Colony-YOLO: A Lightweight Micro-Colony Detection Network Based on Improved YOLOv8n
Abstract
1. Introduction
- (1)
- A dataset of mulberry blight bacterial colonies named MBCD is proposed, including nine species of bacteria, 310 images, and 23,524 colonies.
- (2)
- The StarNet is deployed as the backbone network for Colony-YOLO. StarNet adopts a model design based on star-shaped operations, significantly enhancing the ability to transform input features into high-dimensional feature spaces while effectively reducing computational complexity.
- (3)
- The C2f-MLCA module is designed to significantly enhance the network’s feature extraction capability by integrating local and global features along with channel and spatial information, thereby improving feature extraction capabilities and detection accuracy.
- (4)
- The Shape-IoU is used as the bounding box regression loss to make the model focus on the shape and scale of the bounding box itself, thereby improving its localization ability.
2. Datasets
2.1. Mulberry Bacterial Blight Colony Dataset
- (1)
- Performing horizontal or vertical flips to create mirrored versions of images significantly increases the diversity of the dataset, allowing the model to learn features of colonies from different directions, thereby enhancing its ability to recognize them from various angles.
- (2)
- Randomly adjusting brightness, contrast, and saturation allows the model to better adapt to varying lighting conditions and image qualities during training.
- (3)
- Adding noise to simulate different environmental conditions. Gaussian noise, salt noise, and pepper noise can replicate the noise interference encountered when capturing images in real environments, thus improving resilience to noise, enhancing model performance in complex scenarios.
2.2. Annotated Dataset for Deep-Learning-Based Bacterial Colony Detection
3. Methods
3.1. YOLOv8 Network
3.2. Overall Structure of Colony-YOLO
3.2.1. Lightweight Backbone Network StarNet
3.2.2. C2f-MLCA
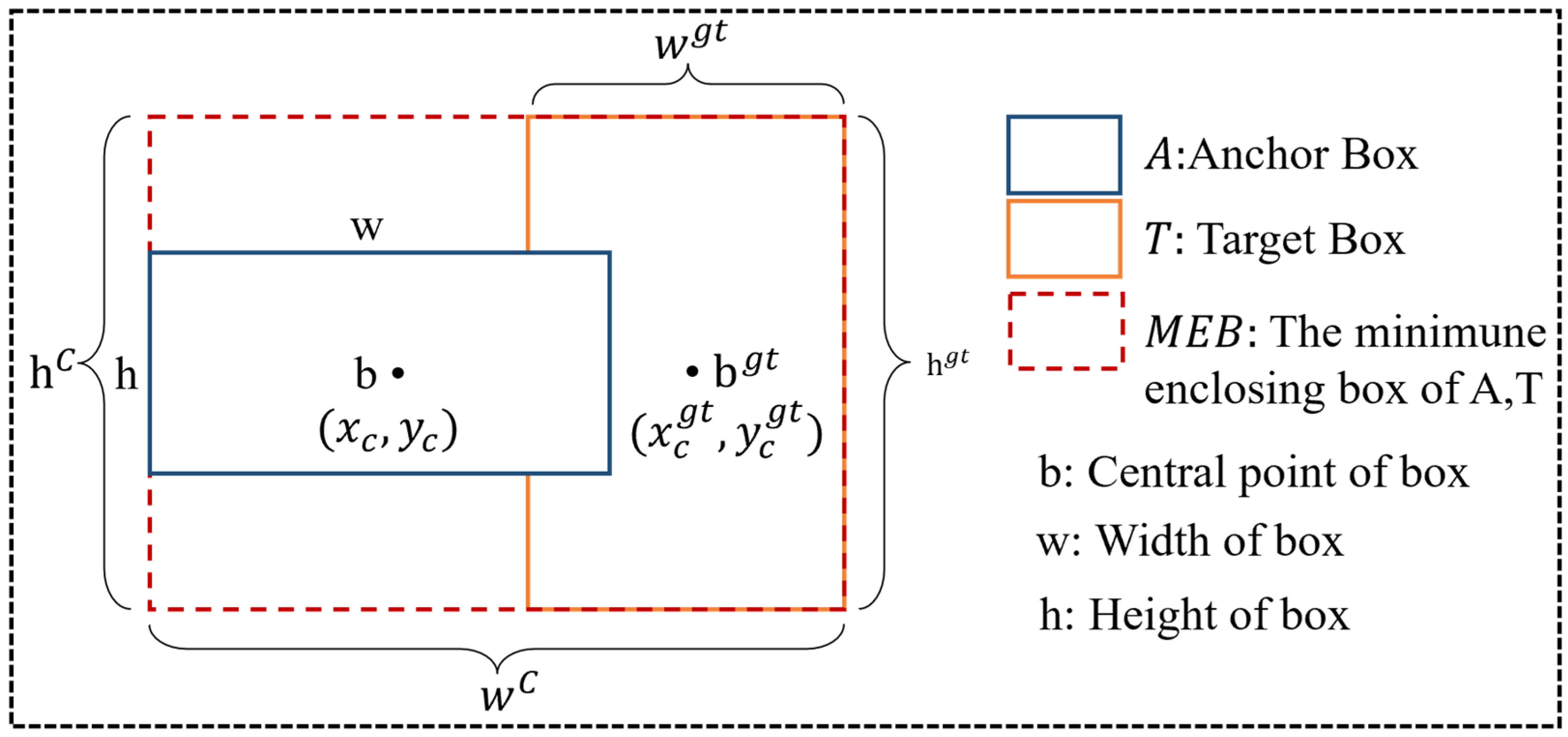
3.2.3. Shape-IoU Loss Function
4. Experiments and Results
4.1. Experiment Environment and Configuration
4.2. Evaluation Metrics
4.3. Ablation Experiment of Colony–YOLO
4.4. Comparative Experiments
4.4.1. Analysis of Lightweight Improvements in Feature Extraction Backbone Networks
4.4.2. Analysis of Loss Function Comparison Results
4.4.3. Comparison of Model Performance on ADBCs and MBCDs
4.4.4. Visualization of Model Detection
5. A Smartphone App for Colony Detection
6. Discussion
6.1. The Advantages of the Proposed Approach
6.2. Analysis of Limitations and Future Work
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Memete, A.R.; Timar, A.V.; Vuscan, A.N.; Miere Groza, F.; Venter, A.C.; Vicas, S.I. Phytochemical Composition of Different Botanical Parts of Morus Species, Health Benefits and Application in Food Industry. Plants 2022, 11, 152. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Shuang, F.; Fu, Q.; Ju, Y.; Zong, C.; Zhao, W.; Zhang, D.; Yao, X.; Cao, F. Evaluation of the Chemical Composition and Antioxidant Activity of Mulberry (Morus alba L.) Fruits from Different Varieties in China. Molecules 2022, 27, 2688. [Google Scholar] [CrossRef]
- Xie, J.; Shu, P.; Strobel, G.; Chen, J.; Wei, J.; Xiang, Z.; Zhou, Z. Pantoea agglomerans SWg2 colonizes mulberry tissues, promotes disease protection and seedling growth. Biol. Control 2017, 113, 9–17. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Huang, Y.; Qazi, I.H.; Liu, J. Analysis of causal pathogens of mulberry bacterial blight in samples collected from eight provinces of China using culturomics and metagenomic sequencing methods. Front. Plant Sci. 2025, 16, 1517050. [Google Scholar] [CrossRef]
- Ansari, A.H.; Akshay, W.; Sanyukta, B.; Vaishnavi, U. Deep Learning-Based Recognition of Plant Leaf Diseases. Int. J. Sci. Res. Eng. Manag. 2024, 8, 1–5. [Google Scholar] [CrossRef]
- Baiyewu, R.A.; Amusa, N.A.; Idowu, G.O.; Smith, A. The occurrence of leaf rust disease of mulberry plant (Morus alba) in lowland humid forest of southwestern Nigeria. Plant Pathol. J. 2005, 4, 107–109. [Google Scholar] [CrossRef]
- Huang, Y.; Wei, J.; Liu, P.; Zhu, Y.; Lin, T.; Lv, Z.; Li, Y.; Zong, M.; Zhou, Y.; Gao, J.; et al. Identification of Mulberry Bacterial Blight Caused by Klebsiella oxytoca in Bazhong, Sichuan, China. Phyton-Ann. Rei Bot. A 2024, 93, 1995–2008. [Google Scholar] [CrossRef]
- Luo, L.; Huang, Y.; Liu, J. Genome Sequence Resources of Klebsiella michiganensis AKKL-001, Which Causes Bacterial Blight of Mulberry. Mol. Plant-Microbe Interact. 2022, 35, 349–351. [Google Scholar] [CrossRef]
- Zhu, B.; Wang, G.; Xie, G.; Zhou, Q.; Zhao, M.; Praphat, K.; Li, B.; Tian, W. Enterobacter spp.: A new evidence causing bacterial wilt on mulberry. Sci. China Life Sci. 2010, 53, 292–300. [Google Scholar] [CrossRef]
- Maji, M.D.; Qadri, S.; Pal, S.C. Evaluation of mulberry phylloplane microorganisms for biological control of bacterial leaf spot of mulberry caused by Xanthomonas campestris pv. mori. Indian J. Agric. Res. 2003, 37, 307–309. [Google Scholar]
- Choudhry, P. High-Throughput Method for Automated Colony and Cell Counting by Digital Image Analysis Based on Edge Detection. PLoS ONE 2016, 11, e0148469. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Rahaman, M.M.; Yao, Y.; Ma, P.; Zhang, J.; Zhao, X.; Jiang, T.; Grzegorzek, M. A comprehensive review of image analysis methods for microorganism counting: From classical image processing to deep learning approaches. Artif. Intell. Rev. 2022, 55, 2875–2944. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Zheng, X.; Chen, X.; Que, Y.; Ma, B. An Automated Method for Bacterial Flora Counting Based on Image Analysis. J. Phys. Conf. Ser. 2020, 1673, 12016. [Google Scholar] [CrossRef]
- Mukherjee, D.P.; Pal, A.; Sarma, S.E.; Majumder, D.D. Bacterial colony counting using distance transform. Int. J. Biomed. Comput. 1995, 38, 131–140. [Google Scholar] [CrossRef]
- Coulthard, M.G. Defining urinary tract infection by bacterial colony counts: A case for 100,000 colonies/ml as the best threshold. Pediatr. Nephrol. 2019, 34, 1639–1649. [Google Scholar] [CrossRef]
- Ferrari, A.; Signoroni, A. Multistage classification for bacterial colonies recognition on solid agar images. In Proceedings of the 2014 IEEE International Conference on Imaging Systems and Techniques (IST) Proceedings, Santorini, Greece, 14–17 October 2014; pp. 101–106. [Google Scholar]
- Brugger, S.D.; Baumberger, C.; Jost, M.; Jenni, W.; Brugger, U.; Muhlemann, K. Automated counting of bacterial colony forming units on agar plates. PLoS ONE 2012, 7, e33695. [Google Scholar] [CrossRef]
- Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 2013, 8, e54072. [Google Scholar] [CrossRef]
- Yoon, S.; Lawrence, K.C.; Park, B. Automatic Counting and Classification of Bacterial Colonies Using Hyperspectral Imaging. Food Bioprocess. Tech. 2015, 8, 2047–2065. [Google Scholar] [CrossRef]
- Zhang, L. Machine learning for enumeration of cell colony forming units. Vis. Comput. Ind. Biomed. Art. 2022, 5, 26. [Google Scholar] [CrossRef]
- Khan, A.U.M.; Torelli, A.; Wolf, I.; Gretz, N. AutoCellSeg: Robust automatic colony forming unit (CFU)/cell analysis using adaptive image segmentation and easy-to-use post-editing techniques. Sci. Rep 2018, 8, 7302. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, C. An automated bacterial colony counting and classification system. Inform. Syst. Front. 2009, 11, 349–368. [Google Scholar] [CrossRef]
- Carl, S.H.; Duempelmann, L.; Shimada, Y.; Buhler, M. A fully automated deep learning pipeline for high-throughput colony segmentation and classification. Biol. Open 2020, 9, bio052936. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.L.; Burton, R.L.; Hill, A.N.; Litorja, M.; Nahm, M.H.; Hwang, J. Low-cost, high-throughput, automated counting of bacterial colonies. Cytom. Part A 2010, 77, 790–797. [Google Scholar] [CrossRef]
- Wang, H.; Ceylan Koydemir, H.; Qiu, Y.; Bai, B.; Zhang, Y.; Jin, Y.; Tok, S.; Yilmaz, E.C.; Gumustekin, E.; Rivenson, Y.; et al. Early detection and classification of live bacteria using time-lapse coherent imaging and deep learning. Light-Sci. Appl. 2020, 9, 118. [Google Scholar] [CrossRef]
- Nagy, S.Á.; Makrai, L.; Csabai, I.; Tőzsér, D.; Szita, G.; Solymosi, N. Bacterial colony size growth estimation by deep learning. BMC Microbiol. 2023, 23, 307. [Google Scholar] [CrossRef]
- Cao, L.; Zeng, L.; Wang, Y.; Cao, J.; Han, Z.; Chen, Y.; Wang, Y.; Zhong, G.; Qiao, S. U2-Net and ResNet50-Based Automatic Pipeline for Bacterial Colony Counting. Microorganisms 2024, 12, 201. [Google Scholar] [CrossRef]
- Jumutc, V.; Suponenkovs, A.; Bondarenko, A.; Bļizņuks, D.; Lihachev, A. Hybrid Approach to Colony-Forming Unit Counting Problem Using Multi-Loss U-Net Reformulation. Sensors 2023, 23, 8337. [Google Scholar] [CrossRef]
- Ebert, N.; Stricker, D.; Wasenmüller, O. Transformer-based Detection of Microorganisms on High-Resolution Petri Dish Images. In Proceedings of the 2023 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW), Paris, France, 2–6 October 2023. [Google Scholar]
- Qiu, H.; Zhang, Q.; Li, J.; Rong, J.; Yang, Z. Lightweight Mulberry Fruit Detection Method Based on Improved YOLOv8n for Automated Harvesting. Agronomy 2024, 14, 2861. [Google Scholar] [CrossRef]
- Li, P.; Zhou, J.; Sun, H.; Zeng, J. RDRM-YOLO: A High-Accuracy and Lightweight Rice Disease Detection Model for Complex Field Environments Based on Improved YOLOv5. Agriculture 2025, 15, 479. [Google Scholar] [CrossRef]
- Xiao, D.; Wang, H.; Liu, Y.; Li, W.; Li, H. DHSW-YOLO: A duck flock daily behavior recognition model adaptable to bright and dark conditions. Comput. Electron. Agric. 2024, 225, 109281. [Google Scholar] [CrossRef]
- Luo, Y.; Lin, K.; Xiao, Z.; Lv, E.; Wei, X.; Li, B.; Lu, H.; Zeng, Z. PBR-YOLO: A lightweight piglet multi-behavior recognition algorithm based on improved yolov8. Smart Agric. Technol. 2025, 10, 100785. [Google Scholar] [CrossRef]
- Meihua, W.; Anbang, W. Detection of small intestinal villus of pigs from pathological images using improved YOLOv5s. Trans. CSAE 2024, 40, 207–215. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Z.; Cao, W.; Qi, X.; Xu, C.; Wen, W. A New Few-Shot Learning Method of Bacterial Colony Counting Based on the Edge Computing Device. Biology 2022, 11, 156. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yi, J.; Wisuthiphaet, N.; Earles, M.; Nitin, N. Accelerating the Detection of Bacteria in Food Using Artificial Intelligence and Optical Imaging. Appl. Environ. Microb. 2023, 89, e01822–e01828. [Google Scholar] [CrossRef]
- Whipp, J.; Dong, A. YOLO-based Deep Learning to Automated Bacterial Colony Counting. In Proceedings of the 2022 IEEE Eighth International Conference on Multimedia Big Data (BigMM), Naples, Italy, 5–7 December 2022; pp. 120–124. [Google Scholar]
- Liu, C. Design and Implementation of Colony Recognition and Analysis System Based on Image Processing. Master’s Thesis, Xi’an Shiyou University, Xi’an, China, 2023. [Google Scholar] [CrossRef]
- Krawczyk, K.; Łochyńska, M. Identification and characterization of Pseudomonas syringae pv. mori affecting white mulberry (Morus alba) in Poland. Eur. J. Plant Pathol. 2020, 158, 281–291. [Google Scholar] [CrossRef]
- Yuan, T.; Huang, Y.; Luo, L.; Wang, J.; Li, J.; Chen, J.; Qin, Y.; Liu, J. Complete Genome Sequence of Pantoea ananatis Strain LCFJ-001 Isolated from Bacterial Wilt Mulberry. Plant Dis. 2023, 107, 2500–2505. [Google Scholar] [CrossRef]
- Wang, J.; Han, W.; Pirhonen, M.; Pan, Y.; Zhang, D.; Zhao, D.; Li, Q.; Cheng, J.; Yang, Z.; Zhu, J. High-Quality Complete Genome Resource of Pectobacterium parvum Isolate FN20211 Causing Aerial Stem Rot of Potato. Mol. Plant-Microbe Interact. 2022, 35, 488–491. [Google Scholar] [CrossRef]
- Merla, C.; Rodrigues, C.; Passet, V.; Corbella, M.; Thorpe, H.A.; Kallonen, T.V.S.; Zong, Z.; Marone, P.; Bandi, C.; Sassera, D.; et al. Description of Klebsiella spallanzanii sp. nov. and of Klebsiella pasteurii sp. nov. Front. Microbiol. 2019, 10, 2360. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.; Jafra, S.; Lojkowska, E.; Potrykus, M.; Van Der Wolf, J.M.; Sledz, W. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015, 166, 18–38. [Google Scholar] [CrossRef]
- Heidari, M.; Aeini, M. Stenotrophomonas maltophilia causing maize (Zea mays L.) seedling soft rot disease, an emerging threat to maize crop. J. Phytopathol. 2024, 172, e13299. [Google Scholar] [CrossRef]
- Shaw, R.K.; Shen, Y.; Wang, J.; Sheng, X.; Zhao, Z.; Yu, H.; Gu, H. Advances in Multi-Omics Approaches for Molecular Breeding of Black Rot Resistance in Brassica oleracea L. Front. Plant Sci. 2021, 12, 742553. [Google Scholar] [CrossRef]
- Jiao, W.; Wen, J.; Li, N.; Ou, T.; Qiu, C.; Ji, Y.; Lin, K.; Liu, X.; Xie, J. The biocontrol potentials of rhizospheric bacterium Bacillus velezensis K0T24 against mulberry bacterial wilt disease. Arch. Microbiol. 2024, 206, 213. [Google Scholar] [CrossRef]
- Makrai, L.; Fodróczy, B.; Nagy, S.Á.; Czeiszing, P.; Csabai, I.; Szita, G.; Solymosi, N. Annotated dataset for deep-learning-based bacterial colony detection. Sci. Data 2023, 10, 497. [Google Scholar] [CrossRef] [PubMed]
- Terven, J.; Córdova-Esparza, D.; Romero-González, J. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Know Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Lin, T.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1612–3144. [Google Scholar] [CrossRef]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1534–1803. [Google Scholar] [CrossRef]
- Ma, X.; Dai, X.; Bai, Y.; Wang, Y.; Fu, Y. Rewrite the Stars. arXiv 2024, arXiv:2403.19967. [Google Scholar] [CrossRef]
- Wan, D.; Lu, R.; Shen, S.; Xu, T.; Lang, X.; Ren, Z. Mixed local channel attention for object detection. Eng. Appl. Artif. Intel. 2023, 123, 106442. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, S. Shape-IoU: More Accurate Metric considering Bounding Box Shape and Scale. arXiv 2023, arXiv:2312.17663. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. arXiv 2017, arXiv:1707.01083. [Google Scholar] [CrossRef]
- Chen, J.; Kao, S.; He, H.; Zhuo, W.; Wen, S.; Lee, C.; Chan, S.H.G. Run, Don’t Walk: Chasing Higher FLOPS for Faster Neural Networks. arXiv 2023, arXiv:2303.03667. [Google Scholar] [CrossRef]
- Gevorgyan, Z. SIoU Loss: More Powerful Learning for Bounding Box Regression. arXiv 2022, arXiv:2205.12740. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection over Union: A Metric and A Loss for Bounding Box Regression. arXiv 2019, arXiv:1902.09630. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, W.; Zhang, Z.; Jia, Z.; Wang, L.; Tan, T. Focal and efficient IOU loss for accurate bounding box regression. Neurocomputing 2022, 506, 146–157. [Google Scholar] [CrossRef]

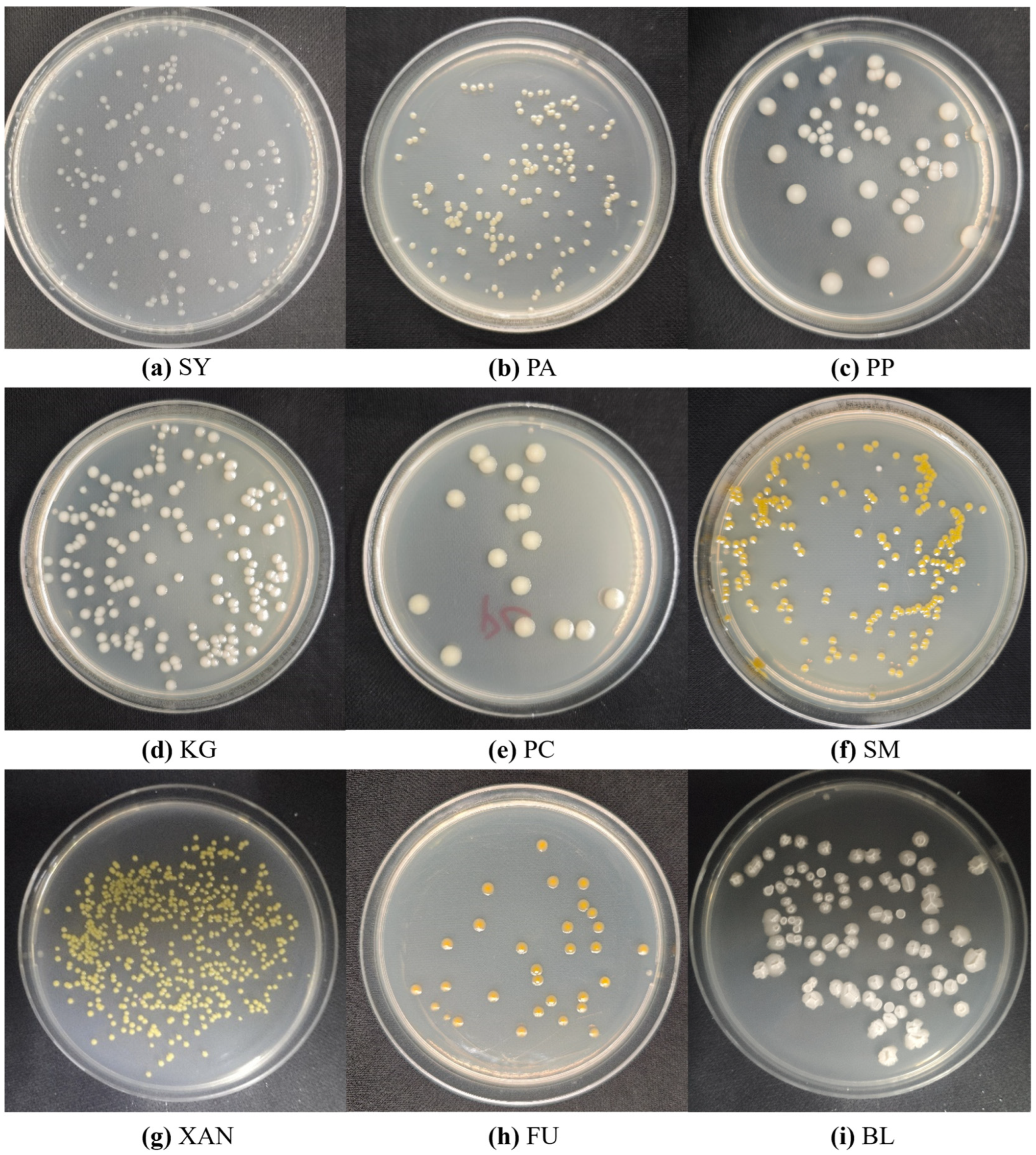







| Bacterial Species (Abbr.) | Reported Infection |
|---|---|
| Pseudomonas syringae (SY) | Mulberry bacterial blight [39] |
| Pantoea ananatis (PA) | Mulberry bacterial wilt [40] |
| Pectobacterium parvum (PP) | Potato bacterial soft rot [41] |
| Klebsiella grimontii (KG) | Hemorrhagic diarrhea [42] |
| Pectobacterium carotovorum (PC) | Cruciferous plants tuber soft rot [43] |
| Stenotrophomonas maltophilia (SM) | Zea mays L. seedling soft rot disease [44] |
| Xanthomonas campestris (XAN) | Cruciferous vegetables black rot [45] |
| Pseudomonas fuwa (FU) | Zanthoxylum spp. black rot |
| Bacillus sp. (BL) | Mulberry rhizosphere bacteria [46] |
| Hyperparameters | Value |
|---|---|
| Training epoch | 400 |
| Batch size | 8 |
| Learning rate | 0.001 |
| IoU | 0.7 |
| Optimizer | SGD |
| Image size | 640 × 640 |
| Weight_decay | 0.005 |
| Momentum | 0.937 |
| Warmup_momentum | 0.8 |
| Workspace | 4 |
| Models | StarNet | C2f-MLCA | Shape-IoU | P/% | R/% | mAP/% | FLOPs/G | Params/M | Weights/MB |
|---|---|---|---|---|---|---|---|---|---|
| M0 | - | - | - | 89.4 | 89.8 | 91.3 | 8.1 | 3.01 | 6.3 |
| M1 | ✓ | - | - | 89.9 | 90.7 | 91.4 | 6.5 | 2.22 | 4.7 |
| M2 | - | ✓ | - | 91.6 | 91.3 | 92.7 | 8.1 | 3.01 | 6.3 |
| M3 | - | - | ✓ | 90.5 | 91.1 | 91.6 | 8.1 | 3.01 | 6.3 |
| M4 | ✓ | ✓ | - | 93.4 | 91.5 | 94.5 | 6.5 | 2.21 | 4.7 |
| M5 | ✓ | - | ✓ | 92.8 | 91.2 | 93.8 | 6.5 | 2.22 | 4.7 |
| M6 | - | ✓ | ✓ | 94.3 | 92.4 | 95.4 | 8.1 | 3.01 | 6.3 |
| M7 | ✓ | ✓ | ✓ | 95.6 | 93.7 | 96.1 | 6.5 | 2.21 | 4.7 |
| Backbone | mAP/% | FLOPs/G | Params/M | Weights/MB |
|---|---|---|---|---|
| Original (yolov8) | 91.3 | 8.1 | 3.01 | 6.3 |
| MobileNet | 88.4 | 22.5 | 8.72 | 16.7 |
| ShuffleNet | 84.6 | 16.4 | 6.38 | 12.9 |
| FasterNet | 86.7 | 10.7 | 4.17 | 8.6 |
| StarNet | 91.4 | 6.5 | 2.22 | 4.7 |
| IoU | P/% | R/% | mAP/% |
|---|---|---|---|
| CIoU | 89.4 | 89.8 | 91.3 |
| SIoU | 87.2 | 86.4 | 90.9 |
| GIoU | 85.6 | 85.4 | 86.7 |
| EIoU | 90.0 | 87.5 | 90.1 |
| Shape-IoU | 90.5 | 91.1 | 91.6 |
| Datasets | Models | P/% | R/% | mAP/% | FLOPs/G | Params/M |
|---|---|---|---|---|---|---|
| Public dataset (ADBC) | Faster R-CNN | 75.2 | 74.0 | 76.5 | 170.2 | 59.13 |
| YOLOv5n | 81.2 | 79.9 | 82.6 | 5.4 | 2.56 | |
| YOLOv8n | 85.1 | 83.4 | 86.7 | 8.2 | 3.03 | |
| YOLOv10n | 86.3 | 82.1 | 87.4 | 7.1 | 2.75 | |
| Colony-YOLO (Ours) | 90.3 | 88.5 | 91.1 | 6.5 | 2.21 | |
| Private dataset (MBCD) | Faster R-CNN | 76.1 | 73.8 | 78.5 | 165.9 | 58.72 |
| YOLOv5n | 84.9 | 80.7 | 84.1 | 5.4 | 2.55 | |
| YOLOv8n | 89.4 | 89.8 | 91.3 | 8.3 | 3.01 | |
| YOLOv10n | 88.2 | 85.4 | 91.2 | 7.1 | 2.75 | |
| Colony-YOLO (Ours) | 95.6 | 93.7 | 96.1 | 6.5 | 2.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Luo, J.; Lin, K.; Chen, Y.; Huang, X.; Liu, J.; Wang, A.; Xiao, D. Colony-YOLO: A Lightweight Micro-Colony Detection Network Based on Improved YOLOv8n. Microorganisms 2025, 13, 1617. https://doi.org/10.3390/microorganisms13071617
Wang M, Luo J, Lin K, Chen Y, Huang X, Liu J, Wang A, Xiao D. Colony-YOLO: A Lightweight Micro-Colony Detection Network Based on Improved YOLOv8n. Microorganisms. 2025; 13(7):1617. https://doi.org/10.3390/microorganisms13071617
Chicago/Turabian StyleWang, Meihua, Junhui Luo, Kai Lin, Yuankai Chen, Xinpeng Huang, Jiping Liu, Anbang Wang, and Deqin Xiao. 2025. "Colony-YOLO: A Lightweight Micro-Colony Detection Network Based on Improved YOLOv8n" Microorganisms 13, no. 7: 1617. https://doi.org/10.3390/microorganisms13071617
APA StyleWang, M., Luo, J., Lin, K., Chen, Y., Huang, X., Liu, J., Wang, A., & Xiao, D. (2025). Colony-YOLO: A Lightweight Micro-Colony Detection Network Based on Improved YOLOv8n. Microorganisms, 13(7), 1617. https://doi.org/10.3390/microorganisms13071617






