Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands
Abstract
1. Introduction
2. Case Description
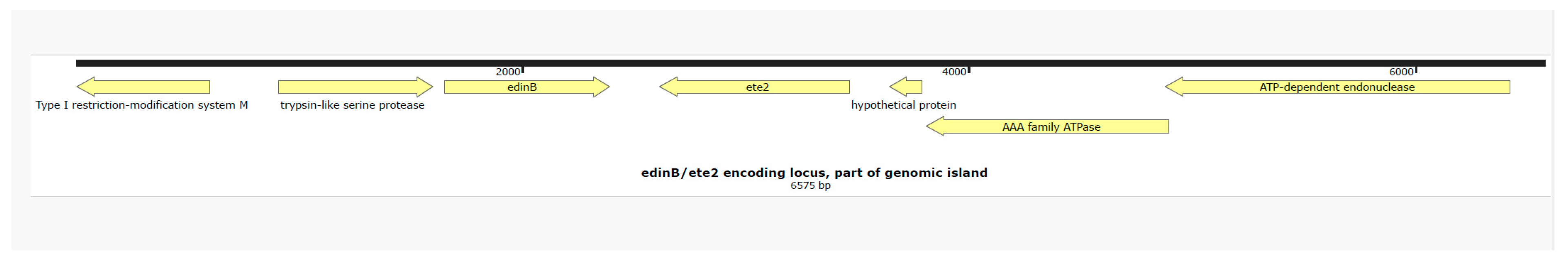
Isolate Characterization
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.-C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [PubMed]
- Sabat, A.J.; Wouthuyzen-Bakker, M.; Rondags, A.; Hughes, L.; Akkerboom, V.; Koutsopetra, O.; Friedrich, A.W.; Bathoorn, E. Case Report: Necrotizing fasciitis caused by Staphylococcus aureus positive for a new sequence variant of exfoliative toxin E. Front. Genet. 2002, 13, 964358. [Google Scholar]
- Lisotto, P.; Raangs, E.C.; Couto, N.; Rosema, S.; Lokate, M.; Zhou, X.; Friedrich, A.W.; Rossen, J.W.A.; Harmsen, H.J.M.; Bathoorn, E.; et al. Long-read sequencing-based in silico phage typing of vancomycin-resistant Enterococcus faecium. BMC Genom. 2021, 22, 758. [Google Scholar]
- Strauß, L.; Ruffing, U.; Abdulla, S.; Alabi, A.; Akulenko, R.; Garrine, M.; Germann, A.; Grobusch, M.P.; Helms, V.; Herrmann, M.; et al. Detecting Staphylococcus aureus Virulence and Resistance Genes: A Comparison of Whole-Genome Sequencing and DNA Microarray Technology. J. Clin. Microbiol. 2016, 54, 1008–1016. [Google Scholar] [PubMed]
- Baig, S.; Larsen, A.R.; Simões, P.M.; Laurent, F.; Johannesen, T.B.; Lilje, B.; Tristan, A.; Schaumburg, F.; Egyir, B.; Cirkovic, I.; et al. Evolution and Population Dynamics of Clonal Complex 152 Community-Associated Methicillin-Resistant Staphylococcus aureus. mSphere 2020, 5, e00226-20. [Google Scholar] [PubMed]
- Imanishi, I.; Nicolas, A.; Caetano, A.B.; Castro, T.L.P.; Tartaglia, N.R.; Mariutti, R.; Guédon, E.; Even, S.; Berkova, N.; Arni, R.K.; et al. Exfoliative toxin E, a new Staphylococcus aureus virulence factor with host-specific activity. Sci. Rep. 2019, 9, 16336. [Google Scholar]
- Bukowski, M.; Wladyka, B.; Dubin, G. Exfoliative toxins of Staphylococcus aureus. Toxins 2010, 2, 1148–1165. [Google Scholar] [CrossRef] [PubMed]
- Wiegers, W.; Just, I.; Müller, H.; Hellwig, A.; Traub, P.; Aktories, K. Alteration of the cytoskeleton of mammalian cells cultured in vitro by Clostridium botulinum C2 toxin and C3 ADP-ribosyltransferase. Eur. J. Cell Biol. 1991, 54, 237–245. [Google Scholar] [PubMed]
- Boyer, L.; Doye, A.; Rolando, M.; Flatau, G.; Munro, P.; Gounon, P.; ClémEnt, R.; Pulcini, C.; Popoff, M.R.; Mettouchi, A.; et al. Induction of transient macroapertures in endothelial cells through RhoA inhibition by Staphylococcus aureus factors. J. Cell Biol. 2006, 173, 809–819. [Google Scholar] [PubMed]
- Courjon, J.; Munro, P.; Benito, Y.; Visvikis, O.; Bouchiat, C.; Boyer, L.; Doye, A.; Lepidi, H.; Ghigo, E.; Lavigne, J.P.; et al. EDIN-B Promotes the Translocation of Staphylococcus aureus to the Bloodstream in the Course of Pneumonia. Toxins 2015, 7, 4131–4142. [Google Scholar] [CrossRef] [PubMed]
- Czech, A.; Yamaguchi, T.; Bader, L.; Linder, S.; Kaminski, K.; Sugai, M.; Aepfelbacher, M. Prevalence of Rho-inactivating epidermal cell differentiation inhibitor toxins in clinical Staphylococcus aureus isolates. J. Infect. Dis. 2001, 184, 785–788. [Google Scholar] [PubMed]
- Yamaguchi, T.; Nishifuji, K.; Sasaki, M.; Fudaba, Y.; Aepfelbacher, M.; Takata, T.; Ohara, M.; Komatsuzawa, H.; Amagai, M.; Sugai, M.; et al. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 2002, 70, 5835–5845. [Google Scholar] [PubMed]
- Egyir, B.; Owusu-Nyantakyi, C.; Bortey, A.; Amuasi, G.R.; Owusu, F.A.; Boateng, W.; Ahmed, H.; Danso, J.K.; Oclu, A.A.G.; Mohktar, Q.; et al. Whole genome sequencing revealed high proportions of ST152 MRSA among clinical Staphylococcus aureus isolates from ten hospitals in Ghana. mSphere 2024, 9, e00446-24. [Google Scholar] [PubMed]
- Shittu, A.O.; Okon, K.; Adesida, S.; Oyedara, O.; Witte, W.; Strommenger, B.; Layer, F.; Nübel, U. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011, 11, 92. [Google Scholar]
- Newell, R.; El-Shakankery, K.; Bhowmik, A.; Rajakulasingam, R.K. Panton–Valentine leucocidin Staphylococcus aureus necrotising pneumonia in a clinically well patient. Br. J. Hosp. Med. 2023, 29, 1–4. [Google Scholar]
- Allou, N.; Allyn, J.; Traversier, N.; Baron, M.; Blondé, R.; Dupieux, C.; Coolen-Allou, N.; Jabot, J.; Miltgen, G. SARS-CoV-2 with Panton-Valentine leukocidin-producing Staphylococcus aureus healthcare-associated pneumonia in the Indian Ocean. Heliyon 2022, 8, e10422. [Google Scholar] [PubMed]
- Larsen, S.A.H.; Kyhl, K.; Baig, S.; Petersen, A.; Av Steinum, M.R.; Clemmensen, S.; Jensen, E.; Á Steig, T.; Gaini, S. Life-Threatening Necrotizing Pneumonia with Panton-Valentine Leukocidin-Producing, Methicillin-Sensitive Staphylococcus aureus in a Healthy Male Co-Infected with Influenza B. Infect. Dis. Rep. 2021, 14, 12–19. [Google Scholar] [PubMed]
- Löffler, B.; Niemann, S.; Ehrhardt, C.; Horn, D.; Lanckohr, C.; Lina, G.; Ludwig, S.; Peters, G. Pathogenesis of Staphylococcus aureus necrotizing pneumonia: The role of PVL and an influenza coinfection. Expert. Rev. Anti Infect. Ther. 2013, 11, 1041–1051. [Google Scholar] [PubMed]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [PubMed]
- Ragle, B.E.; Bubeck Wardenburg, J. Anti-alpha-hemolysin monoclonal antibodies mediate protection against Staphylococcus aureus pneumonia. Infect. Immun. 2009, 77, 2712–2718. [Google Scholar] [PubMed]

| Gene | Virulence Factor |
|---|---|
| aur | ACME |
| cap5H | capsule |
| cap5J | capsule |
| cap5K | capsule |
| ebpS | adhesion |
| edinB | exotoxin |
| etE2 | exfoliative toxin |
| eno | adhesion |
| hla | hemolysin |
| hlb-intact | hemolysin |
| hlgA | hemolysin |
| hlgB | hemolysin |
| hlIII | hemolysin |
| icaA | adhesion |
| icaD | biofilm |
| isaB | immunodominant antigen |
| isdA | MSCRAMM |
| lukF-PV | leukotoxin (PVL) |
| lukS-PV | leukotoxin (PVL) |
| sak | immune-evasion |
| scn | immune-evasion |
| setB1 | superantigen-like |
| setB3 | superantigen-like |
| ssl01 | superantigen-like |
| ssl02 | superantigen-like |
| ssl10 | superantigen-like |
| sspA | serine protease |
| sspB | cystein protease |
| sspP | cystein protease |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Steen, W.J.; Fliss, M.A.; Metz, E.; Filoda, K.; van den Berg, C.H.S.B.; Sinha, B.; Bathoorn, E. Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands. Microorganisms 2025, 13, 1618. https://doi.org/10.3390/microorganisms13071618
van Steen WJ, Fliss MA, Metz E, Filoda K, van den Berg CHSB, Sinha B, Bathoorn E. Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands. Microorganisms. 2025; 13(7):1618. https://doi.org/10.3390/microorganisms13071618
Chicago/Turabian Stylevan Steen, Wouter J., Monika A. Fliss, Ethel Metz, Klaus Filoda, Charlotte H. S. B. van den Berg, Bhanu Sinha, and Erik Bathoorn. 2025. "Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands" Microorganisms 13, no. 7: 1618. https://doi.org/10.3390/microorganisms13071618
APA Stylevan Steen, W. J., Fliss, M. A., Metz, E., Filoda, K., van den Berg, C. H. S. B., Sinha, B., & Bathoorn, E. (2025). Case Report: Fatal Necrotizing Pneumonia by Exfoliative Toxin etE2-Producing Staphylococcus aureus Belonging to MLST ST152 in The Netherlands. Microorganisms, 13(7), 1618. https://doi.org/10.3390/microorganisms13071618







