An Updated Perspective on the Aromatic Metabolic Pathways of Plant-Derived Homocyclic Aromatic Compounds in Aspergillus niger
Abstract
1. Introduction
2. Metabolism of Hydroxycinnamic Acids
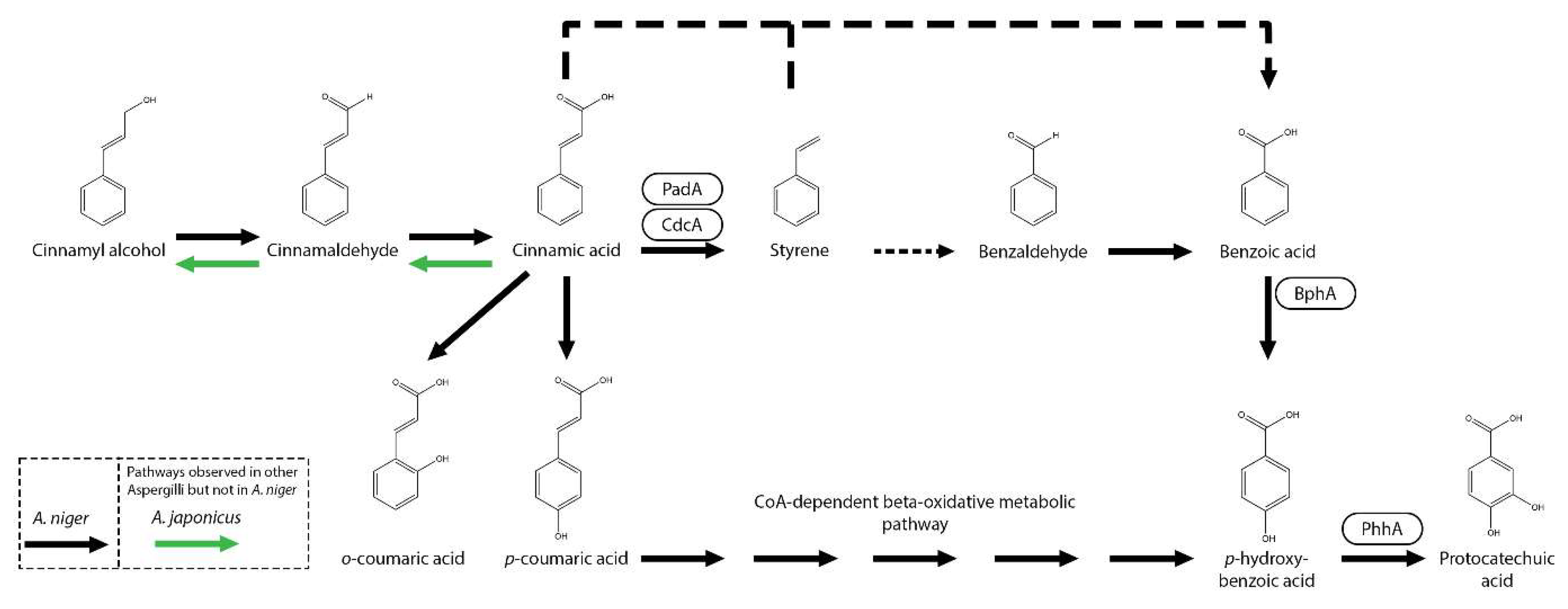
3. Metabolism of Cinnamic Acid
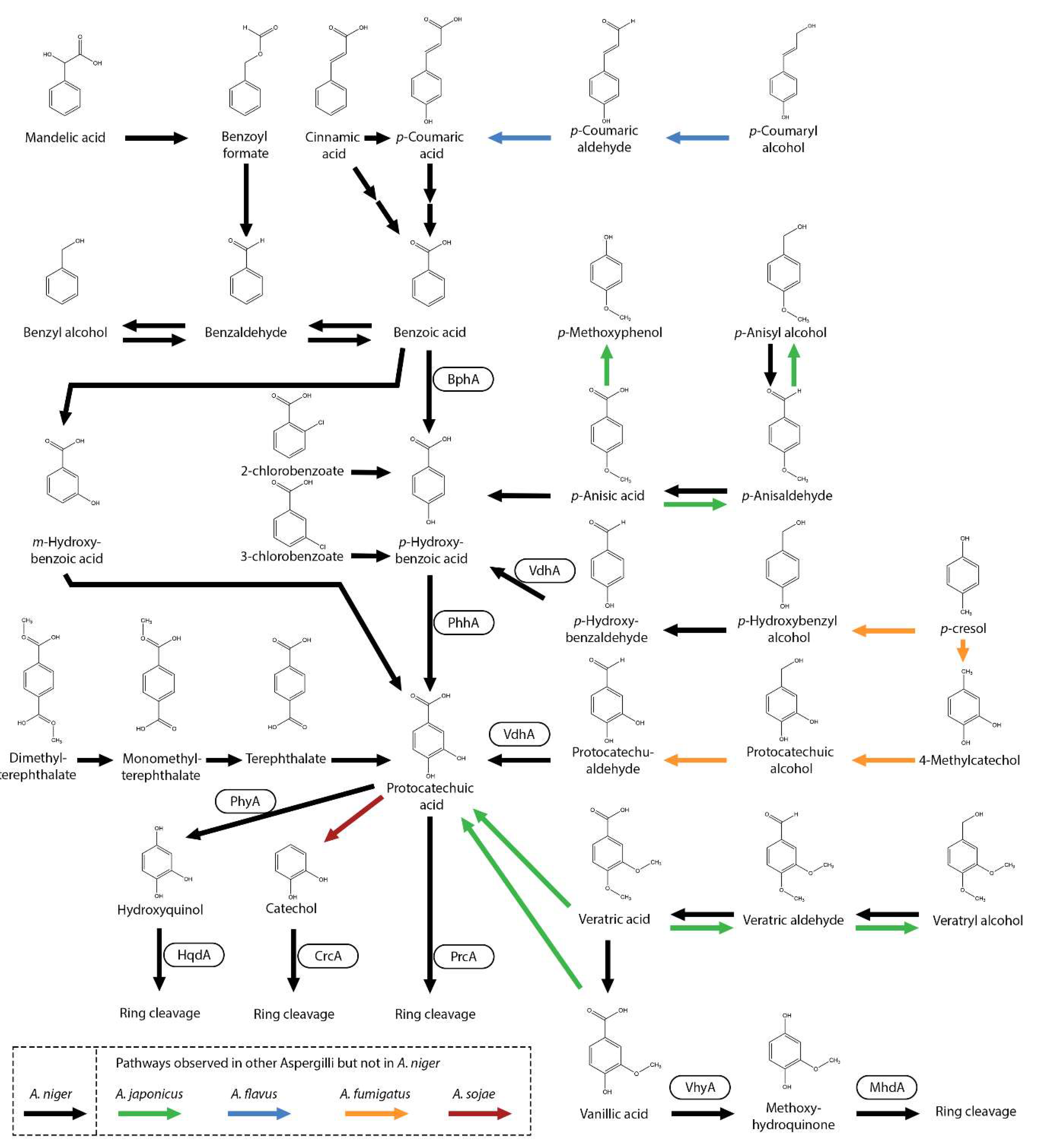
4. Metabolism of Benzoic Acid and Related Aromatic Compounds
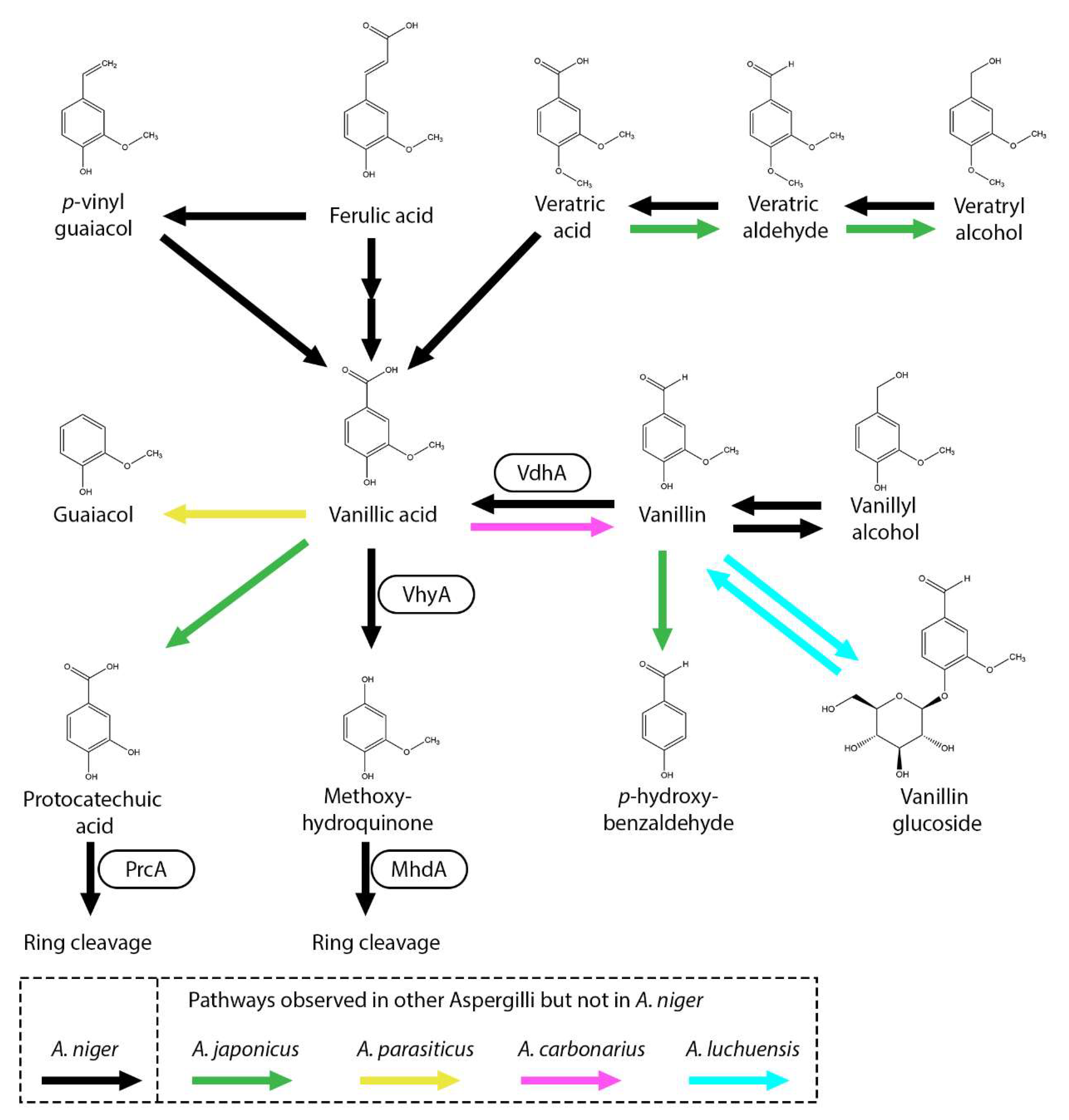
5. Metabolism of Guaiacyl Units and Related Aromatic Compounds
6. Conversion of Syringyl Units
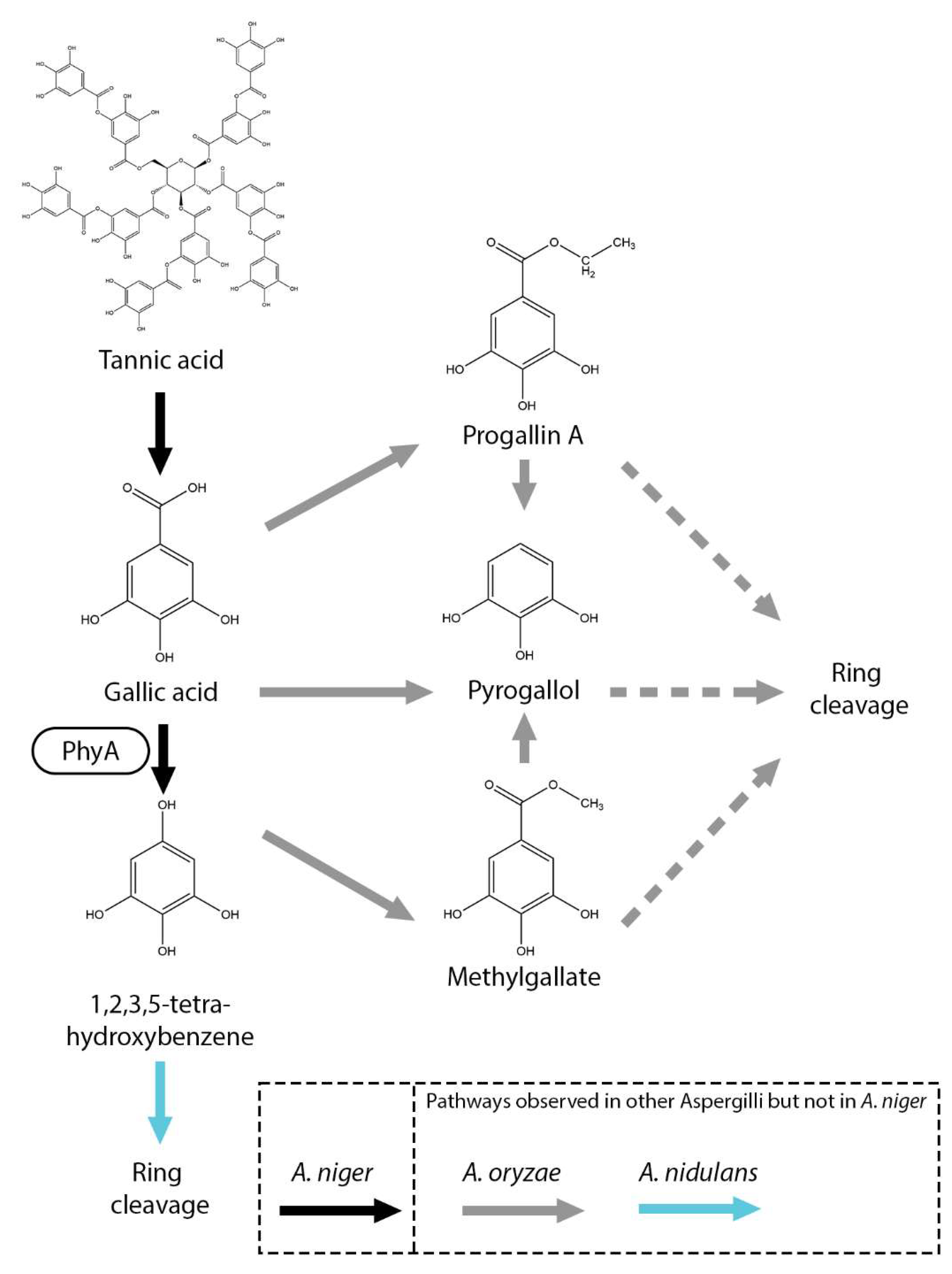
7. Gallic Acid Metabolic Pathways
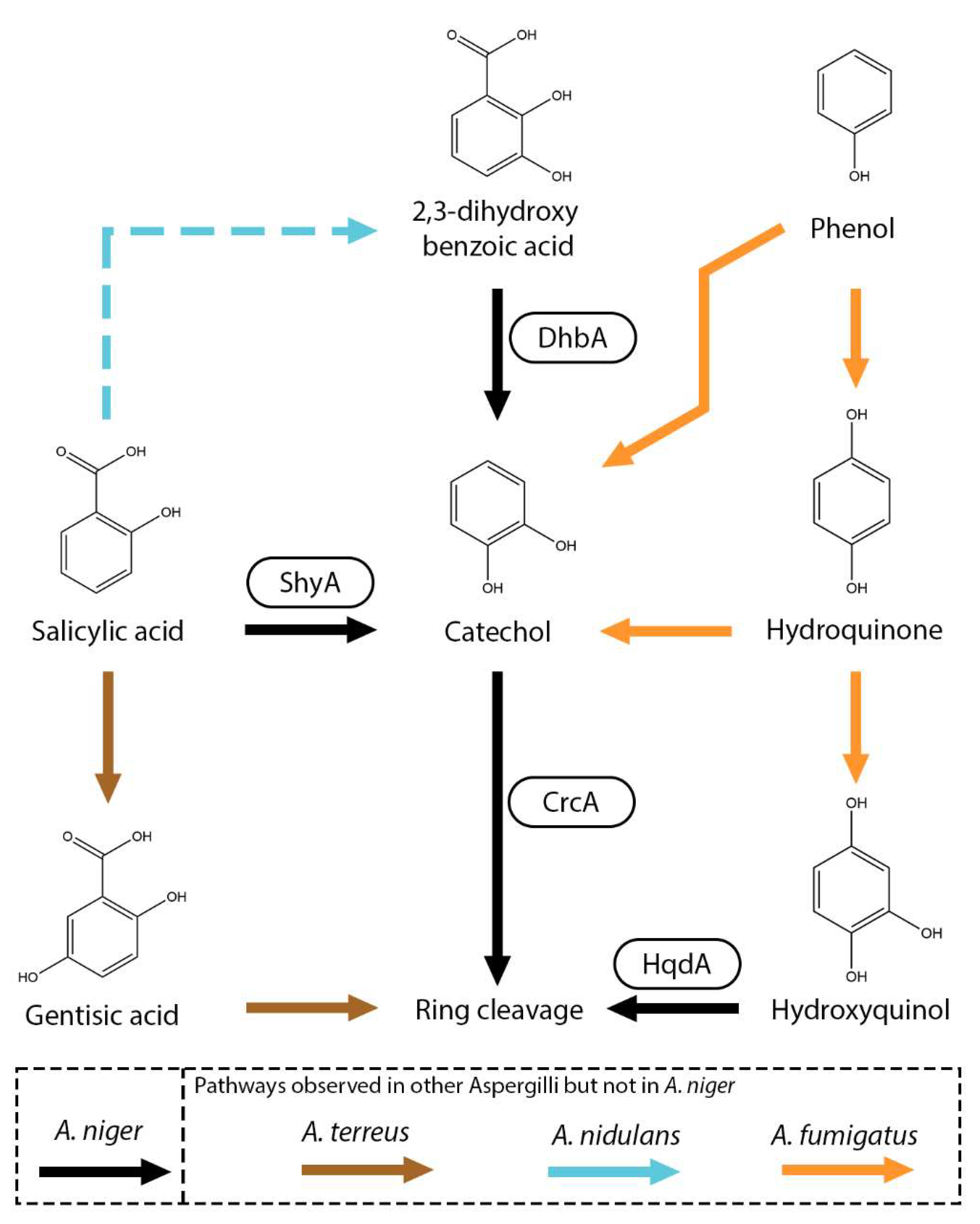
8. Salicylic Acid Metabolic Pathways
9. Ring Cleavage Pathways
10. Regulation of Aromatic Metabolic Pathways
11. Future Perspectives: Strategies for Studying and Identifying Aromatic Metabolic Pathways in Filamentous Fungi
12. Future Perspectives: Production and Accumulation of Aromatic Compounds Using Fungi
13. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Z.H.; Wang, Q.; Ruan, X.; Pan, C.D.; Jiang, D.A. Phenolics and plant allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef]
- Myers, R.L. The 100 Most Important Chemical Compounds, 2007th ed.; Bloomsbury Publishing: New York, NY, USA, 2007; Volume 1. [Google Scholar]
- Chen, F.; Tobimatsu, Y.; Havkin-Frenkel, D.; Dixon, R.A.; Ralph, J. A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 2012, 109, 1772–1777. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, J.; Zhao, D.; Jia, L.; Qin, B.; Cao, X.; Zang, L.; Lu, F.; Liu, F. Biological degradation of lignin: A critical review on progress and perspectives. Ind. Crops Prod. 2022, 188, 115715. [Google Scholar] [CrossRef]
- Atiwesh, G.; Parrish, C.C.; Banoub, J.; Le, T.A.T. Lignin degradation by microorganisms: A review. Biotechnol. Prog. 2022, 38, e3226. [Google Scholar] [CrossRef] [PubMed]
- Madadi, M.; Abbas, A. Lignin Degradation by Fungal Pretreatment: A Review. J. Plant Pathol. Microbiol. 2017, 8, 1–6. [Google Scholar]
- Pollegioni, L.; Tonin, F.; Rosini, E. Lignin-degrading enzymes. FEBS J. 2015, 282, 1190–1213. [Google Scholar] [CrossRef] [PubMed]
- Ralet, M.C.; Thibault, J.F.; Faulds, C.B.; Williamson, G. Isolation and purification of feruloylated oligosaccharides from cell walls of sugar-beet pulp. Carbohydr. Res. 1994, 263, 227–241. [Google Scholar] [CrossRef]
- Colquhoun, I.J.; Ralet, M.C.; Thibault, J.F.; Faulds, C.B.; Williamson, G. Structure identification of feruloylated oligosaccharides from sugar-beet pulp by NMR spectroscopy. Carbohydr. Res. 1994, 263, 243–256. [Google Scholar] [CrossRef]
- Ralet, M.C.; Faulds, C.B.; Williamson, G.; Thibault, J.F. Degradation of feruloylated oligosaccharides from sugar-beet pulp and wheat bran by ferulic acid esterases from Aspergillus niger. Carbohydr. Res. 1994, 263, 257–269. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Dilokpimol, A.; Visser, J.; Mäkelä, M.R.; Hildén, K.S.; de Vries, R.P. A comparison between the homocyclic aromatic metabolic pathways from plant-derived compounds by bacteria and fungi. Biotechnol. Adv. 2019, 37, 107396. [Google Scholar] [CrossRef]
- Milstein, O.; Vered, Y.; Shragina, L.; Gressel, J.; Flowers, H.M.; Hüttermann, A. Metabolism of lignin related aromatic compounds by Aspergillus japonicus. Arch. Microbiol. 1983, 135, 147–154. [Google Scholar] [CrossRef]
- Mäkelä, M.R.; Marinović, M.; Nousiainen, P.; Liwanag, A.J.M.; Benoit, I.; Sipilä, J.; Hatakka, A.; de Vries, R.P.; Hildén, K.S. Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv. Appl. Microbiol. 2015, 91, 63–137. [Google Scholar]
- Kasirajan, L.; Kamaraj, K.; Maupin-Furlow, J.A.; Uthandi, S. Isolation, purification, and identification of novel lignin-degrading Aspergillus caespitosus strain S2. Biomass Convers. Biorefin 2024, 14, 28685–28699. [Google Scholar] [CrossRef]
- Virmani, S.; Arora, A.; Kaushik, S.; Suman, A. Lignin degradation by isolated lignolytic Acinetobacter baumanii S2, Aspergillus niger SF4 and Rhodotorula glutinis and profiling products from bio-valorization perspective. Waste Biomass Valorization 2024, 15, 101–114. [Google Scholar] [CrossRef]
- Yang, Y.S.; Zhou, J.T.; Lu, H.; Yuan, Y.L.; Zhao, L.H. Isolation and characterization of a fungus Aspergillus sp. strain F-3 capable of degrading alkali lignin. Biodegradation 2011, 22, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.M.; Batista, L.R.; Rezende, E.F.; Fungaro, M.H.P.; Sartori, D.; Alves, E. Identification of fungi of the genus Aspergillus section Nigri using polyphasic taxonomy. Braz. J. Microbiol. 2011, 42, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Boschloo, J.G.; Moonen, E.; van Gorcom, R.F.M.; Hermes, H.F.M.; Bos, C.J. Genetic analysis of Aspergillus niger mutants defective in benzoate-4-hydroxylase function. Curr. Genet. 1991, 19, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Faber, B.W.; Van Gorcom, R.F.M.; Duine, J.A. Purification and characterization of benzoate-parahydroxylase, a cytochrome P450 (CYP53A1), from Aspergillus niger. Arch. Biochem. Biophys. 2001, 394, 245–254. [Google Scholar] [CrossRef]
- Boschloo, J.G.; Paffen, A.; Koot, T.; van den Tweel, W.J.J.; van Gorcom, R.F.M.; Cordewener, J.H.G.; Bos, C.J. Genetic analysis of benzoate metabolism in Aspergillus niger. Appl. Microbiol. Biotechnol. 1990, 34, 225–228. [Google Scholar] [CrossRef]
- Martins, T.M.; Hartmann, D.O.; Planchon, S.; Martins, I.; Renaut, J.; Silva Pereira, C. The old 3-oxoadipate pathway revisited: New insights in the catabolism of aromatics in the saprophytic fungus Aspergillus nidulans. Fungal Genet. Biol. 2015, 74, 32–44. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Dilokpimol, A.; Visser, J.; Hildén, K.S.; Mäkelä, M.R.; de Vries, R.P. Discovery and functional analysis of a salicylic acid hydroxylase from Aspergillus niger. Appl. Environ. Microbiol. 2021, 87, e02701-20. [Google Scholar] [CrossRef]
- Santha, R.; Savithri, H.S.; Rao, N.A.; Vaidyanathan, C.S. 2,3-Dihydroxybenzoic acid decarboxylase from Aspergillus niger: A novel decarboxylase. Eur. J. Biochem. 1995, 230, 104–110. [Google Scholar] [CrossRef]
- Subba Rao, P.V.; Moore, K.; Towers, G.H.N. O-Pyrocatechuic acid carboxy-lysase from Aspergillus niger. Arch. Biochem. Biophys. 1967, 122, 466–473. [Google Scholar]
- Plumridge, A.; Melin, P.; Stratford, M.; Novodvorska, M.; Shunburne, L.; Dyer, P.S.; Roubos, J.A.; Menke, H.; Stark, J.; Stam, H.; et al. The decarboxylation of the weak-acid preservative, sorbic acid, is encoded by linked genes in Aspergillus spp. Fungal Genet. Biol. 2010, 47, 683–692. [Google Scholar]
- Lubbers, R.J.M.; Dilokpimol, A.; Navarro, J.; Peng, M.; Wang, M.; Lipzen, A.; Ng, V.; Grigoriev, I.V.; Visser, J.; Hildén, K.S.; et al. Cinnamic acid and sorbic acid conversion are mediated by the same transcriptional regulator in Aspergillus niger. Front. Bioeng. Biotechnol. 2019, 7, 249. [Google Scholar] [CrossRef] [PubMed]
- Stratford, M.; Plumridge, A.; Pleasants, M.W.; Novodvorska, M.; Baker-Glenn, C.A.G.; Pattenden, G.; Archer, D.B. Mapping the structural requirements of inducers and substrates for decarboxylation of weak acid preservatives by the food spoilage mould Aspergillus niger. Int. J. Food Microbiol. 2012, 157, 375–383. [Google Scholar] [CrossRef][Green Version]
- Lubbers, R.J.M.; de Vries, R.P. Production of protocatechuic acid from p-hydroxyphenyl (H) units and related aromatic compounds using an Aspergillus niger cell factory. mBio 2021, 12, e00391-21. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Dilokpimol, A.; Peng, M.; Visser, J.; Mäkelä, M.R.; Hildén, K.S.; De Vries, R.P. Discovery of novel p-hydroxybenzoate-m-hydroxylase, protocatechuate 3,4 ring-cleavage dioxygenase, and hydroxyquinol 1,2 ring-cleavage dioxygenase from the filamentous fungus Aspergillus niger. ACS Sustain. Chem. Eng. 2019, 7, 19081–19089. [Google Scholar] [CrossRef]
- Semana, P.; Powlowski, J. Four aromatic intradiol ring cleavage dioxygenases from Aspergillus niger. Appl. Environ. Microbiol. 2019, 85, e01786-19. [Google Scholar] [CrossRef]
- Maeda, M.; Tokashiki, M.; Tokashiki, M.; Uechi, K.; Ito, S.; Taira, T. Characterization and induction of phenolic acid decarboxylase from Aspergillus luchuensis. J. Biosci. Bioeng. 2018, 126, 162–168. [Google Scholar] [CrossRef]
- Maeda, M.; Motosoko, M.; Tokashiki, T.; Tokashiki, J.; Mizutani, O.; Uechi, K.; Goto, M.; Taira, T. Phenolic acid decarboxylase of Aspergillus luchuensis plays a crucial role in 4-vinylguaiacol production during awamori brewing. J. Biosci. Bioeng. 2020, 130, 352–359. [Google Scholar] [CrossRef]
- Taira, J.; Toyoshima, R.; Ameku, N.; Iguchi, A.; Tamaki, Y. Vanillin production by biotransformation of phenolic compounds in fungus, Aspergillus luchuensis. AMB Express 2018, 8, 40. [Google Scholar] [CrossRef]
- Jones, K.H.; Trudgill, P.W.; Hopper, D.J. Metabolism of p-cresol by the fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 1993, 59, 1125–1130. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Martínez-Reyes, N.; Rhanama, N.; Nair, R.; Prieto, I.; Ihalainen, P.; Heikkilä, M.; de Vries, R.P. Vanillin dehydrogenase (VhdA) from Aspergillus niger is active on depolymerized lignin. Sustain. Chem. Environ. 2024, 8, 100179. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Dilokpimol, A.; Visser, J.; de Vries, R.P. Aspergillus niger uses the peroxisomal CoA-dependent β-oxidative genes to degrade the hydroxycinnamic acids caffeic acid, ferulic acid, and p-coumaric acid. Appl. Microbiol. Biotechnol. 2021, 105, 4199–4211. [Google Scholar] [CrossRef] [PubMed]
- Faulds, C.B.; Williamson, G. Release of ferulic acid from wheat bran by a ferulic acid esterase (FAE-III) from Aspergillus niger. Appl. Microbiol. Biotechnol. 1995, 43, 1082–1087. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, E.; Saulnier, L.; Brunel, M.; Marot, C.; Lesage-Meessen, L.; Asther, M.; Thibault, J.F. Release of ferulic acid from agroindustrial by-products by the cell wall-degrading enzymes produced by Aspergillus niger I-1472. Enzyme Microb. Technol. 2002, 31, 1000–1005. [Google Scholar] [CrossRef]
- Williamson, G.; Faulds, C.B.; Kroon, P.A. Specificity of ferulic acid (feruloyl) esterases. Biochem. Soc. Trans. 1998, 26, 205–209. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Mäkelä, M.R.; Aguilar-Pontes, M.V.; Benoit-Gelber, I.; Hildén, K.S.; de Vries, R.P. Diversity of fungal feruloyl esterases: Updated phylogenetic classification, properties, and industrial applications. Biotechnol. Biofuels 2016, 9, 231. [Google Scholar] [CrossRef]
- Dilokpimol, A.; Mäkelä, M.R.; Mansouri, S.; Belova, O.; Waterstraat, M.; Bunzel, M.; de Vries, R.P.; Hildén, K.S. Expanding the feruloyl esterase gene family of Aspergillus niger by characterization of a feruloyl esterase, FaeC. New Biotechnol. 2017, 37, 200–209. [Google Scholar] [CrossRef]
- de Vries, R.P.; Visser, J. Regulation of the feruloyl esterase (faeA) gene from Aspergillus niger. Appl. Environ. Microbiol. 1999, 65, 5500–5503. [Google Scholar] [CrossRef]
- de Vries, R.P.; vanKuijk, P.A.; Kester, H.C.M.; Visser, J. The Aspergillus niger faeB gene encodes a second feruloyl esterase involved in pectin and xylan degradation and is specifically induced in the presence of aromatic compounds. Biochem. J. 2002, 386, 377–386. [Google Scholar] [CrossRef]
- Todokoro, T.; Negoro, H.; Kotaka, A.; Hata, Y.; Ishida, H. Aspergillus oryzae FaeA is responsible for the release of ferulic acid, a precursor of off-odor 4-vinylguaiacol in sake brewing. J. Biosci. Bioeng. 2022, 133, 140–145. [Google Scholar] [CrossRef]
- Edlin, D.A.N.; Narbad, A.; Dickinson, J.R.; Lloyd, D. The biotransformation of simple phenolic compounds by Brettanomyces anomalus. FEMS Microbiol. Lett. 1995, 125, 311–315. [Google Scholar] [CrossRef]
- Linke, D.; Riemer, S.J.L.; Schimanski, S.; Nieter, A.; Krings, U.; Berger, R.G. Cold generation of smoke flavour by the first phenolic acid decarboxylase from a filamentous ascomycete—Isaria farinosa. Fungal Biol. 2017, 121, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Baqueiro-Peña, I.; Rodríguez-Serrano, G.; González-Zamora, E.; Augur, C.; Loera, O.; Saucedo-Castañeda, G. Biotransformation of ferulic acid to 4-vinylguaiacol by a wild and a diploid strain of Aspergillus niger. Bioresour. Technol. 2010, 101, 4721–4724. [Google Scholar] [CrossRef] [PubMed]
- Nazareth, S.; Mavinkurve, S. Degradation of ferulic acid via 4-vinylguaiacol by Fusarium solani (Mart.) Sacc. Can. J. Microbiol. 1986, 32, 494–497. [Google Scholar] [CrossRef]
- Zhang, P.H.; Yu, X.Y.; Weng, L.X.; Sun, L.L.; Mao, Z.C.; Zhang, Y.L. Degradation of ferulic acid by the endophytic fungus Colletotrichum gloeosporioides TMTM-13 associated with Ostrya rehderiana Chun. ACS Omega 2019, 4, 21000–21004. [Google Scholar] [CrossRef]
- Plumridge, A.; Stratford, M.; Lowe, K.C.; Archer, D.B. The weak-acid preservative sorbic acid is decarboxylated and detoxified by a phenylacrylic acid decarboxylase, PadA1, in the spoilage mold Aspergillus niger. Appl. Environ. Microbiol. 2008, 74, 550–552. [Google Scholar] [CrossRef]
- Tian, G.; Liu, Y. Mechanistic insights into the catalytic reaction of ferulic acid decarboxylase from Aspergillus niger: A QM/MM study. Phys. Chem. Chem. Phys. 2017, 19, 7733–7742. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Dudareva, N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Fosså, A.; Beyer, A.; Pfitzner, E.; Wenzel, B.; Kunau, W.H. Molecular cloning, sequencing and sequence analysis of the fox-2 gene of Neurospora crassa encoding the multifunctional β-oxidation protein. MGG Mol. General Genet. 1995, 247, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Requena, N.; Füller, P.; Franken, P. Molecular characterization of GmFOX2, an evolutionarily highly conserved gene from the mycorrhizal fungus Glomus mosseae, down-regulated during interaction with rhizobacteria. Mol. Plant-Microbe Interact. 1999, 12, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Maggio-Hall, L.A.; Keller, N.P. Mitochondrial β-oxidation in Aspergillus nidulans. Mol. Microbiol. 2004, 54, 1173–1185. [Google Scholar] [CrossRef]
- Klose, J.; Kronstad, J.W. The multifunctional β-oxidation enzyme is required for full symptom development by the biotrophic maize pathogen Ustilago maydis. Eukaryot. Cell 2006, 5, 2047–2061. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Soanes, D.M.; Kershaw, M.J.; Talbot, N.J. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid β-oxidation during appressorium-mediated plant infection. Mol. Plant-Microbe Interact. 2007, 20, 475–491. [Google Scholar] [CrossRef]
- Boisnard, S.; Espagne, E.; Zickler, D.; Bourdais, A.; Riquet, A.L.; Berteaux-Lecellier, V. Peroxisomal ABC transporters and β-oxidation during the life cycle of the filamentous fungus Podospora anserina. Fungal Genet. Biol. 2009, 46, 55–66. [Google Scholar] [CrossRef]
- Bocks, S.M. Fungal Metabolism-I. the transformations of coumarin, o-coumaric acid and trans-cinnamic acid by Aspergillus niger. Phytochemistry 1967, 6, 127–130. [Google Scholar] [CrossRef]
- Bye, A.; King, H.K. The biosynthesis of 4-hydroxycoumarin and dicoumarol by Aspergillus fumigatus Fresenius. Biochem. J. 1970, 117, 237–245. [Google Scholar] [CrossRef]
- Bye, A.; Ashton, W.M.; King, H.K. A proposed mechanism for the conversion of o-coumaric acid to 4-hydroxy-coumarin in Aspergillus fumigatus fresenius. Biochem. Biophys. Res. Commun. 1968, 32, 94–97. [Google Scholar] [CrossRef]
- Iyayi, C.B.; Dart, R.K. The degradation of p-coumaryl alcohol by Aspergillus flavus. J. Gen. Microbiol. 1982, 128, 1473–1482. [Google Scholar] [CrossRef]
- Catarine Santos Rodrigues, C.; Latércia Tranches Dias, A.; de Oliveira Silva, E. Unprecedented derivatization of ferulic acid through selective methoxylation by Aspergillus brasiliensis ATCC 16404. Biocatal. Biotransform. 2019, 37, 233–237. [Google Scholar] [CrossRef]
- Pinches, S.E.; Apps, P. Production in food of 1,3-pentadiene and styrene by Trichoderma species. Int. J. Food Microbiol. 2007, 116, 182–185. [Google Scholar] [CrossRef]
- Pagot, Y.; Belin, J.M.; Husson, F.; Spinnler, H.E. Metabolism of phenylalanine and biosynthesis of styrene in Penicillium camemberti. J. Dairy Res. 2007, 74, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Lafeuille, J.L.; Buniak, M.L.; Vioujas, M.C.; Lefevre, S. Natural formation of styrene by cinnamon mold flora. J. Food Sci. 2009, 74, M276–M283. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.G.; Dai, C.C. Biodegradation of a model allelochemical cinnamic acid by a novel endophytic fungus Phomopsis liquidambari. Int. Biodeterior. Biodegradation 2015, 104, 498–507. [Google Scholar] [CrossRef]
- Konzock, O.; Tous-Mohedano, M.; Cibin, I.; Chen, Y.; Norbeck, J. Cinnamic acid and p-coumaric acid are metabolized to 4-hydroxybenzoic acid by Yarrowia lipolytica. AMB Express 2023, 13, 84. [Google Scholar] [CrossRef]
- Said, S.; Neves, F.M.; Griffiths, A.J.F. Cinnamic acid inhibits the growth of the fungus Neurospora crassa, but is eliminated as acetophenone. Int. Biodeterior. Biodegrad. 2004, 54, 1–6. [Google Scholar] [CrossRef]
- Ma, L.; Liu, X.; Liang, J.; Zhang, Z. Biotransformations of cinnamaldehyde, cinnamic acid and acetophenone with Mucor. World J. Microbiol. Biotechnol. 2011, 27, 2133–2137. [Google Scholar] [CrossRef]
- Nimura, Y.; Tsujiyama, S.-i.; Ueno, M. Bioconversion of cinnamic acid derivatives by Schizophyllum commune. J. General Appl. Microbiol. 2010, 56, 381–387. [Google Scholar] [CrossRef]
- Krebs, H.A.; Wiggins, D.; Stubbs, M. Studies on the mechanism of the antifungal action of benzoate. Biochem. J. 1983, 214, 657–663. [Google Scholar] [CrossRef]
- del Olmo, A.; Calzada, J.; Nuñez, M. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017, 57, 3084–3103. [Google Scholar] [CrossRef]
- van Gorcom, R.F.M.; Boschloo, J.G.; Kuijvenhoven, A.; Lange, J.; van Vark, A.J.; Bos, C.J.; van Balken, J.A.M.; Pouwels, P.H.; van den Hondel, C.A.M.J.J. Isolation and molecular characterisation of the benzoate-para-hydroxylase gene (bphA) of Aspergillus niger: A member of a new gene family of the cytochrome P450 superfamily. MGG Mol. General Genet. 1990, 223, 192–197. [Google Scholar] [CrossRef]
- Yuasa, K.; Ishlzuka, K.; Kaburaki, S.; Sakasal, T. Metabolism of phenylalanine in Aspergillus sojae. Agric. Biol. Chem. 1975, 39, 2199–2206. [Google Scholar] [CrossRef]
- Martins, T.M.; Bento, A.; Martins, C.; Tomé, A.S.; Moreira, C.J.S.; Silva Pereira, C. Bringing up to date the toolkit for the catabolism of aromatic compounds in fungi: The unexpected 1,2,3,5-tetrahydroxybenzene central pathway. Microb. Biotechnol. 2024, 17, e14371. [Google Scholar] [CrossRef]
- Bocks, S.M. Fungal metabolism-III. The hydroxylation of anisole, phenoxyacetic acid, phenylacetic acid and benzoic acid by Aspergillus niger. Phytochemistry 1967, 6, 785–789. [Google Scholar] [CrossRef]
- Shailubhai, K.; Sahasrabudhe, S.R.; Vora, K.A.; Modi, V.V. Degradation of chlorinated derivatives of phenoxyacetic acid and benzoic acid by Aspergillus niger. FEMS Microbiol. Lett. 1983, 18, 279–282. [Google Scholar] [CrossRef]
- Jamaluddin, M.; Rao, P.V.; Vaidyanathan, C.S. Involvement of the protocatechuate pathway in the metabolism of mandelic acid by Aspergillus niger. J. Bacteriol. 1970, 101, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Ganji, S.H.; Karigar, C.S.; Pujar, B.G. Metabolism of dimethylterephthalate by Aspergillus niger. Biodegradation 1995, 6, 61–66. [Google Scholar] [CrossRef]
- Lubbers, R.J.M.; Dilokpimol, A.; Nousiainen, P.A.; Cioc, R.C.; Visser, J.; Bruijnincx, P.C.A.; de Vries, R.P. Vanillic acid and methoxyhydroquinone production from guaiacyl units and related aromatic compounds using Aspergillus niger cell factories. Microb. Cell Fact. 2021, 20, 151. [Google Scholar] [CrossRef]
- Palazzolo, M.A.; Mascotti, M.L.; Lewkowicz, E.S.; Kurina-Sanz, M. Self-sufficient redox biotransformation of lignin-related benzoic acids with Aspergillus flavus. J. Ind. Microbiol. Biotechnol. 2015, 42, 1581–1589. [Google Scholar] [CrossRef]
- Raman, T.S.; Shanmugasundaram, E.R. Metabolism of some aromatic acids by Aspergillus niger. J. Bacteriol. 1962, 84, 1339–1340. [Google Scholar] [CrossRef] [PubMed]
- Buswell, J.A.; Eriksson, K.E. Vanillate hydroxylase from Sporotrichum pulverulentum. Methods Enzymol. 1988, 161, 274–281. [Google Scholar] [PubMed]
- Rahouti, M.; Seigle-Murandi, F.; Steiman, R.; Eriksson, K.E. Metabolism of ferulic acid by Paecilomyces variotii and Pestalotia palmarum. Appl. Environ. Microbiol. 1989, 55, 2391–2398. [Google Scholar] [CrossRef] [PubMed]
- Guiraud, P.; Steiman, R.; Seigle-Murandi, F.; Benoit-Guyod, J.L. Metabolism of vanillic acid by Micromycetes. World J. Microbiol. Biotechnol. 1992, 8, 270–275. [Google Scholar] [CrossRef]
- Seth, M.; Chand, S. Biosynthesis of tannase and hydrolysis of tannins to gallic acid by Aspergillus awamori—Optimisation of process parameters. Process Biochem. 2000, 36, 39–44. [Google Scholar] [CrossRef]
- Lokeswari, N.; Jaya Raju, K. Optimization of gallic acid production from Terminalia chebula by Aspergillus niger. E-J. Chem. 2007, 4, 287–293. [Google Scholar] [CrossRef][Green Version]
- Mukherjee, G.; Banerjee, R. Biosynthesis of tannase and gallic acid from tannin rich substrates by Rhizopus oryzae and Aspergillus foetidus. J. Basic Microbiol. 2004, 44, 42–48. [Google Scholar] [CrossRef]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic acid: A versatile antioxidant with promising therapeutic and industrial applications. R. Soc. Chem. Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Guo, D.; Zhang, Z.; Liu, D.; Zheng, H.; Chen, H.; Chen, K. A comparative study on the degradation of gallic acid by Aspergillus oryzae and Phanerochaete chrysosporium. Water Sci. Technol. 2014, 70, 175–181. [Google Scholar] [CrossRef]
- Arentshorst, M.; Falco, M.D.; Moisan, M.C.; Reid, I.D.; Spaapen, T.O.M.; van Dam, J.; Demirci, E.; Powlowski, J.; Punt, P.J.; Tsang, A.; et al. Identification of a conserved transcriptional activator-repressor module controlling the expression of genes involved in tannic acid degradation and gallic acid utilization in Aspergillus niger. Front. Fungal Biol. 2021, 2, 681631. [Google Scholar] [CrossRef]
- Shah, J. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 2003, 6, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Martins, T.M.; Martins, C.; Guedes, P.; Silva Pereira, C. Twists and turns in the salicylate catabolism of Aspergillus terreus, revealing new roles of the 3-hydroxyanthranilate pathway. mSystems 2021, 6, e00230-20. [Google Scholar] [CrossRef] [PubMed]
- Penn, C.D.; Daniel, S.L. Salicylate degradation by the fungal plant pathogen Sclerotinia sclerotiorum. Curr. Microbiol. 2013, 67, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, K.V.; Tian, Z.; Wang, Y.; Smith, J.; Zylstra, G.; Huang, B.; Belanger, F.C. Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci. Rep. 2015, 5, 10939. [Google Scholar] [CrossRef]
- Rocheleau, H.; Al-harthi, R.; Ouellet, T. Degradation of salicylic acid by Fusarium graminearum. Fungal Biol. 2019, 123, 77–86. [Google Scholar] [CrossRef]
- Choi, S.; Lee, H.N.; Park, E.; Lee, S.J.; Kim, E.S. Recent advances in microbial production of cis,cis-muconic acid. Biomolecules 2020, 10, 1238. [Google Scholar] [CrossRef]
- Bachman, D.M.; Dragoon, B.; John, S. Reduction of salicylate to saligenin by Neurospora. Arch. Biochem. Biophys. 1960, 91, 326. [Google Scholar] [CrossRef]
- Sgro, M.; Chow, N.; Olyaei, F.; Arentshorst, M.; Geoffrion, N.; Ram, A.F.J.; Powlowski, J.; Tsang, A. Functional analysis of the protocatechuate branch of the β-ketoadipate pathway in Aspergillus niger. J. Biol. Chem. 2023, 299, 105003. [Google Scholar] [CrossRef]
- Vaillancourt, F.; Bolin, J.; Eltis, L. The ins and outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 241–267. [Google Scholar] [CrossRef]
- Kowalczyk, J.E.; Benoit, I.; De Vries, R.P. Regulation of Plant Biomass Utilization in Aspergillus, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2014; Volume 88. [Google Scholar]
- Seekles, S.J.; van Dam, J.; Arentshorst, M.; Ram, A.F.J. Natural variation and the role of Zn2 Cys6 Transcription factors SdrA, WarA and WarB in sorbic acid resistance of Aspergillus niger. Microorganisms 2022, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Hynes, M.J.; Murray, S.L.; Duncan, A.; Khew, G.S.; Davis, M.A. Regulatory genes controlling fatty acid catabolism and peroxisomal functions in the filamentous fungus Aspergillus nidulans. Eukaryot. Cell 2006, 5, 794–805. [Google Scholar] [CrossRef]
- Hynes, M.J.; Murray, S.L.; Khew, G.S.; Davis, M.A. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 2008, 178, 1355–1369. [Google Scholar] [CrossRef]
- Arentshorst, M.; Reijngoud, J.; van Tol, D.J.C.; Reid, I.D.; Arendsen, Y.; Pel, H.J.; van Peij, N.N.M.E.; Visser, J.; Punt, P.J.; Tsang, A.; et al. Utilization of ferulic acid in Aspergillus niger requires the transcription factor FarA and a newly identified Far-like protein (FarD) that lacks the canonical Zn(II)2Cys6 domain. Front. Fungal Biol. 2022, 3, 978845. [Google Scholar] [CrossRef]
- Lesage-Meessen, L.; Stentelaire, C.; Lomascolo, A.; Couteau, D.; Asther, M.; Moukha, S.; Record, E.; Sigoillot, J.C.; Asther, M. Fungal transformation of ferulic acid from sugar beet pulp to natural vanillin. J. Sci. Food Agric. 1999, 79, 487–490. [Google Scholar] [CrossRef]
- Fleige, C.; Meyer, F.; Steinbüchel, A. Metabolic engineering of the actinomycete Amycolatopsis sp. strain ATCC 39116 towards enhanced production of natural vanillin. Appl. Environ. Microbiol. 2016, 82, 3410–3419. [Google Scholar] [CrossRef]

| Enzyme | Aspergillus Species | Year Identified | Evidence 1 | Phenotype Observed on Aromatic Compounds When the Gene Is Deleted | Observed Enzymatic Activity of the Recombinant Protein Toward Aromatic Compounds | References |
|---|---|---|---|---|---|---|
| Benzoate-p-hydroxylase (BphA) | A. niger, A. nidulans | 1990 | IDA, IEP, IMP | Benzoic acid, cinnamic acid, benzaldehyde, benzyl alcohol | Benzoic acid, 2-fluorobenzoic acid, 2-chlorobenzoic acid, salicylic acid, 2-methylbenzoic acid, 3-fluorobenzoic acid, 3-chlorobenzoic acid, 3-methylbenzoic acid, 3-methoxybenzoic acid | [18,19,20,21] |
| 2,3-dihydroxybenzoate decarboxylase (DhbA) | A. niger, A. nidulans | 1995 | IDA, IEP, IMP | 2,3-Dihydroxybenzoic acid | 2,3-Dihydroxybenzoic acid | [21,22,23,24] |
| Cinnamic acid decarboxylase (CdcA) Ferulic acid decarboxylase (FdcA) | A. niger | 2010 | IDA, IEP, IMP | Cinnamic acid | Cinnamic acid, caffeic acid, ferulic acid, p-coumaric acid | [25,26,27] |
| p-hydroxybenzoate-m-hydroxylase (PhhA) | A. niger, A. nidulans | 2015 | IDA, IEP, IMP | Benzoic acid, benzaldehyde, benzyl alcohol, cinnamic acid, p-hydroxybenzoic acid, p-coumaric acid, p-anisic acid, p-anisyl alcohol, | p-Hydroxybenzoic acid | [21,28,29] |
| Protocatechuate 3,4-dioxygenase (PrcA) | A. niger, A. nidulans | 2015 | EXP, IDA, IEP, IMP | Benzoic acid, benzaldehyde, benzyl alcohol, cinnamic acid, m-hydroxybenzoic acid, p-anisic acid, p-anisyl alcohol, p-coumaric acid, p-hydroxybenzoic acid, protocatechuic acid, protocatechuic aldehyde | Protocatechuic acid | [21,28,29,30] |
| Phenolic acid decarboxylase (PadA) | A. luchuensis | 2018 | IDA, IEP | n.d. | Caffeic acid, ferulic acid, p-coumaric acid | [31,32,33] |
| Protocatechuate hydroxylase (PhyA) | A. niger, A. nidulans | 2021 | IDA, IEP, IMP | Protocatechuic acid, gallic acid | Protocatechuic acid | [28,29] |
| Vanilliate hydroxylase (VhyA) | A. niger | 2021 | EXP, IDA, IEP, IMP | Ferulic acid, vanillic acid, vanillin | Vanillic acid | [34] |
| Vanillin dehydrogenase (VdhA) | A. niger | 2021 | IDA, IEP, IMP | Vanillin, p-hydroxybenzaldehyde, protocatechuic aldehyde | Vanillin, benzaldehyde, p-hydroxybenzaldehyde, protocatechuic aldehyde, syringic aldehyde, cinnamyl aldehyde, p-anisyl aldehyde, veratryl aldehyde | [34,35] |
| Methoxyhydroquinone 1,2-dioxygenase (MhdA) | A. niger | 2021 | IDA, IEP, IMP | Coniferyl alcohol, ferulic acid, vanillic acid, vanillin, vanillyl alcohol, veratric acid | Methoxyhydroquinone | [34] |
| Salicylate hydroxylase (ShyA) | A. niger, A. nidulans | 2021 | EXP, IDA, IEP, IMP | Salicylic acid | 4-Aminosalicylic acid, 2,3-dihydroxybenzoic acid, gentisic acid, salicylic acid | [21,22] |
| Catechol 1,2-dioxygenase (CrcA) | A. niger, A. nidulans | 2021 | EXP, IDA, IEP, IMP | Salicylic acid, 2,3-dihydroxybenzoic acid, catechol | Catechol, hydroxyquinol, 3-methylcatechol, 4-methylcatechol | [21,22,30] |
| Hydroxycinnamate-CoA synthase (HcsA) | A. niger | 2021 | IEP, IMP | Ferulic acid, p-coumaric acid, caffeic acid, m-coumaric acid, dihydroferulic acid, dihydrocaffeic acid, phloretic acid | n.d. | [36] |
| Fatty acid oxidase (FoxA) | A. niger | 2021 | IEP, IMP | Ferulic acid, p-coumaric acid, caffeic acid, m-coumaric acid, dihydroferulic acid, dihydrocaffeic acid, phloretic acid | n.d. | [36] |
| 3-ketoacyl-CoA thiolase (KatA) | A. niger | 2021 | IEP, IMP | Ferulic acid, p-coumaric acid, caffeic acid, m-coumaric acid, dihydroferulic acid, dihydrocaffeic acid, phloretic acid, 4-methoxycinnamic acid | n.d. | [36] |
| Thioesterases (TheA, TheB, TheC, TheD) | A. niger | 2021 | IEP, IMP | Ferulic acid, p-coumaric acid, caffeic acid, m-coumaric acid, dihydroferulic acid, dihydrocaffeic acid, phloretic acid | n.d. | [36] |
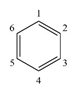 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aromatic Compound | 1 | 2 | 3 | 4 | 5 | 6 | ΔprcA [65] | ΔhqdA [65] | ΔprcA/hqdA [65] | ΔcrcA [34] | ΔmhdA [81] | Δ5330 [34] | Δ17 [92] |
| H-unit-related compounds | |||||||||||||
| Cinnamic acid | CH=CHC=OOH | H | H | H | H | H | ++ | − | ++ | − | − | − | n.d. |
| p-coumaric acid | CH=CHC=OOH | H | H | OH | H | H | + | − | ++ | − | − | − | ++ |
| Caffeic acid | CH=CHC=OOH | H | OH | OH | H | H | + | − | ++ | − | − | − | ++ |
| Benzaldehyde | CH=O | H | H | H | H | H | ++ | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| Benzoic acid | C=OOH | H | H | H | H | H | ++ | − | ++ | n.d. | − | n.d. | n.d. |
| Benzyl alcohol | CH2OH | H | H | H | H | H | ++ | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| p-hydroxybenzoic acid | C=OOH | H | H | OH | H | H | ++ | − | ++ | n.d. | − | n.d. | ++ |
| p-hydroxybenzaldehyde | CH=O | H | H | OH | H | H | ++ | − | ++ | n.d. | n.d. | n.d. | n.d. |
| m-hydroxybenzoic acid | C=OOH | H | OH | H | H | H | ++ | n.d. | n.d. | n.d. | − | n.d. | n.d. |
| Protocatechuic acid | C=OOH | H | OH | OH | H | H | + | − | ++ | − | − | − | ++ |
| Protocatechuic aldehyde | CH=O | H | OH | OH | H | H | + | − | ++ | n.d. | − | n.d. | n.d. |
| p-anisic acid | C=OOH | H | H | OCH3 | H | H | ++ | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| p-anisyl alcohol | CH2OH | H | H | OCH3 | H | H | ++ | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| G-unit-related compounds | |||||||||||||
| Coniferyl alcohol | CH=CHCH2=OH | H | OCH3 | OH | H | H | n.d. | n.d. | n.d. | n.d. | ++ | n.d. | n.d. |
| Ferulic acid | CH=CHC=OOH | H | OCH3 | OH | H | H | − | − | − | − | ++ | − | − |
| Vanillic acid | C=OOH | H | OCH3 | OH | H | H | − | − | − | n.d. | ++ | n.d. | − |
| Vanillin | CH=O | H | OCH3 | OH | H | H | n.d. | n.d. | n.d. | n.d. | ++ | n.d. | n.d. |
| Vanillyl alcohol | CH2OH | H | OCH3 | OH | H | H | n.d. | n.d. | n.d. | n.d. | ++ | n.d. | n.d. |
| Veratric acid | C=OOH | H | OCH3 | OCH3 | H | H | − | n.d. | n.d. | n.d. | + | n.d. | n.d. |
| S-unit-related compounds | |||||||||||||
| Gallic acid | C=OOH | H | OH | OH | OH | H | − | − | − | n.d. | n.d. | n.d. | − |
| Others | |||||||||||||
| Salicylic acid | C=OOH | OH | H | H | H | H | − | − | n.d. | ++ | n.d. | − | ++ |
| 2,3-dihydroxybenzoic acid | C=OOH | OH | OH | H | H | H | − | − | n.d. | ++ | n.d. | − | n.d. |
| Catechol | OH | OH | H | H | H | H | − | − | − | ++ | n.d. | − | ++ |
| Gentisic acid | C=OOH | OH | H | H | OH | H | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lubbers, R.J.M. An Updated Perspective on the Aromatic Metabolic Pathways of Plant-Derived Homocyclic Aromatic Compounds in Aspergillus niger. Microorganisms 2025, 13, 1718. https://doi.org/10.3390/microorganisms13081718
Lubbers RJM. An Updated Perspective on the Aromatic Metabolic Pathways of Plant-Derived Homocyclic Aromatic Compounds in Aspergillus niger. Microorganisms. 2025; 13(8):1718. https://doi.org/10.3390/microorganisms13081718
Chicago/Turabian StyleLubbers, Ronnie J. M. 2025. "An Updated Perspective on the Aromatic Metabolic Pathways of Plant-Derived Homocyclic Aromatic Compounds in Aspergillus niger" Microorganisms 13, no. 8: 1718. https://doi.org/10.3390/microorganisms13081718
APA StyleLubbers, R. J. M. (2025). An Updated Perspective on the Aromatic Metabolic Pathways of Plant-Derived Homocyclic Aromatic Compounds in Aspergillus niger. Microorganisms, 13(8), 1718. https://doi.org/10.3390/microorganisms13081718






