Virulence, Antibiotic Resistance and Cytotoxic Effects of Lactococcus lactis Isolated from Chinese Cows with Clinical Mastitis on MAC-T Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Farm Distribution and Clinical Mastitis Sample Collection
2.2. Bacterial Isolation and Culture
2.3. 16S rDNA Sequencing for Identification and Biochemical Detection
2.4. Growth Curve of Lactococcus lactis
2.5. Determination of Antimicrobial Resistance
2.6. Primary Screening for Virulence Factors
2.7. Genome Sequencing, Assembly, Annotation and Bioinformatics Analysis
2.8. Cell Culture
2.9. Cell Viability Using the CCK-8 Method
2.10. Lactate Dehydrogenase (LDH) Release Assay
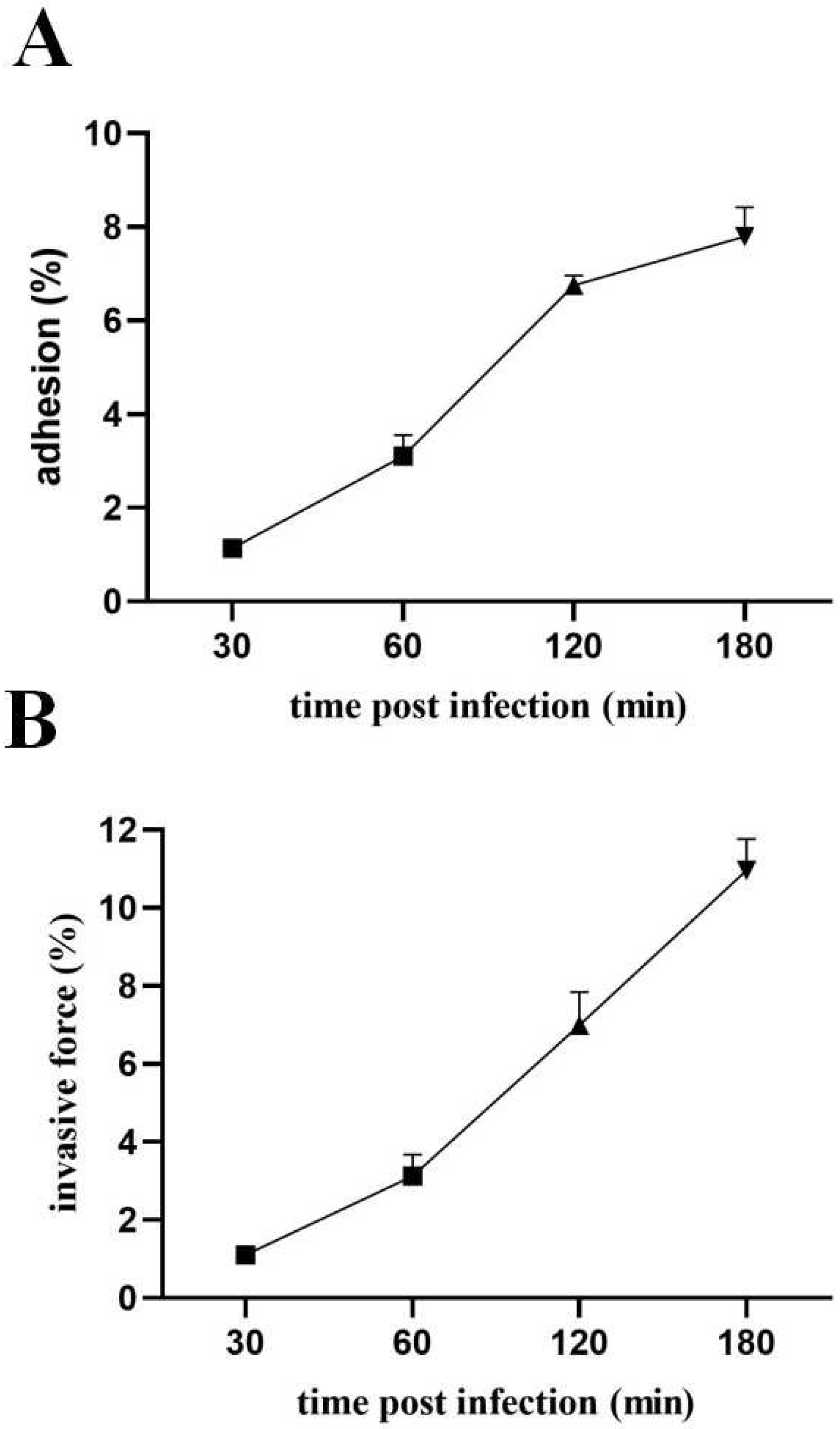
2.11. Adhesion and Invasiveness Measurements
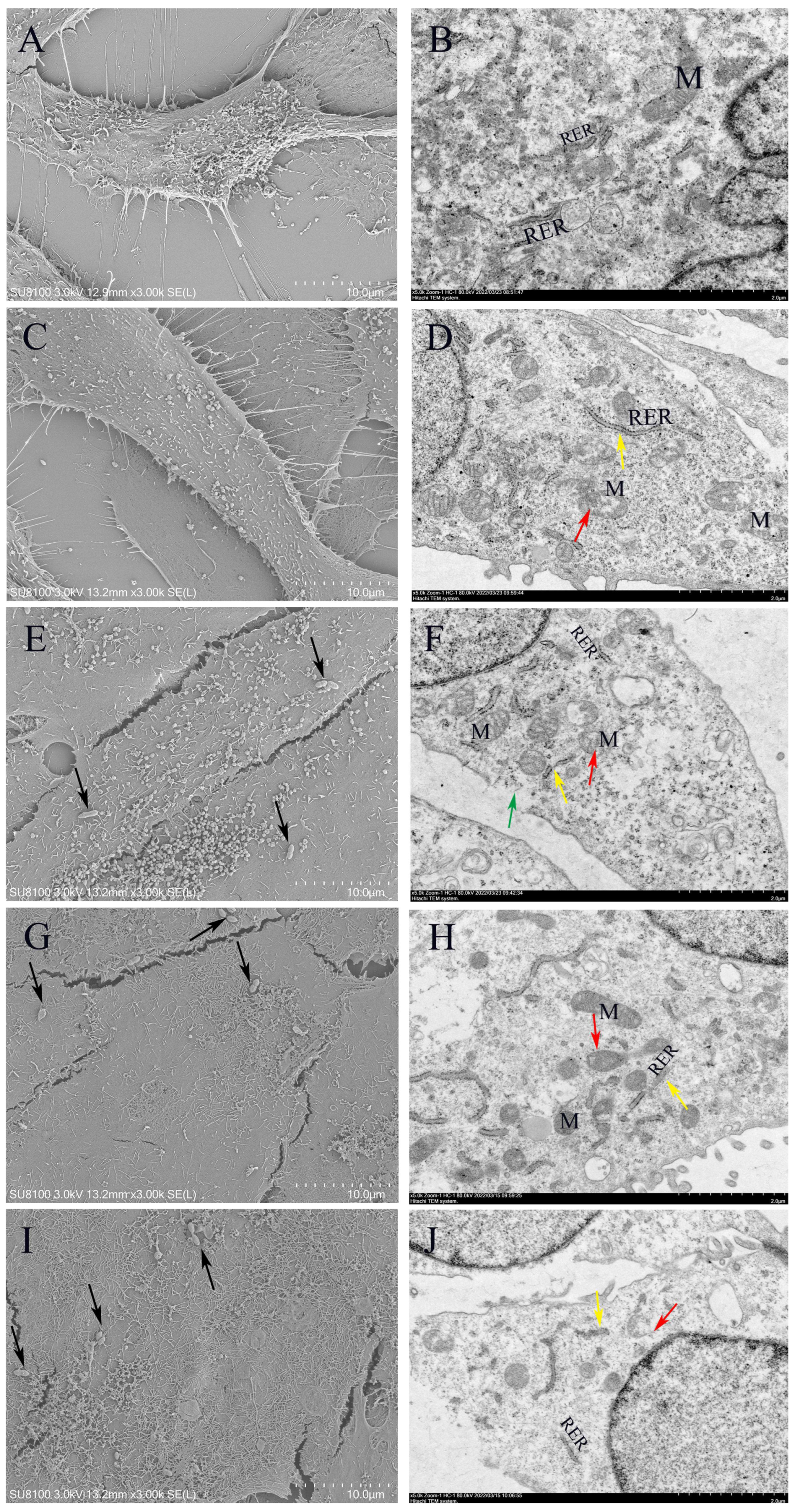
2.12. Cell Morphology and Ultrastructure Analysis
3. Results
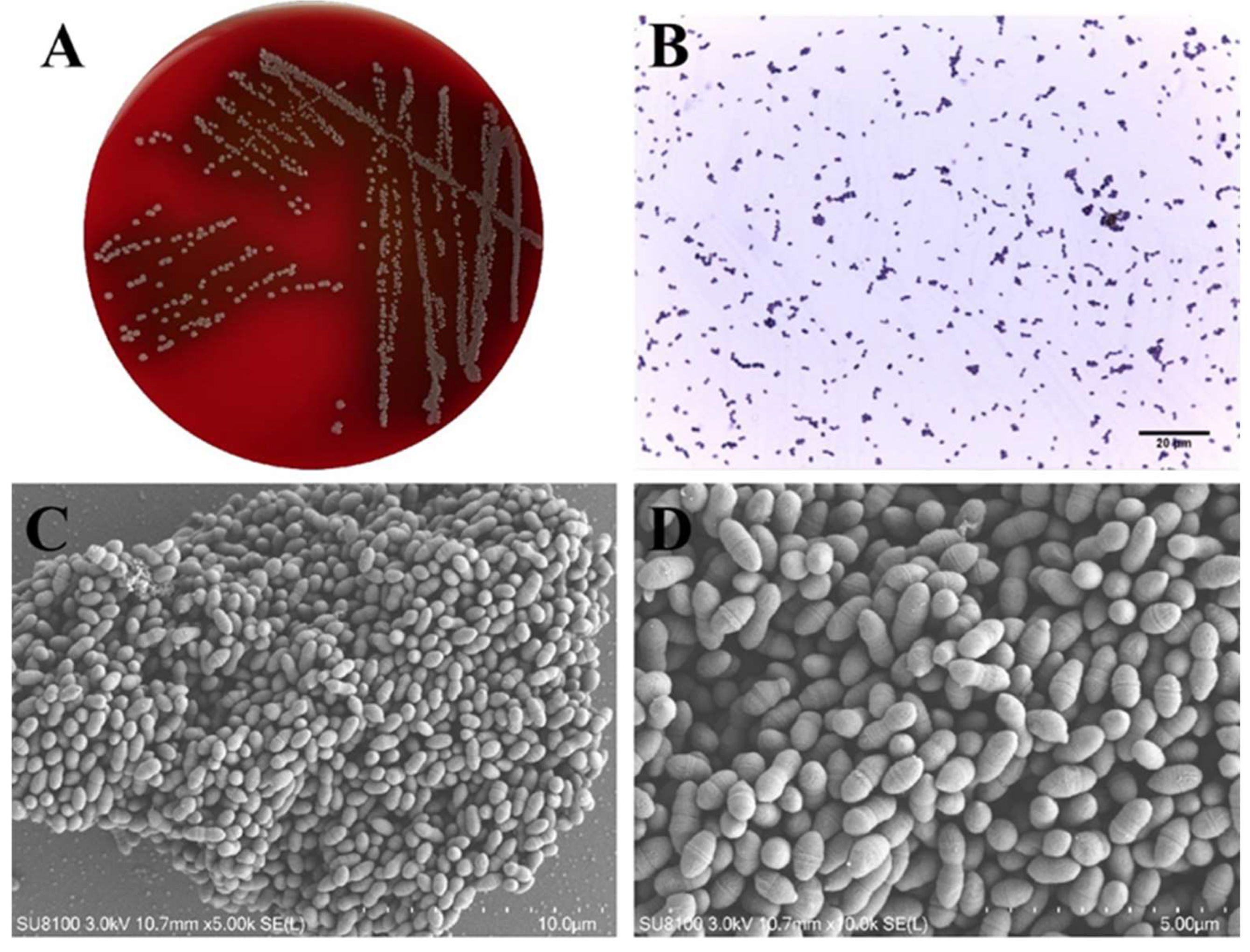
3.1. Morphological Characteristics of Lactococcus lactis
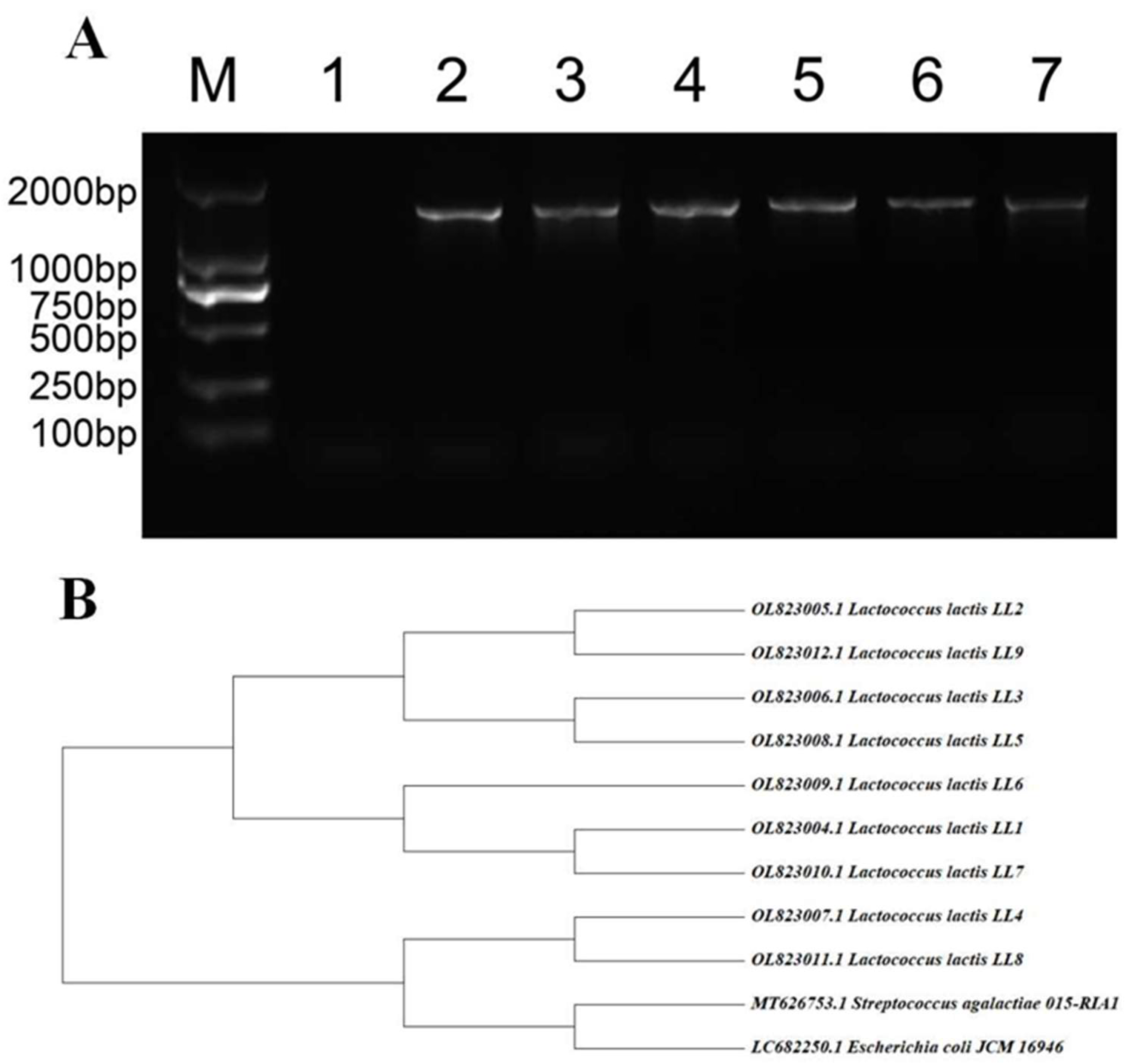
3.2. Results of 16S rRNA Identification and Homology Analysis
3.3. Biochemical Detection of Lactococcus lactis
3.4. Growth Ability of Lactococcus lactis
3.5. Antimicrobial Resistance Profiles of Lactococcus lactis
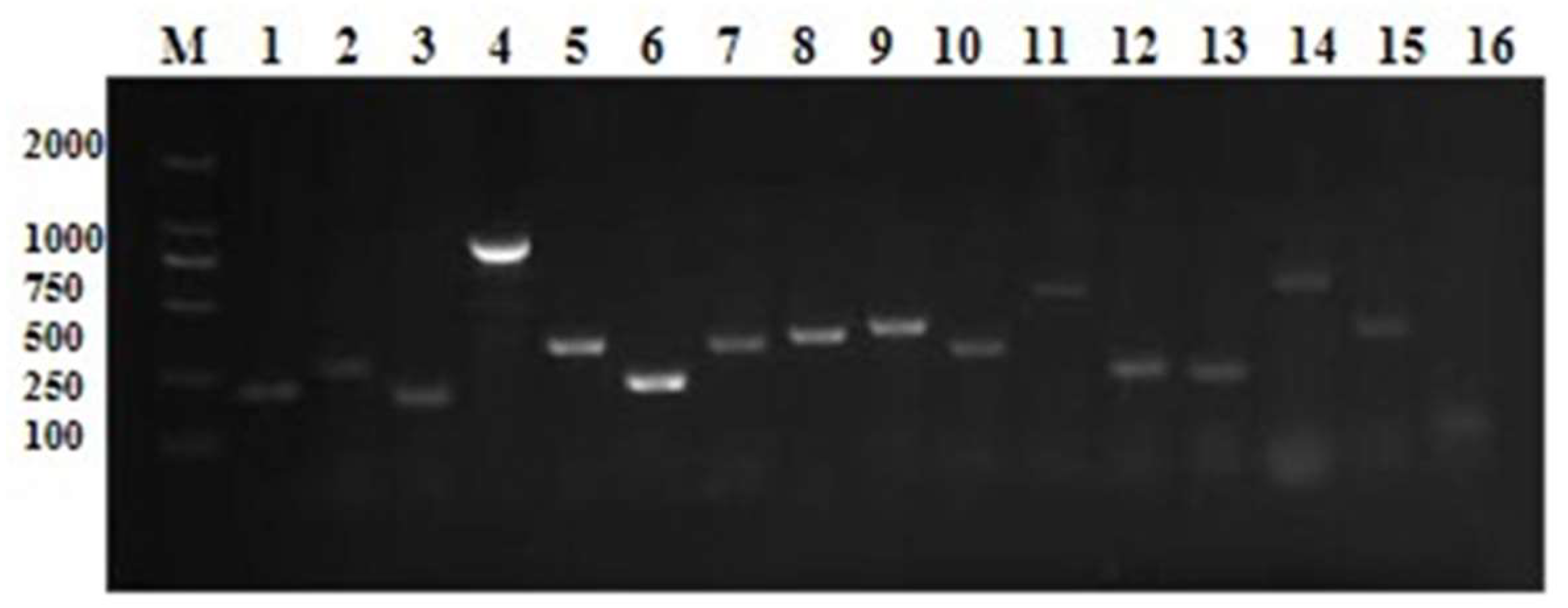
3.6. Lactococcus lactis Virulence Factor Primary Screening
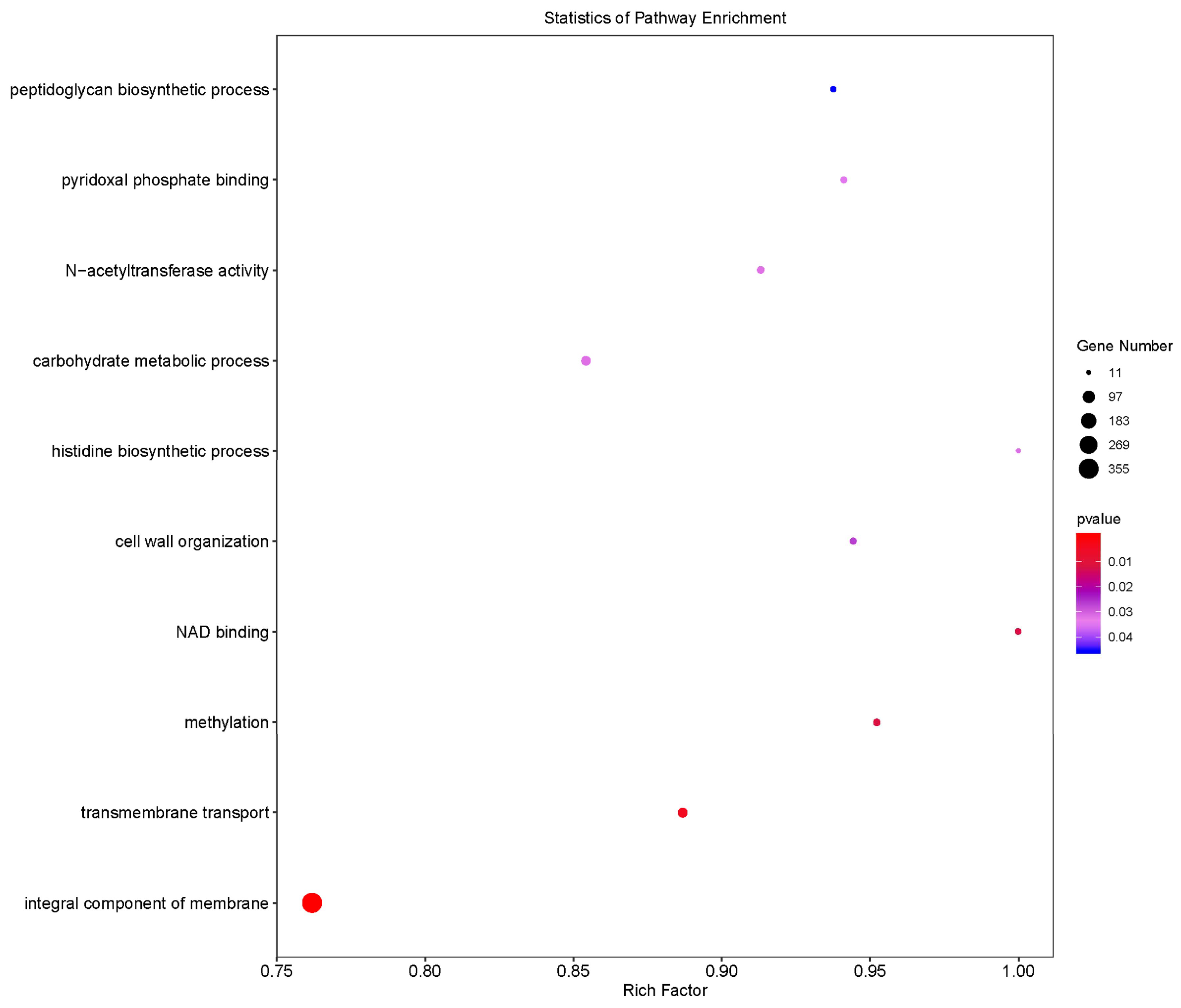
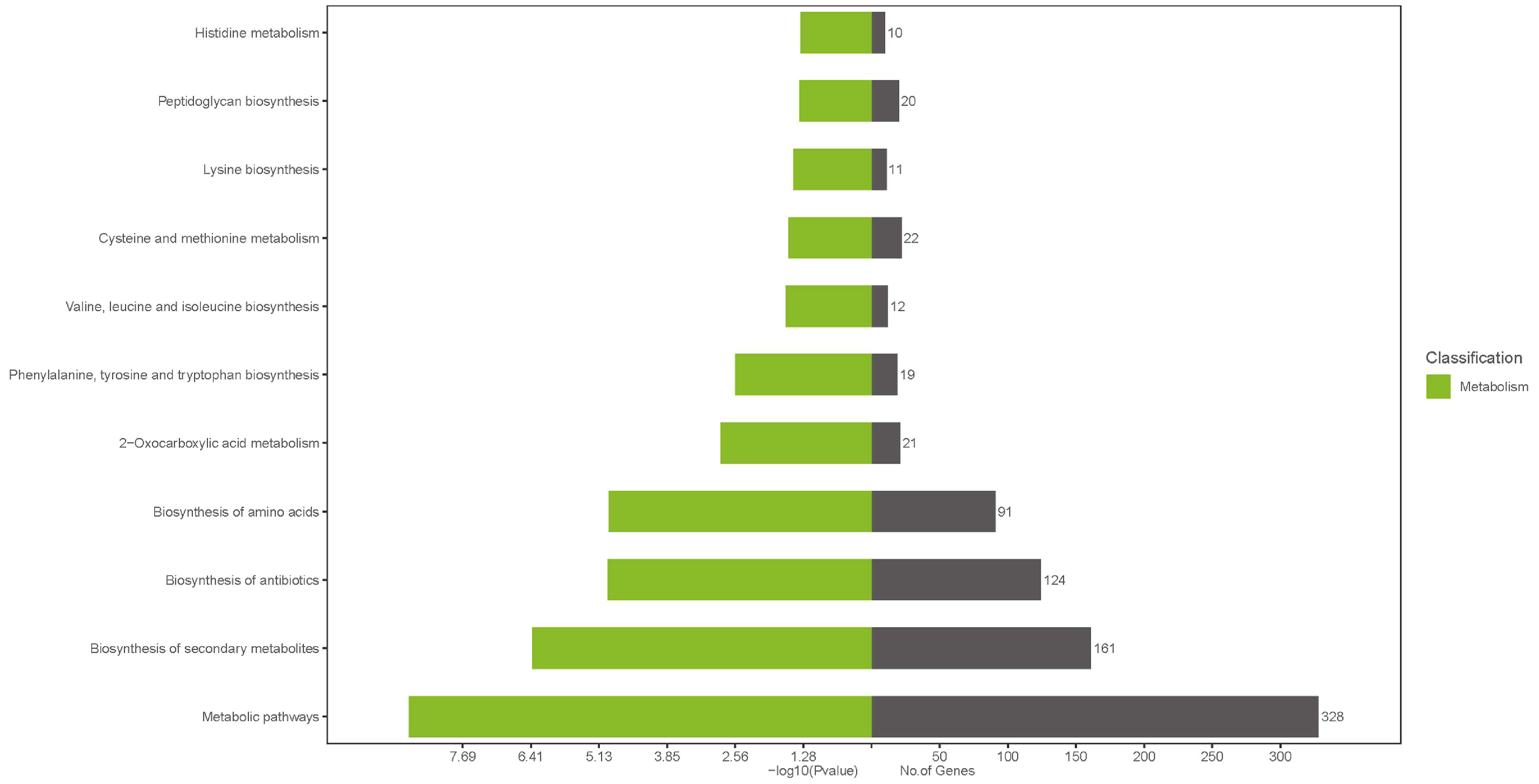
3.7. Bioinformatics Analysis of the Lactococcus lactis Genome
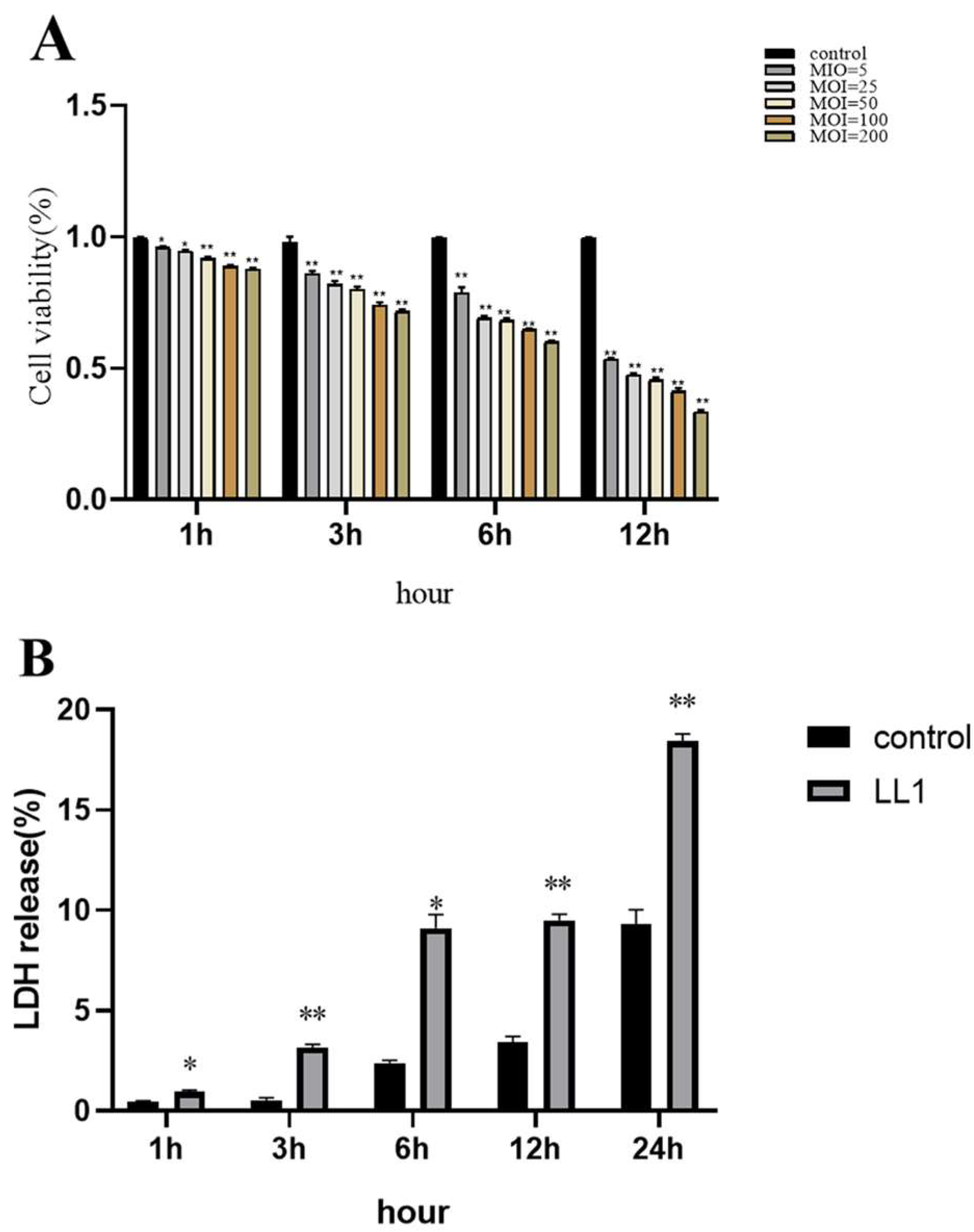
3.8. Effect of Lactococcus lactis on MAC-T Cell Viability
3.9. Pathogenic Effects of Lactococcus lactis on MAC-T
3.10. Morphology of Lactococcus lactis on MAC-T
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, D.; Beck, B.R.; Heo, S.B.; Kim, J.; Kim, H.D.; Lee, S.M.; Kim, Y.; Oh, S.Y.; Lee, K.; Do, H.; et al. Lactococcus lactis Bfe920 Activates the Innate Immune System of Olive Flounder (Paralichthys olivaceus), Resulting in Protection against Streptococcus iniae Infection and Enhancing Feed Efficiency and Weight Gain in Large-Scale Field Studies. Fish Shellfish. Immunol. 2013, 35, 1585–1590. [Google Scholar] [CrossRef] [PubMed]
- Haapala, V.; Pohjanvirta, T.; Vähänikkilä, N.; Halkilahti, J.; Simonen, H.; Pelkonen, S.; Soveri, T.; Simojoki, H.; Autio, T. Semen as a Source of Mycoplasma Bovis Mastitis in Dairy Herds. Vet. Microbiol. 2018, 216, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Devriese, L.A.; Hommez, J.; Laevens, H.; Pot, B.; Vandamme, P.; Haesebrouck, F. Identification of Aesculin-Hydrolyzing Streptococci, Lactococci, Aerococci and Enterococci from Subclinical Intramammary Infections in Dairy Cows. Vet. Microbiol. 1999, 70, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Werner, B.; Moroni, P.; Gioia, G.; Lavín-Alconero, L.; Yousaf, A.; Charter, M.E.; Carter, B.M.; Bennett, J.; Nydam, D.V.; Welcome, F.; et al. Short Communication: Genotypic and Phenotypic Identification of Environmental Streptococci and Association of Lactococcus lactis ssp. Lactis with Intramammary Infections among Different Dairy Farms. J. Dairy Sci. 2014, 97, 6964–6969. [Google Scholar] [CrossRef] [PubMed]
- Kok, J.; van Gijtenbeek, L.A.; de Jong, A.; van der Meulen, S.B.; Solopova, A.; Kuipers, O.P. The Evolution of Gene Regulation Research in Lactococcus lactis. FEMS Microbiol. Rev. 2017, 41 (Suppl. S1), S220–S243. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; McAuliffe, O.E.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P. Plasmids of Lactococci—Genetic Accessories or Genetic Necessities? FEMS Microbiol. Rev. 2006, 30, 243–273. [Google Scholar] [CrossRef] [PubMed]
- Casalta, E.; Montel, M.C. Safety Assessment of Dairy Microorganisms: The Lactococcus Genus. Int. J. Food Microbiol. 2008, 126, 271–273. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, A.; Hase, R.; Suzuki, D.; Toguchi, A.; Otsuka, Y.; Hirata, N.; Hosokawa, N. Lactococcus lactis Cholangitis and Bacteremia Identified by Maldi-Tof Mass Spectrometry: A Case Report and Review of the Literature on Lactococcus lactis Infection. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2019, 25, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xie, X.; Du, T.; Jiang, X.; Miao, W.; Wang, T. Lactococcus lactis, a Bacterium with Probiotic Functions and Pathogenicity. World J. Microbiol. Biotechnol. 2023, 39, 325. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.X.; Cao, H.; Jiang, S.; Li, X.; Fung, K.K.; Lee, C.H.; Sridhar, S.; Chen, J.-K.; Ho, P.L. Genomic Investigation of Lactococcus Formosensis, Lactococcus Garvieae, and Lactococcus Petauri Reveals Differences in Species Distribution by Human and Animal Sources. Microbiol. Spectr. 2024, 12, e0054124. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Pan, Z.; Yu, Y.; Yu, L.; Wu, F.; Dong, J.; Wang, T.; Li, L. Prevalence, Virulence, and Antibiotics Gene Profiles in Lactococcus Garvieae Isolated from Cows with Clinical Mastitis in China. Microorganisms 2023, 11, 379. [Google Scholar] [CrossRef] [PubMed]
- Vanaporn, M.; Wand, M.; Michell, S.L.; Sarkar-Tyson, M.; Ireland, P.; Goldman, S.; Kewcharoenwong, C.; Rinchai, D.; Lertmemongkolchai, G.; Titball, R.W. Superoxide Dismutase C Is Required for Intracellular Survival and Virulence of Burkholderia Pseudomallei. Microbiology 2011, 157 Pt 8, 2392–2400. [Google Scholar] [CrossRef] [PubMed]
- Stipcevic, T.; Piljac, T.; Isseroff, R.R. Di-Rhamnolipid from Pseudomonas Aeruginosa Displays Differential Effects on Human Keratinocyte and Fibroblast Cultures. J. Dermatol. Sci. 2005, 40, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Wear, S.S.; Sande, C. Assembly of Bacterial Capsular Polysaccharides and Exopolysaccharides. Annu. Rev. Microbiol. 2020, 74, 521–543. [Google Scholar] [CrossRef] [PubMed]
- Gale, R.T.; Li, F.K.K.; Sun, T.; Strynadka, N.C.J.; Brown, E.D.B. subtilis LytR-CpsA-Psr Enzymes Transfer Wall Teichoic Acids from Authentic Lipid-Linked Substrates to Mature Peptidoglycan In Vitro. Cell Chem. Biol. 2017, 24, 1537–1546.e4. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial Biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Toh, H.; Oshima, K.; Yoshizaki, M.; Kawanishi, M.; Nakaya, K.; Suzuki, T.; Miyauchi, E.; Ishii, Y.; Tanabe, S.; et al. Complete Genome Sequence and Comparative Analysis of the Fish Pathogen Lactococcus Garvieae. PLoS ONE 2011, 6, e23184. [Google Scholar] [CrossRef] [PubMed]
- Eraclio, G.; Ricci, G.; Moroni, P.; Santisteban, C.; Plumed-Ferrer, C.; Bennett, J.; Fortina, M.G. Sand Bedding as a Reservoir for Lactococcus Garvieae Dissemination in Dairy Farms. Can. J. Microbiol. 2019, 65, 84–89. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.; Kamel, M. Bovine Mastitis Prevention and Control in the Post-Antibiotic Era. Trop. Anim. Health Prod. 2021, 53, 236. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Fan, C.; Zhang, Z.; Li, S.; Xu, C.; Zhao, Y.; Han, L.; Zhang, D.; Liu, M. Enterococcal Isolates from Bovine Subclinical and Clinical Mastitis: Antimicrobial Resistance and Integron-Gene Cassette Distribution. Microb. Pathog. 2019, 129, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Poutrel, B.; Stegemann, M.R.; Roy, O.; Pothier, F.; Tilt, N.; Payne-Johnson, M. Evaluation of the Efficacy of Systemic Danofloxacin in the Treatment of Induced Acute Escherichia Coli Bovine Mastitis. J. Dairy Res. 2008, 75, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ma, H.; Song, C.; Fan, S.; Fan, H.; Zhou, W.; Cao, J. Prevalence and Molecular Characterization of Multi-Resistant Escherichia Coli Isolates from Clinical Bovine Mastitis in China. Anim. Biotechnol. 2024, 35, 2322541. [Google Scholar] [CrossRef] [PubMed]
- Plumed-Ferrer, C.; Uusikylä, K.; Korhonen, J.; von Wright, A. Characterization of Lactococcus lactis Isolates from Bovine Mastitis. Vet. Microbiol. 2013, 167, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Pires, A.J.; Pereira, G.; Fangueiro, D.; Bexiga, R.; Oliveira, M. When the Solution Becomes the Problem: A Review on Antimicrobial Resistance in Dairy Cattle. Future Microbiol. 2024, 19, 903–929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, Z.; Huang, C.; Gao, X.; Wang, Z.; Liu, Y.; Tian, C.; Hong, W.; Niu, S.; Liu, M. The Phylogenetic Group, Antimicrobial Susceptibility, and Virulence Genes of Escherichia Coli from Clinical Bovine Mastitis. J. Dairy Sci. 2018, 101, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hou, S.; Zhang, Q.; Ma, Y.; Zhang, Y.; Kan, W.; Zhao, X. Prevalence of Virulence and Resistance to Antibiotics in Pathogenic Enterococci Isolated from Mastitic Cows. J. Vet. Med. Sci. 2016, 78, 1663–1668. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Chen, Y.; Qiu, C.; Guo, M.Y. Lncrnas Transcriptome Analysis Revealed Potential Mechanisms of Selenium to Mastitis in Dairy Cows. Biol. Trace Elem. Res. 2022, 200, 4316–4324. [Google Scholar] [CrossRef] [PubMed]
- Timonen, A.A.E.; Katholm, J.; Petersen, A.; Orro, T.; Mõtus, K.; Kalmus, P. Elimination of Selected Mastitis Pathogens During the Dry Period. J. Dairy Sci. 2018, 101, 9332–9338. [Google Scholar] [CrossRef] [PubMed]
- Galgano, M.; Pellegrini, F.; Mrenoshki, D.; Addante, L.; Sposato, A.; Del Sambro, L.; Capozzi, L.; Catalano, E.; Solito, M.; D’Amico, F.; et al. Inhibition of Biofilm Production and Determination of in Vitro Time-Kill Thymus vulgaris L. Essential Oil (Teo) for the Control of Mastitis in Small Ruminants. Pathogens 2025, 14, 412. [Google Scholar] [CrossRef] [PubMed]
- Bausewein, M.; Mansfeld, R.; Doherr, M.G.; Harms, J.; Sorge, U.S. Sensitivity and Specificity for the Detection of Clinical Mastitis by Automatic Milking Systems in Bavarian Dairy Herds. Animals 2022, 12, 2131. [Google Scholar] [CrossRef] [PubMed]
- Su, F.J.; Chen, M.M. Protective Efficacy of Novel Oral Biofilm Vaccines against Lactococcus Garvieae Infection in Mullet, Mugil Cephalus. Vaccines 2021, 9, 844. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.S.; Frola, I.D.; Natanael, B.; Gobelli, D.; Nader-Macias, M.E.F.; Bogni, C.I. In Vitro Characterization of Lactic Acid Bacteria Isolated from Bovine Milk as Potential Probiotic Strains to Prevent Bovine Mastitis. Probiotics Antimicrob. Proteins 2019, 11, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Stoffels, K.; Broos, J.; Kuipers, O.P. Altering Specificity and Enhancing Stability of the Antimicrobial Peptides Nisin and Rombocin through Dehydrated Amino Acid Residue Engineering. Peptides 2024, 174, 171152. [Google Scholar] [CrossRef] [PubMed]
- Ture, M.; Altinok, I. Detection of Putative Virulence Genes of Lactococcus Garvieae. Dis. Aquat. Org. 2016, 119, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Hwanhlem, N.; Biscola, V.; El-Ghaish, S.; Jaffrès, E.; Dousset, X.; Haertlé, T.; H-Kittikun, A.; Chobert, J.M. Bacteriocin-Producing Lactic Acid Bacteria Isolated from Mangrove Forests in Southern Thailand as Potential Bio-Control Agents: Purification and Characterization of Bacteriocin Produced by Lactococcus lactis subsp. Lactis Kt2w2l. Probiotics Antimicrob. Proteins 2013, 5, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Tenea, G.N. Metabiotics Signature through Genome Sequencing and in Vitro Inhibitory Assessment of a Novel Lactococcus lactis Strain Utncys6-1 Isolated from Amazonian Camu-Camu Fruits. Int. J. Mol. Sci. 2023, 24, 6127. [Google Scholar] [CrossRef] [PubMed]
- Sylvere, N.; Mustopa, A.Z.; Budiarti, S.; Meilina, L.; Hertati, A.; Handayani, I. Whole-Genome Sequence Analysis and Probiotic Characteristics of Lactococcus lactis subsp. Lactis Strain Lac3 Isolated from Traditional Fermented Buffalo Milk (Dadih). J. Genet. Eng. Biotechnol. 2023, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Feito, J.; Contente, D.; Ponce-Alonso, M.; Díaz-Formoso, L.; Araújo, C.; Peña, N.; Borrero, J.; Gómez-Sala, B.; Del Campo, R.; Muñoz-Atienza, E.; et al. Draft Genome Sequence of Lactococcus lactis subsp. Cremoris Wa2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm. Microorganisms 2022, 10, 521. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Du, T.; Jiang, X.; Liu, S.; Cheng, Y.; Zhang, Z.; Miao, W.; Wang, T. Lactococcus Garvieae Exerts a Critical Role in Inducing Inflammation in Dairy Mastitis by Triggering Nlrp3 Inflammasome-Mediated Pyroptosis in Mac-T Cells. World J. Microbiol. Biotechnol. 2024, 40, 132. [Google Scholar] [CrossRef] [PubMed]
- Maichomo, M.; Gachogo, R.; Masila, E.; Ogali, I.; Otieno, L.; Langat, N.; Onywera, R.; Malonza, V.; Inguyesi, C.; Onyambu, F.; et al. Draft Genome Sequences of Enterococcus faecium, Enterococcus gallinarum, and Lactococcus lactis Strains Isolated from a Mastitis-Infected Camel in Isiolo County, Kenya. Microbiol. Resour. Announc. 2023, 12, e0108322. [Google Scholar] [CrossRef] [PubMed]
- Plumed-Ferrer, C.; Barberio, A.; Franklin-Guild, R.; Werner, B.; McDonough, P.; Bennett, J.; Gioia, G.; Rota, N.; Welcome, F.; Nydam, D.V.; et al. Antimicrobial Susceptibilities and Random Amplified Polymorphic DNA-Pcr Fingerprint Characterization of Lactococcus lactis ssp. Lactis and Lactococcus Garvieae Isolated from Bovine Intramammary Infections. J. Dairy Sci. 2015, 98, 6216–6225. [Google Scholar] [CrossRef] [PubMed]
- Soltani, M.; Baldisserotto, B.; Hosseini Shekarabi, S.P.; Shafiei, S.; Bashiri, M. Lactococcosis a Re-Emerging Disease in Aquaculture: Disease Significant and Phytotherapy. Vet. Sci. 2021, 8, 181. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P. Mammary Microbiota of Dairy Ruminants: Fact or Fiction? Vet. Res. 2017, 48, 25. [Google Scholar] [CrossRef] [PubMed]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef] [PubMed]
- Bayili, G.R.; Johansen, P.G.; Hougaard, A.B.; Diawara, B.; Ouedraogo, G.A.; Jespersen, L.; Sawadogo-Lingani, H. Technological Properties of Indigenous Lactococcus lactis Strains Isolated from Lait Caillé, a Spontaneous Fermented Milk from Burkina Faso. J. Dairy Res. 2020, 87, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.X.; Lima, S.F.; Higgins, C.H.; Canniatti-Brazaca, S.G.; Bicalho, R.C. The Lactococcus Genus as a Potential Emerging Mastitis Pathogen Group: A Report on an Outbreak Investigation. J. Dairy Sci. 2016, 99, 9864–9874. [Google Scholar] [CrossRef] [PubMed]
- Hansen, W.; Kietzmann, M. Comments on the Guidelines for the Prudent Use of Antibacterial Veterinary Pharmaceuticals (Guidelines for Antibiotics). Tierärztliche Prax. Ausg. G Großtiere Nutztiere 2012, 40, 182–185. [Google Scholar]
- Angelopoulou, A.; Warda, A.K.; Hill, C.; Ross, R.P. Non-Antibiotic Microbial Solutions for Bovine Mastitis-Live Biotherapeutics, Bacteriophage, and Phage Lysins. Crit. Rev. Microbiol. 2019, 45, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Forde, A.; Fitzgerald, G.F. Molecular Organization of Exopolysaccharide (Eps) Encoding Genes on the Lactococcal Bacteriophage Adsorption Blocking Plasmid, Pci658. Plasmid 2003, 49, 130–142. [Google Scholar] [CrossRef] [PubMed]
- van Kranenburg, R.; Marugg, J.D.; Van Swam, I.I.; Willem, N.J.; de Vos, W.M. Molecular Characterization of the Plasmid-Encoded Eps Gene Cluster Essential for Exopolysaccharide Biosynthesis in Lactococcus lactis. Mol. Microbiol. 1997, 24, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, L.P.; Huang, T.; Lei, A.Y.; Huang, Y.; Luo, F.G.; Wang, D.Y.; Huang, W.Y.; Chen, M.; Huang, J. Genomic Comparison of Virulent and Non-Virulent Serotype V St1 Streptococcus Agalactiae in Fish. Vet. Microbiol. 2017, 207, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Reiß, S.; Schlüter, R.; Mäder, U.; Beyer, A.; Reiß, W.; Marles-Wright, J.; Lewis, R.J.; Pförtner, H.; Völker, U.; et al. Deletion of Membrane-Associated Asp23 Leads to Upregulation of Cell Wall Stress Genes in Staphylococcus aureus. Mol. Microbiol. 2014, 93, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Feltcher, M.E.; Braunstein, M. Emerging Themes in Seca2-Mediated Protein Export. Nat. Rev. Microbiol. 2012, 10, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Ponomarev, V.A.; Koonin, E.V. Two C or Not Two C: Recurrent Disruption of Zn-Ribbons, Gene Duplication, Lineage-Specific Gene Loss, and Horizontal Gene Transfer in Evolution of Bacterial Ribosomal Proteins. Genome Biol. 2001, 2, research0033.1. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, D.; Liu, L.; Zheng, Q.; Song, Y.; Ye, L.; Sha, S.; Kang, J.; Xin, Y.; Ma, Y. Characterization of Mycobacterial Udp-N-Acetylglucosamine Enolpyruvyle Transferase (Mura). Res. Microbiol. 2014, 165, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Fan, Y.; Wang, X.; Li, P.; Huang, Y.; Jiao, J.; Zhao, C.; Li, Y.; Wang, S.; Du, X. Characterization of Molecular Chaperone Groel as a Potential Virulence Factor in Cronobacter Sakazakii. Foods 2023, 12, 3404. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, Y.; Ma, X.; Wei, L.; Wan, X.; Zhang, Z.; Ding, J.; Peng, J.; Liu, G.; Gou, H.; et al. Effect of Two Different Drug-Resistant Staphylococcus aureus Strains on the Physiological Properties of Mac-T Cells and Their Transcriptome Analysis. Front. Vet. Sci. 2022, 9, 818928. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.J.; Li, H.P.; Zhu, H.S.; Sui, S.P.; Chen, P.G.; Deng, Y.; Sui, T.M.; Wang, Y.Y. Nf-Κb Is Involved in the Lps-Mediated Proliferation and Apoptosis of Mac-T Epithelial Cells as Part of the Subacute Ruminal Acidosis Response in Cows. Biotechnol. Lett. 2016, 38, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, E.; Krumrych, W.; Markiewicz, H. The Effect of Low Intensity Laser Irradiation of Inflamed Udders on the Efficacy of Antibiotic Treatment of Clinical Mastitis in Dairy Cows. Vet. Ital. 2019, 55, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis Detection, Management, and Prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Abbreviation of Target Gene | Primer Name | Primer Sequence (5′ to 3′) | Product Size | Annealing Temperature (°C) |
|---|---|---|---|---|---|
| Hemolysin 1 | hly1 | H1 F | CCTCCTCCGACTAGGAACCA | 521 | 54 |
| H1 R | GAAAAGCCAGCTTCTCGTGC | ||||
| Hemolysin 2 | hly2 | H2 F | TCTCGTGCACACCGATGAAA | 492 | 53 |
| H2 R | TGAACTTCGGCTTCTGCGAT | ||||
| Hemolysin 3 | hly3 | H3 F | AACGCGAGAACAGGCAAAAC | 291 | 56 |
| H3 R | CCCACGTCGAGAGCATAGAC | ||||
| NADH oxidase | NADHO | NADHO F | TGCGATGGGTTCAAGACCAA | 331 | 53 |
| NADHO R | GCCTTTAAAAGCCTCGGCAG | ||||
| Superoxide dismutase | SOD | SOD F | GCAGCGATTGAAAAACACCCA | 80 | 54 |
| SOD R | TCTTCTGGCAAACGGTCCAA | ||||
| Phosphoglucomutase | pgm | PG F | AAGTTTACGGCGAAGACGGT | 997 | 53 |
| PG R | TTTTCTGGTGCATTGGCACG | ||||
| Enolase | eno | E F | CAAGAGCGATCATTGCACGG | 201 | 54 |
| E R | CATTCGGACGCGGTATGGTA | ||||
| Adhesin Pav | pav | AP F | CCTGTCGGGCGCTTTTATTG | 232 | 56 |
| AP R | TCCCGGAAGAAGAGTACGGT | ||||
| Adhesin PsaA | PsaA | APSA F | GTTGCAACAGCTGGACACAG | 180 | 54 |
| APSA R | ATACGGTTGAGTTGGGCTGG | ||||
| Adhesin cluster 1 | adhC1 | adhC1 F | TTGGGCACATCAGACTGGAC | 264 | 54 |
| adhC1 R | AGCATCATCAGCTGCCAAGT | ||||
| Adhesin cluster 2 | adhC2 | adhC2F | CTGCGAGTGGCATCTCCATT | 160 | 52 |
| adhC2 R | TCAACACTGCGACCTTCTGT | ||||
| Adhesin | AF | AF F | CAGCCAGCACCAGGTTATGA | 358 | 54 |
| AF R | CTCCTGCGTTGACATGGACT | ||||
| capsule gene cluster A | CGC A | 1020-F | ACCTTCACTTGCATTCATAGGGT | 304 | 54 |
| 1323-R | TTGTCCCAGAGGGTTCTCCT | ||||
| capsule gene cluster B | CGC B | 851-F | TAGGAGGTGTTCCTGGGAGG | 549 | 54 |
| 1399-R | TGTCCCACTCCTACTGTCGT | ||||
| capsule gene cluster C | CGC C | 6329-F | AAAAACGGAGGGCAACAAGC | 785 | 60 |
| 7175-R | CACTTGTACAGGCCACTGGT | ||||
| capsule gene cluster D | CGC D | 5358-F | TGGAGGGTATTGCCTACCGA | 650 | 54 |
| 6007-R | CCACAGCAGCTTCTTCACCT | ||||
| conserved hypothetical protein | CHP | CHP F | CTGCTGATCAAGTCCAAGC | 303 | 52 |
| CHP R | GAGAAACGACCTTAGCTCCA | ||||
| exopolysaccharide A | EpsA | EpsA F | TTATAGCCTCCCCAGTTTACAC | 299 | 52 |
| EpsA R | TTTAGCAGTCTCGTCTGCAATC | ||||
| exopolysaccharideB | EpsB | EpsB F | CGCAAGTGCTAATCTAGCTG | 317 | 52 |
| EpsB R | AGAGAGGCGGAGTATCAATC | ||||
| exopolysaccharide C | EpsC | EpsC F | TAACAACTATCACTGCGACTCC | 343 | 52 |
| EpsC R | TCAGGGTTCTCAATGATTCCAC | ||||
| exopolysaccharide D | EpsD | EpsD F | TTTCTTATTGCGGCTGCATTGC | 270 | 52 |
| EpsD R | CTCATCAATTGAGTGTCGTCTG | ||||
| exopolysaccharide L | EpsL | EpsL F | ACCAATCGTACAGATCAACG | 473 | 52 |
| EpsL R | CTTGAGCCACCACTATCAAG | ||||
| exopolysaccharide R | EpsR | EpsR F | TTTTACCACCGGCTAAAGGAAC | 211 | 52 |
| EpsR R | TTGCAGAACTGTCATTAGGCTC | ||||
| exopolysaccharide X | EpsX | EpsX F | TATTGAAGCAACAGCCTCACTG | 198 | 52 |
| EpsX R | TTTTTGTCTGGGTAACTAGCCC | ||||
| rhamnosyltransferase | RIF | RIF F | TTGATGGTAAATCCTGATGG | 307 | 52 |
| RIF R | GAACAAACCGACCTACAACA | ||||
| 30SrRNA gene | 30S | 30S F | TACGAACACCGTATCCTTGAC | 207 | 52 |
| 30S R | TACGAACACCGTATCCTTGAC | ||||
| LPxTG-1 | LP1 | LP1-F | GTGAACGTGGAGCTTCCAGA | 878 | 54 |
| LP1-R | CCACTCACATGGGGGAGTTC | ||||
| LPxTG-2 | LP2 | LP2-F | GCCAGTGAGAGAACCGTTGA | 767 | 54 |
| LP2-R | CAGGTTCAAGTGCAACTGCC | ||||
| LPxTG-3 | LP3 | LP3-F | TTAAGCACAACGGCAACAGC | 231 | 54 |
| LP3-R | CACGCGAAATGATGGTGCAT | ||||
| LPxTG-4 | LP4 | LP4-F | GGGAGCACCGGATTCACTTT | 928 | 52 |
| LP4-R | ACAAAGCCGCAGACCTTACA |
| Number | Ribose | Sucrose | Lactose | Liquid Gelatin | Sorbitol | Maltose | Esculin | VP | Galactose | Trehalose | Glucose |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LL1 | - | + | + | - | - | + | + | + | + | + | + |
| LL2 | - | + | + | - | - | + | - | + | + | + | + |
| LL3 | - | - | + | - | - | + | + | + | + | + | + |
| LL4 | - | + | + | - | - | + | + | + | + | + | + |
| LL5 | - | + | + | - | - | + | + | + | + | + | + |
| LL6 | - | + | + | - | - | + | + | + | + | + | + |
| LL7 | - | - | + | - | - | + | + | + | + | + | + |
| LL8 | - | - | + | - | - | + | + | + | + | + | + |
| LL9 | - | - | + | - | - | + | + | + | + | + | + |
| MIC (μg/mL) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial | >16 | 16 | 8 | 4 | 2 | 1 | 0.5 | 0.25 | 0.12 | 0.06 | 0.03 | MIC50 (μg/mL) | MIC90 (μg/mL) | Resistance Rate |
| Penicillin | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 4 | 0 | 0 | 0 | 0.25 | 0.50 | 100 |
| Cephalexin | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16 | 16 | 100 |
| Ampicillin | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | 2 | 0 | 0 | 0.25 | 0.50 | 37.5 |
| Ceftiofur | 0 | 1 | 0 | 0 | 0 | 3 | 4 | 0 | 0 | 0 | 0 | 0.5 | 1.0 | 12.5 |
| Cefquinome | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 3 | 0 | 0 | 0.25 | 0.50 | 0 |
| Lincomycin | 1 | 0 | 1 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 50 |
| Oxytetracycline | 1 | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 16 | 100 |
| Marbofloxacin | 0 | 0 | 0 | 0 | 0 | 6 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Rifaximin | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | >16 | >16 | 100 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 1 | 0 | 0 | 0.25 | 0.25 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Wu, F.; Du, T.; Jiang, X.; Liu, S.; Cheng, Y.; Hu, J. Virulence, Antibiotic Resistance and Cytotoxic Effects of Lactococcus lactis Isolated from Chinese Cows with Clinical Mastitis on MAC-T Cells. Microorganisms 2025, 13, 1674. https://doi.org/10.3390/microorganisms13071674
Wang T, Wu F, Du T, Jiang X, Liu S, Cheng Y, Hu J. Virulence, Antibiotic Resistance and Cytotoxic Effects of Lactococcus lactis Isolated from Chinese Cows with Clinical Mastitis on MAC-T Cells. Microorganisms. 2025; 13(7):1674. https://doi.org/10.3390/microorganisms13071674
Chicago/Turabian StyleWang, Tiancheng, Fan Wu, Tao Du, Xiaodan Jiang, Shuhong Liu, Yiru Cheng, and Jianmin Hu. 2025. "Virulence, Antibiotic Resistance and Cytotoxic Effects of Lactococcus lactis Isolated from Chinese Cows with Clinical Mastitis on MAC-T Cells" Microorganisms 13, no. 7: 1674. https://doi.org/10.3390/microorganisms13071674
APA StyleWang, T., Wu, F., Du, T., Jiang, X., Liu, S., Cheng, Y., & Hu, J. (2025). Virulence, Antibiotic Resistance and Cytotoxic Effects of Lactococcus lactis Isolated from Chinese Cows with Clinical Mastitis on MAC-T Cells. Microorganisms, 13(7), 1674. https://doi.org/10.3390/microorganisms13071674






