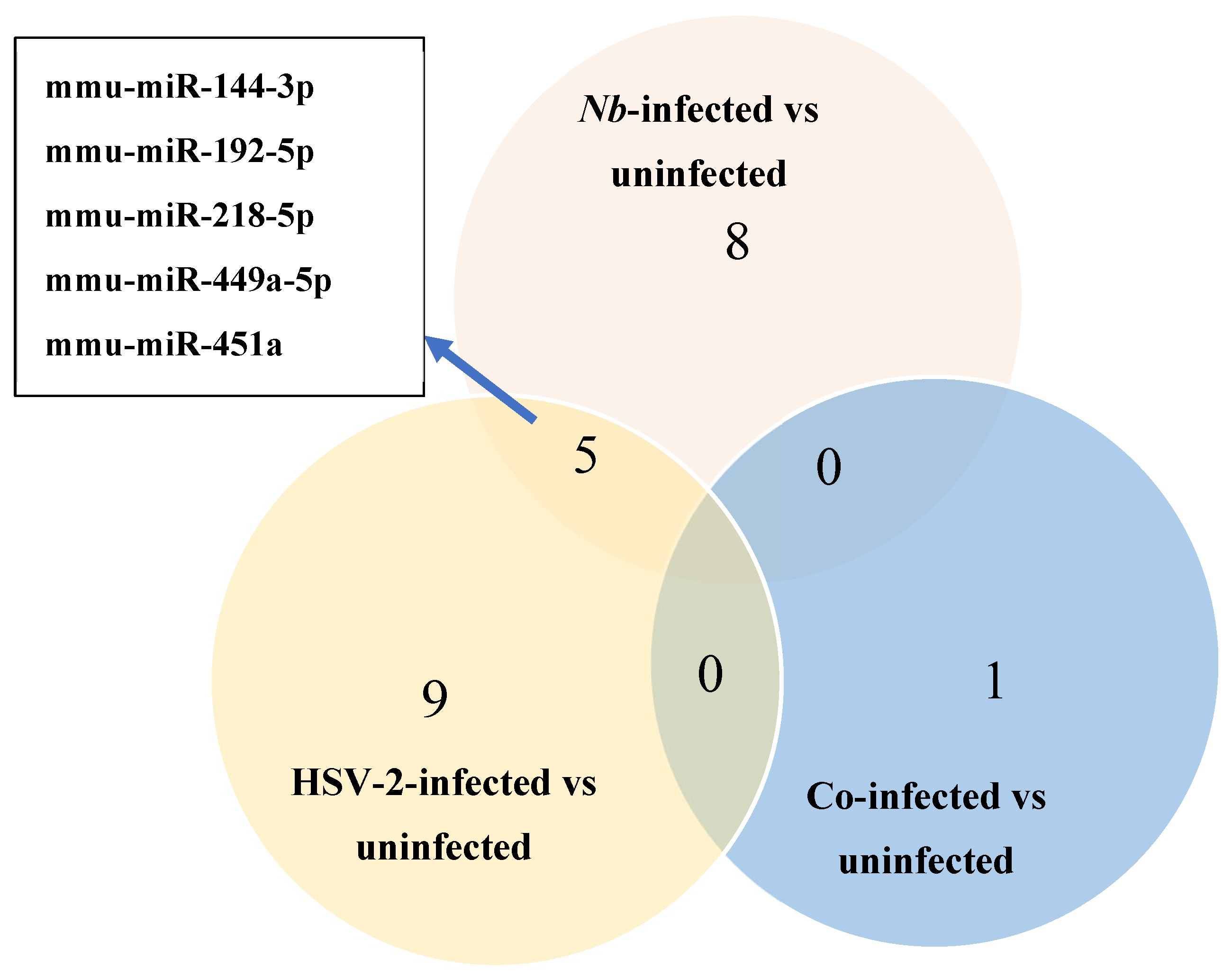
| Nb-Infected Versus Uninfected | |||
|---|---|---|---|
| Name of miRNA | Log Fold Change (LogFC) * | p-Value * | Up/Downregulation |
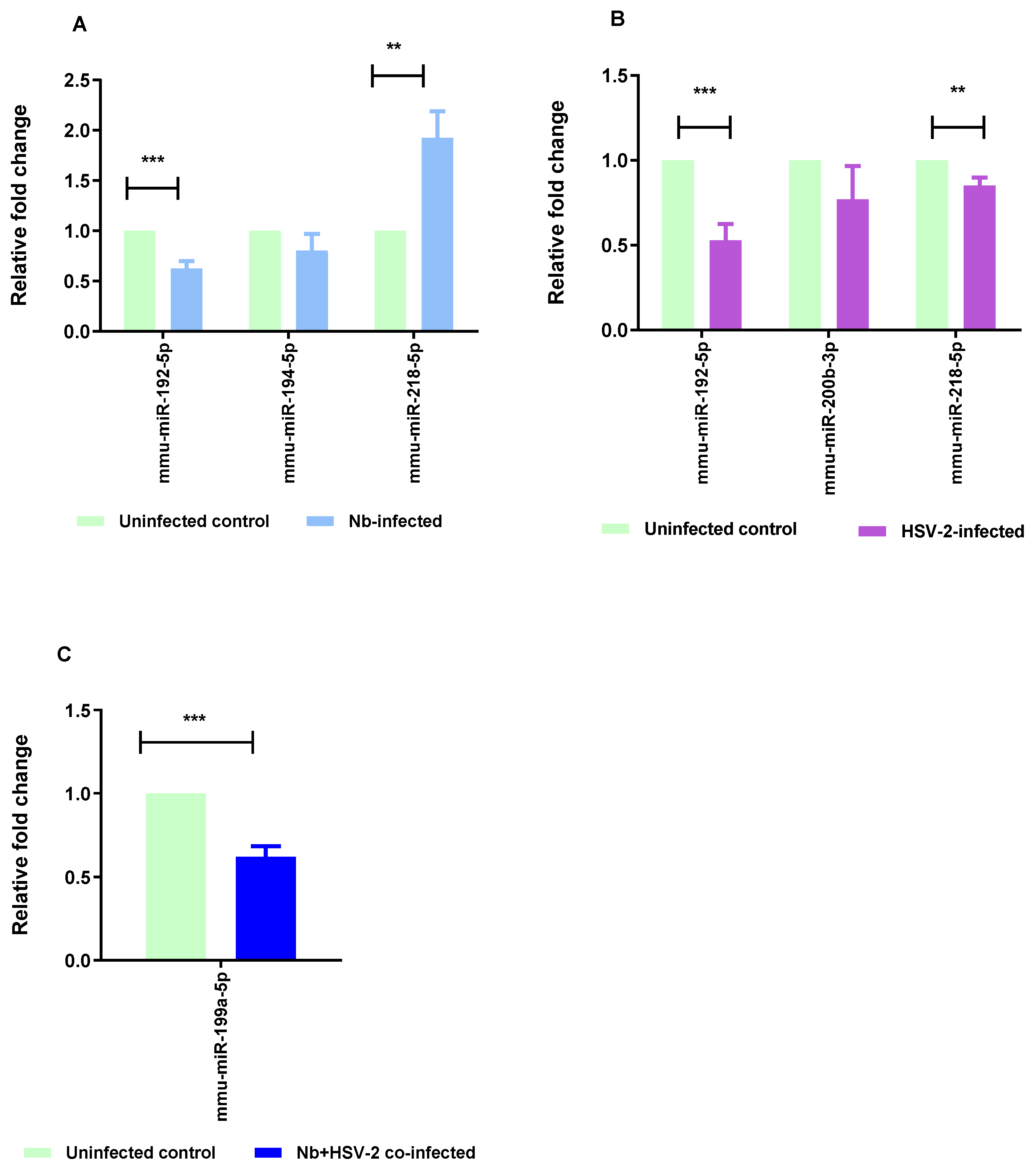
| mmu-miR-194-5p | −3.27 | 8.00 × 10−4 | Down |
| mmu-miR-218-5p | 2.09 | 4.20 × 10−3 | Up |
| mmu-miR-449a-5p | 2.17 | 4.30 × 10−3 | Up |
| mmu-miR-192-5p | −1.92 | 8.38 × 10−3 | Down |
| mmu-miR-497a-3p | 1.43 | 1.57 × 10−2 | Up |
| mmu-miR-144-3p | 1.51 | 2.65 × 10−2 | Up |
| mmu-miR-33-5p | 1.34 | 4.05 × 10−2 | Up |
| mmu-miR-451a | 1.06 | 4.44 × 10−2 | Up |
| HSV-2-infected versus Uninfected | |||
| Name of miRNA | Log Fold Change (LogFC) * | p-Value * | Up/Downregulation |
| mmu-miR-192-5p | −2.94 | 3.48 × 10−4 | Down |
| mmu-miR-451a | 1.88 | 1.37 × 10−3 | Up |
| mmu-miR-449a-5p | 1.72 | 2.88 × 10−3 | Up |
| mmu-miR-218-5p | −1.54 | 9.12 × 10−3 | Down |
| mmu-miR-144-3p | 1.71 | 9.73 × 10−3 | Up |
| mmu-miR-376a-3p | 1.26 | 1.05 × 10−2 | Up |
| mmu-miR-205-3p | −1.46 | 1.91 × 10−2 | Down |
| mmu-miR-103-3p | −1.31 | 2.07 × 10−2 | Down |
| mmu-miR-200b-3p | −1.17 | 3.77 × 10−2 | Down |
| Nb/HSV-2 co-infected versus Uninfected | |||
| Name of miRNA | Log Fold Change (LogFC) * | p-Value * | Up/Downregulation |
| mmu-miR-199a-5p | −2.46 | 4.88 × 10−2 | Down |
| Name of miRNA | Predicted Immune-Related Targets | Predicted Regulatory Effect |
|---|---|---|
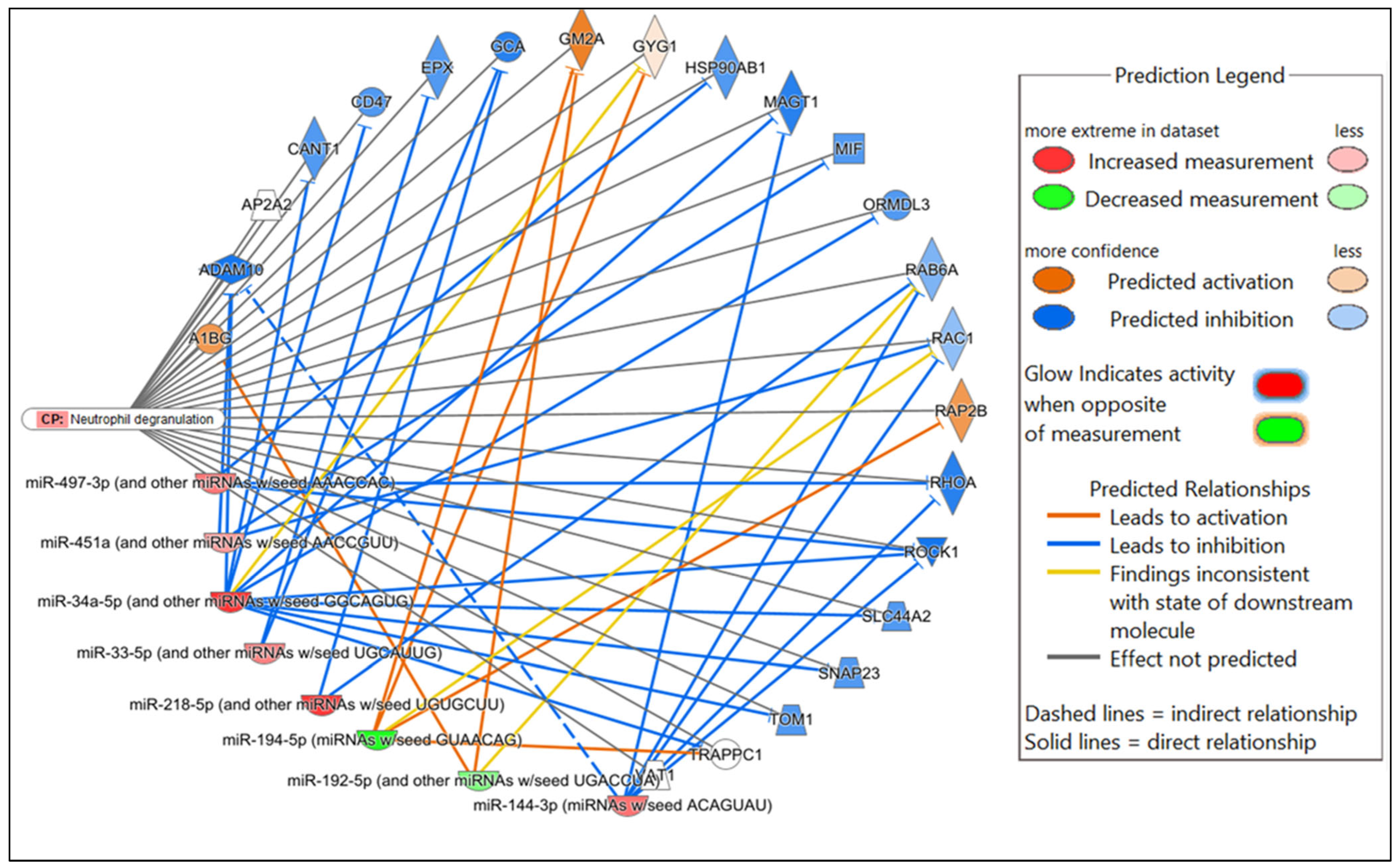
| mmu-miR-144-3p | AP1S3, CTNNB1, EP300, HIF1A, ITCH, JUN, MAGT1, MAP3K8, MEF2A, MTOR, RAC1, RHOA, ROCK1, TNFSF11, UBE2D1, UBE2D3 | Predicted inhibition |
| mmu-miR-192-5p | A1BG, DHX58, GM2A, NLRC5, RSAD2, STK3, ZEB1 | Predicted activation |
| mmu-miR-194-5p | FASLG, GYG1, IL9, RAP2B, SUMO2, TAB3 | Predicted activation |
| mmu-miR-218-5p | C5, CD200R1, COL1A1, DDX41, LMO7, NUP50, PIK3C2A, PLCG1, RAB6A, RICTOR, RNF41, RPS6KA3, SH3KBP1, SOCS3, SPSB1, UBE2H, VAMP7, VAT1 | Predicted inhibition |
| mmu-miR-33-5p | EPX3, GCA | Predicted inhibition |
| mmu-miR-449a-5p | ADAM10, AP2A2, BCL2, BCL6, BTN1A1, C9, CANT1, CASP2, CCL22, CCND1, CD47, CD79A, CREB1, CSF1R, CTNNB1, DEFB124, DNM1L, FBX017, FCER1A, GRAP2, GSTO1, IL23R, IL6R, ISG20, MAP2K1, MAP3K3, MAPT, MUC5B, MYC, MYH9, NECTIN2, ORMDL3, PTPN4, RNF4, ROCK1, SIAH1, SLC44A2, SMAD3, SNAP23, TOM1, TP53 *, TRAPPC1, TRIM21, TXN, UBE2L3, VAMP2, VAT1, VEGFA, WASF1 | Predicted inhibition * Predicted activation (TP53) |
| mmu-miR-451a | ATF2, EDAR, MIF, RAC1, WASF1, YBX1 | Predicted inhibition |
| mmu-miR-497a-3p | HSP90AB1, IL19 | Predicted inhibition |
| Name of miRNA | Predicted Immune-Related Targets | Predicted Regulatory Effect |
|---|---|---|
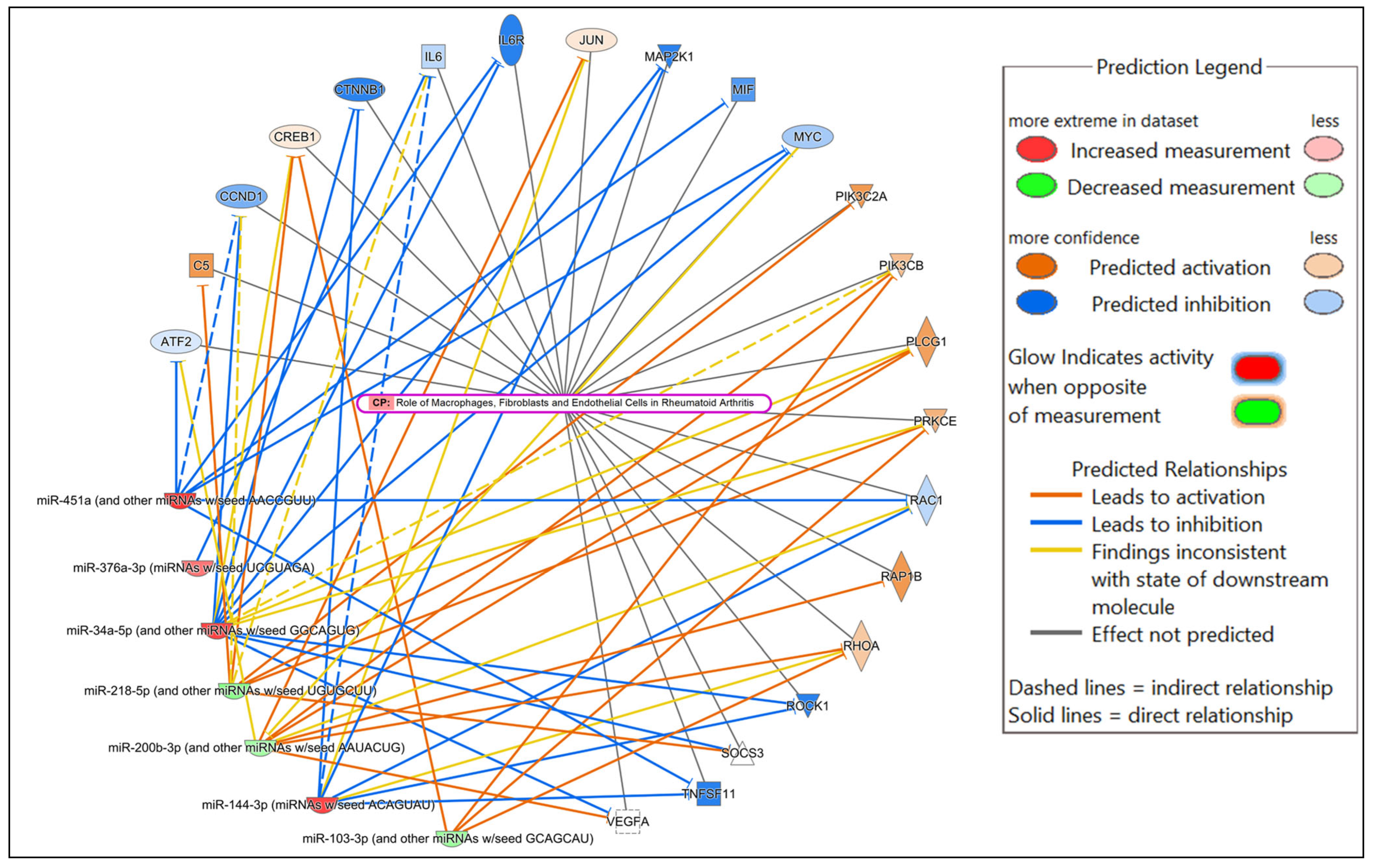
| mmu-miR-103-3p | ARIH1, BTLA, CRKL, EEF2, FBXW11, ICOS, IL10RB, N4BP1, NUP58, PIK3CB, PRKCE, PTGS2, RAB10, SDCBP | Predicted activation |
| mmu-miR-144-3p | AP1S3, CTNNB1, EP300, HIF1A, ITCH, JUN, MAGT1, MAP3K8, MEF2A, MTOR, RAC1, RHOA, ROCK1, TNFSF11, UBE2D1, UBE2D3 | Predicted inhibition |
| mmu-miR-192-5p | A1BG, DHX58, GM2A, NLRC5, RSAD2, STK3, ZEB1 | Predicted activation |
| mmu-miR-200b-3p | AP1S2, ARIH1, CRKL, ELOC, MSN, PLCG1, PTEN, PTPN12, PTPN13, RAP1B, SEC23A, SNAP25, UBE2W, ZEB1 | Predicted activation |
| mmu-miR-205-3p | PSMA5 | Predicted activation |
| mmu-miR-218-5p | C5, CD200R1, COL1A1, DDX41, LMO7, NUP50, PIK3C2A, PLCG1, PRKG1, RAB6A, RICTOR, RNF41, RPS6KA3, SH3KBP1, SOCS3, SPSB1, UBE2H, VAT1 | Predicted activation |
| mmu-miR-449a-5p | ADAM10, AP2A2, BCL2, BCL6, BTN1A1, C9, CANT1, CASP2, CCL22, CCND1, CD47, CD79A, CREB1, CSF1R, CTNNB1, DEFB124, DNM1L, FBXO17, FCER1A, GRAP2, GSTO1, IL23R, IL6R, ISG20, MAP2K1, MAP3K3, MAPT, MUC5B, MYC, MYH9, NECTIN2, ORMDL3, PTPN4, RNF4, ROCK1, SIAH1, SLC44A2, SMAD3, SNAP23, TOM1, TP53 *, TRAPPC1, TRIM21, TXN, UBE2L3, VAMP2, VAT1, VEGFA, WASF1 | Predicted inhibition * Predicted activation (TP53) |
| mmu-miR-376a-3p | CTSO, IL6, KIF5A, TRIM9 | Predicted inhibition |
| mmu-miR-451a | ATF2, EDAR, MIF, RAC1, WASF1, YBX1 | Predicted inhibition |
| Name of miRNA | Predicted Immune-Related Targets | Predicted Regulatory Effect |
|---|---|---|
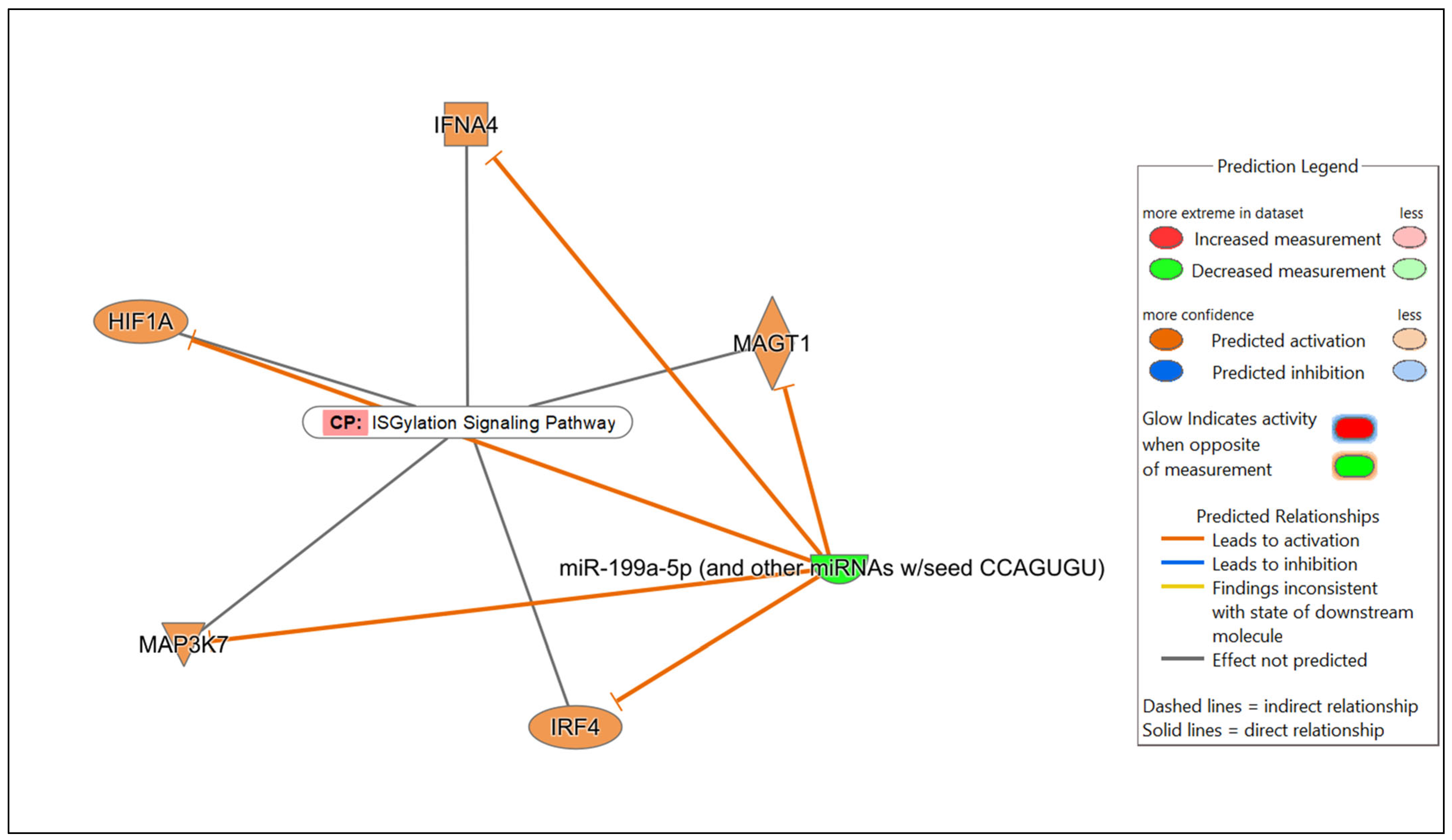
| mmu-miR-199a-5p | AP1G1, CD200R1, CTSZ, GRB10, HGF, HIF1A, HLA-A, IFNA4, IL22RA2, IL2RA, IRF4, KLHL3, MAGT1, MAP3K7, MYH9, NECTIN2, NUP214, PDCD4, SLC2A5, SMARCA4, SOS2, STK19 | Predicted activation |
| Top Canonical Pathways | ||
|---|---|---|
| Name | p-Value * | Overlap † |
| Neutrophil degranulation | 1.60 × 10−18 | 4.8% (23/477) |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 1.16 × 10−16 | 5.6% (19/337) |
| Hypoxia signaling in the cardiovascular system | 3.49 × 10−16 | 15.4% (12/78) |
| Interleukin-4 and interleukin-13 signaling | 7.80 × 10−16 | 11.7% (13/111) |
| NGF Signaling | 2.47 × 10−15 | 10.7% (13/121) |
| Top Diseases and Biological Functions | ||
| Diseases and disorders | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Inflammatory Response | 1.85 × 10−8–2.05 × 10−36 | 87 |
| Infectious Diseases | 2.83 × 10−8–1.73 × 10−29 | 76 |
| Organismal Injury and Abnormalities | 3.24 × 10−8–1.73 × 10−29 | 107 |
| Cancer | 3.24 × 10−8–9.38 × 10−21 | 89 |
| Haematological Disease | 3.10 × 10−8–2.56 × 10−20 | 62 |
| Molecular and Cellular Functions | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Cell-To-Cell Signaling and Interaction | 2.03 × 10−8–3.36 × 10−29 | 74 |
| Cell Death and Survival | 3.13 × 10−8–1.61 × 10−28 | 77 |
| Cellular Development | 3.24 × 10−8–3.51 × 10−27 | 80 |
| Cellular Growth and Proliferation | 3.24 × 10−8–3.51 × 10−27 | 81 |
| Cellular Movement | 3.31 × 10−8–3.05 × 10−26 | 70 |
| Physiological System Development and Function | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Haematological System Development and Function | 3.15 × 10−8–2.36 × 10−32 | 80 |
| Tissue Morphology | 3.15 × 10−8–2.36 × 10−32 | 68 |
| Haematopoiesis | 2.06 × 10−8–3.51 × 10−27 | 47 |
| Lymphoid Tissue Structure and Development | 3.15 × 10−8–3.51 × 10−27 | 60 |
| Tissue Development | 3.24 × 10−8–3.51 × 10−27 | 66 |
| Top Canonical Pathways | ||
|---|---|---|
| Name | p-Value * | Overlap † |
| Role of Macrophages, Fibroblasts, and Endothelial Cells in Rheumatoid Arthritis | 8.68 × 10−19 | 6.5% (22/337) |
| Neutrophil degranulation | 7.97 × 10−18 | 5.0% (24/477) |
| Hepatitis B Chronic Liver Pathogenesis Signaling Pathway | 4.28 × 10−17 | 9.0% (17/188) |
| Hypoxia Signaling in the Cardiovascular System | 6.96 × 10−17 | 16.7% (13/78) |
| Hepatic Fibrosis Signaling Pathway | 7.52 × 10−17 | 5.3% (22/416) |
| Top Diseases and Biological Functions | ||
| Diseases and disorders | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Inflammatory Response | 4.64 × 10−9–4.75 × 10−37 | 100 |
| Infectious Diseases | 8.67 × 10−10–1.66 × 10−31 | 86 |
| Organismal Injury and Abnormalities | 5.03 × 10−9–1.66 × 10−31 | 127 |
| Cancer | 5.03 × 10−9–5.40 × 10−22 | 108 |
| Tumor Morphology | 1.99 × 10−9–3.40 × 10−20 | 38 |
| Molecular and Cellular Functions | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Cell Death and Survival | 4.92 × 10−9–4.28 × 10−30 | 89 |
| Cell-To-Cell Signaling and Interaction | 4.51 × 10−9–1.61 × 10−28 | 85 |
| Cellular Movement | 4.64 × 10−9–4.39 × 10−28 | 82 |
| Cellular Development | 5.00 × 10−9–2.13 × 10−26 | 99 |
| Cellular Growth and Proliferation | 5.00 × 10−9–2.13 × 10−26 | 98 |
| Physiological System Development and Function | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Haematological System Development and Function | 4.64 × 10−9–1.17 × 10−35 | 90 |
| Tissue Morphology | 3.75 × 10−9–1.17 × 10−35 | 80 |
| Immune Cell Trafficking | 4.64 × 10−9–2.46 × 10−27 | 66 |
| Haematopoiesis | 1.06 × 10−9–2.13 × 10−26 | 53 |
| Lymphoid Tissue Structure and Development | 3.75 × 10−9–2.13 × 10−26 | 70 |
| Top Canonical Pathways | ||
|---|---|---|
| Name | p-Value * | Overlap † |
| ISGylation Signaling Pathway | 3.25 × 10−8 | 4.6% (5/109) |
| Glucocorticoid Receptor Signaling | 4.96 × 10−7 | 1.2% (7/600) |
| Hepatitis B Chronic Liver Pathogenesis Signaling Pathway | 1.89 × 10−5 | 2.1% (4/188) |
| Natural Killer Cell Signaling | 2.55 × 10−5 | 2.0% (4/203) |
| Interferon alpha/beta Signaling | 3.75 × 10−5 | 3.9% (3/76) |
| Top Diseases and Biological Functions | ||
| Diseases and disorders | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Cancer | 5.15 × 10−3–5.25 × 10−13 | 21 |
| Haematological Disease | 5.15 × 10−3–5.25 × 10−13 | 17 |
| Immunological Disease | 4.90 × 10−3–5.25 × 10−13 | 18 |
| Organismal Injury and Abnormalities | 5.15 × 10−3–5.25 × 10−13 | 23 |
| Inflammatory Response | 5.15 × 10−3–3.81 × 10−10 | 20 |
| Molecular and Cellular Functions | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Cell-To-Cell Signaling and Interaction | 5.15 × 10−3–3.81 × 10−10 | 14 |
| Cellular Development | 5.15 × 10−3–6.04 × 10−10 | 20 |
| Cellular Growth and Proliferation | 5.15 × 10−3–6.04 × 10−10 | 20 |
| Cellular Function and Maintenance | 5.15 × 10−3–4.91 × 10−8 | 16 |
| Cellular Movement | 5.15 × 10−3–6.64 × 10−8 | 15 |
| Physiological System Development and Function | ||
| Name | p-Value Range ** | No. of Molecules ‡ |
| Haematological System Development and Function | 5.15 × 10−3–3.81 × 10−10 | 18 |
| Immune Cell Trafficking | 4.29 × 10−3–3.81 × 10−10 | 14 |
| Lymphoid Tissue Structure and Development | 5.15 × 10−3–3.63 × 10−9 | 15 |
| Tissue Morphology | 4.94 × 10−3–5.17 × 10−9 | 16 |
| Haematopoiesis | 5.15 × 10−3–3.17 × 10−7 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pillay, R.; Naidoo, P.; Mkhize-Kwitshana, Z.L. Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection. Microorganisms 2025, 13, 1734. https://doi.org/10.3390/microorganisms13081734
Pillay R, Naidoo P, Mkhize-Kwitshana ZL. Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection. Microorganisms. 2025; 13(8):1734. https://doi.org/10.3390/microorganisms13081734
Chicago/Turabian StylePillay, Roxanne, Pragalathan Naidoo, and Zilungile L. Mkhize-Kwitshana. 2025. "Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection" Microorganisms 13, no. 8: 1734. https://doi.org/10.3390/microorganisms13081734
APA StylePillay, R., Naidoo, P., & Mkhize-Kwitshana, Z. L. (2025). Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection. Microorganisms, 13(8), 1734. https://doi.org/10.3390/microorganisms13081734





