Single Mutation in iolT1 in ptsG-Deficient Corynebacterium glutamicum Enables Growth Boost in Xylose-Containing Media
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Recombinant DNA Work
2.3. Preculture and Media
2.4. BioLector Micro-Cultivations
2.5. Continuous Adaptive Laboratory Evolution
2.6. Shake Flask Cultivations
2.7. Analytical Methods and Whole-Genome Sequencing
2.8. Determination of Kinetic Parameters
3. Results
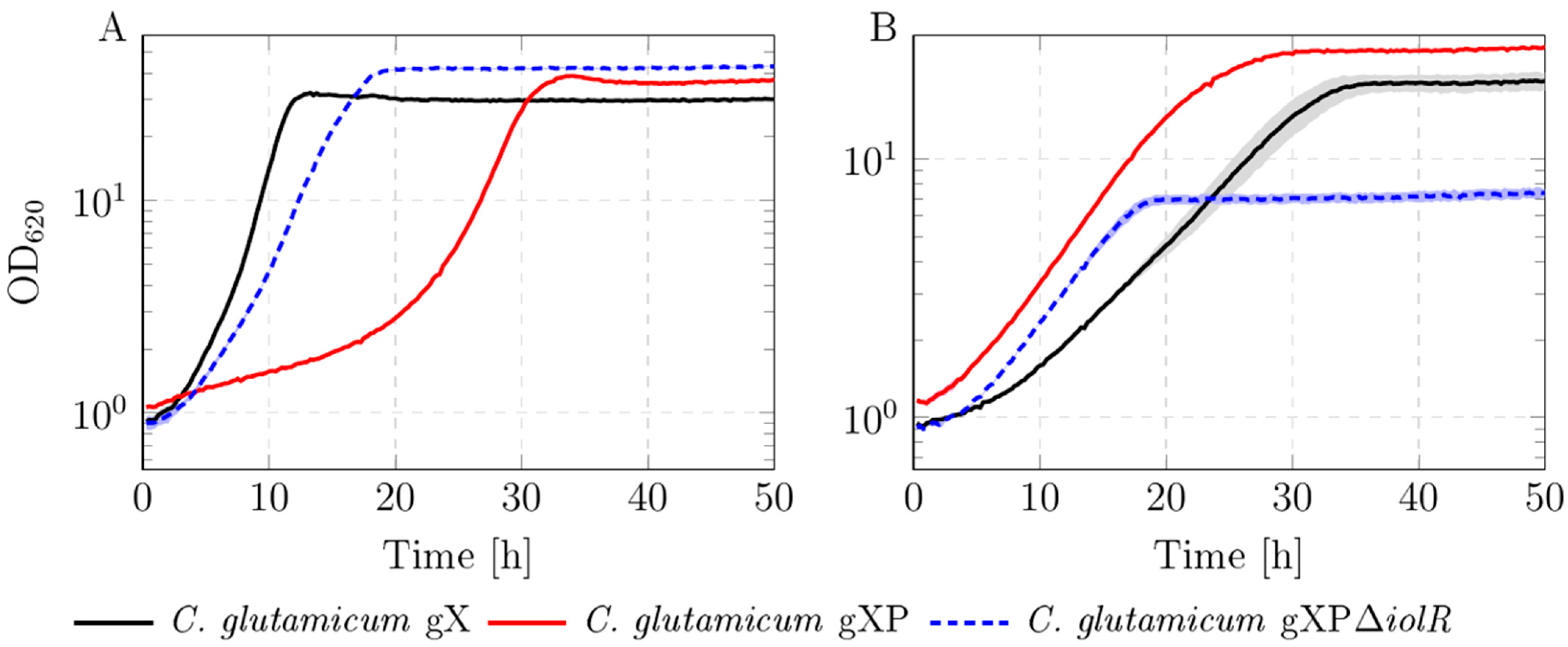
3.1. Strain Engineering
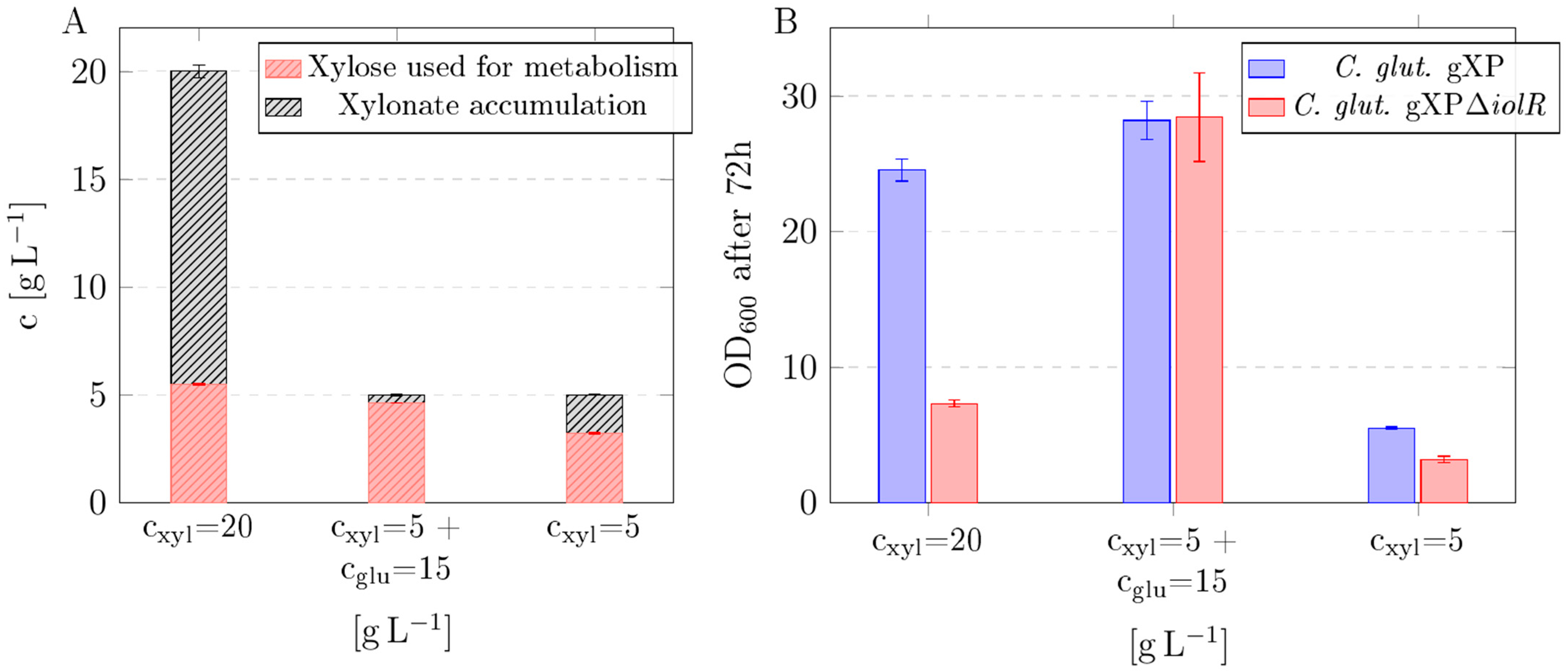
3.2. Impact of Substrate Composition on Xylose Utilization and Xylonate Accumulation
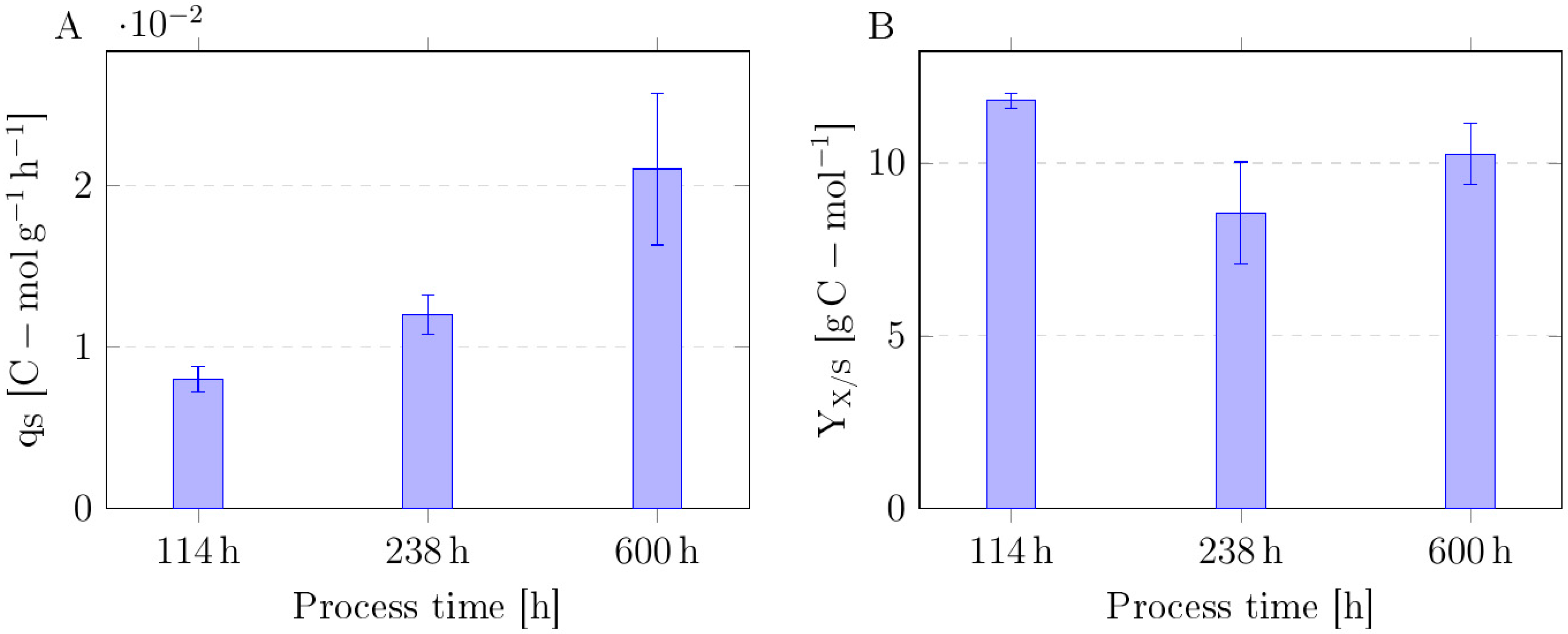
3.3. Continuous Adaptive Laboratory Evolution
3.4. Mutations in Evolved Populations ALE_1 and ALE_2
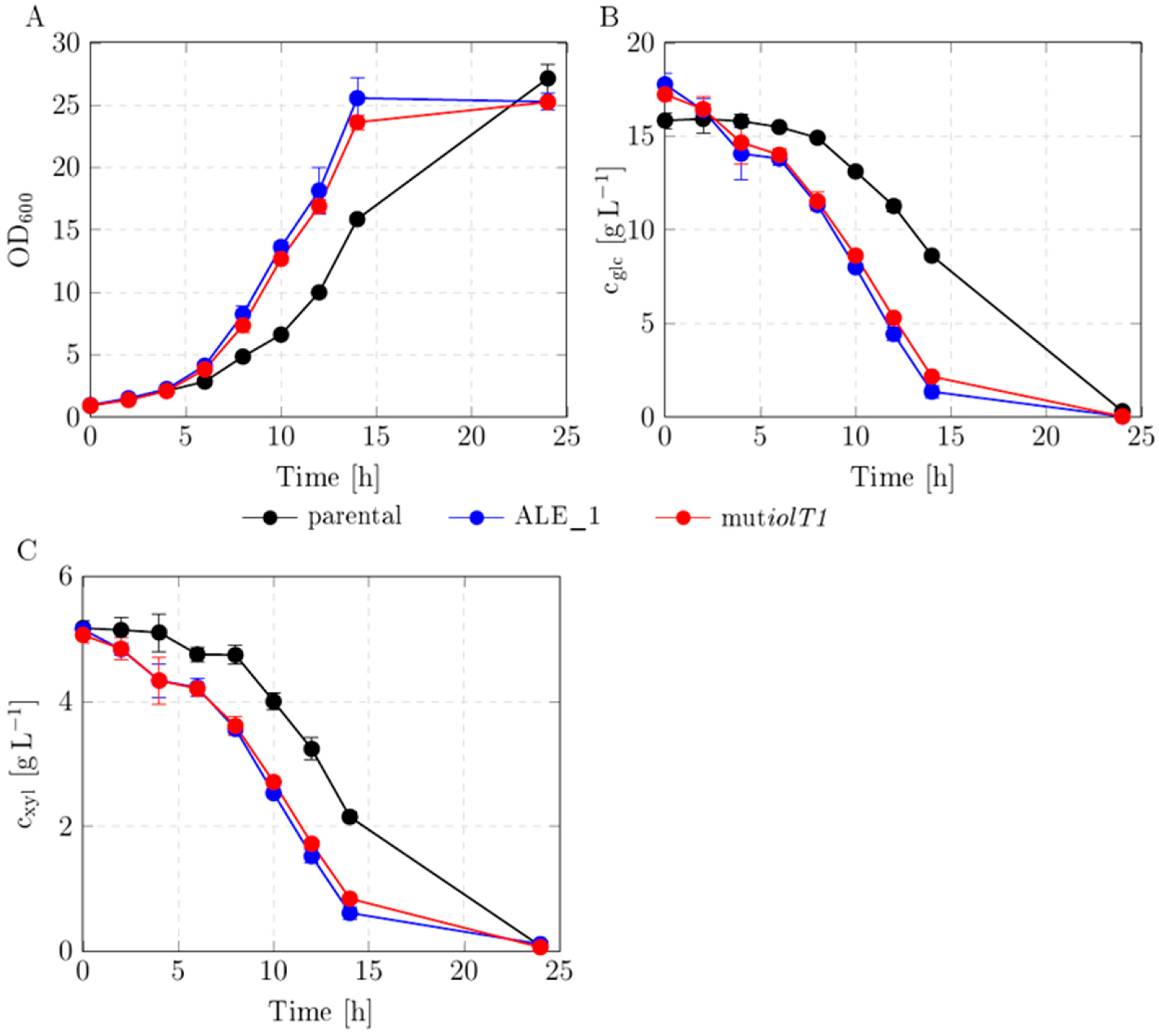
3.5. Reverse Engineering of One Mutation into Parental Background and Growth Characterization
4. Discussion
4.1. Deletion of ptsG Leads to Better Growth in Xylose-Containing Medium
4.2. Carbon Uptake Dynamics During ALE Cultivation Reveal Improved Co-Utilization with Stable Sugar Preference
4.3. Preliminary Characterization of iolT1 Mutation Reveals Growth and Uptake Boost
- C2689275T and G2689278A in 16srRNA Copy E
- G1992621A in cg2103 (DtxRR103H)
- C2769644T in cg2910 (IpsAD141N)
- C190823G in cg0223 (IolT1A452P)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALE | adaptive laboratory evolution |
| C. glutamicum | Corynebacterium glutamicum |
| DO | dissolved oxygen |
| dYT | double yeast tryptone |
| E. coli | Escherichia coli |
| glc | glucose |
| GRAS | generally recognized as safe |
| LB | lysogeny broth |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| OD | optical density |
| PCA | protocatechuic acid |
| PEP | phosphoenolpyruvate |
| PTS | phosphotransferase system |
| WGS | whole-genome sequencing |
| XI | xylose isomerase |
| xyl | xylose |
References
- Isikgor, F.H.; Becer, C.R. Lignocellulosic Biomass: A Sustainable Platform for the Production of Bio-Based Chemicals and Polymers. Polym. Chem. 2015, 6, 4497–4559. [Google Scholar] [CrossRef]
- Pérez-García, F.; Brito, L.F.; Irla, M.; Jorge, J.M.P.; Sgobba, E. Editorial: Promising and Sustainable Microbial Feedstocks for Biotechnological Processes. Front. Microbiol. 2022, 13, 973723. [Google Scholar] [CrossRef] [PubMed]
- Cazier, E.A.; Pham, T.-N.; Cossus, L.; Abla, M.; Ilc, T.; Lawrence, P. Exploring Industrial Lignocellulosic Waste: Sources, Types, and Potential as High-Value Molecules. Waste Manag. 2024, 188, 11–38. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Feng, Y.; Fu, J.; Guo, H.; Guo, Y.; Han, B.; Jiang, Z.; Kong, L.; Li, C.; Liu, H.; et al. Catalytic Conversion of Lignocellulosic Biomass into Chemicals and Fuels. Green Energy Environ. 2023, 8, 10–114. [Google Scholar] [CrossRef]
- Xu, J.-Z.; Ruan, H.-Z.; Yu, H.-B.; Liu, L.-M.; Zhang, W. Metabolic Engineering of Carbohydrate Metabolism Systems in Corynebacterium glutamicum for Improving the Efficiency of L-Lysine Production from Mixed Sugar. Microb. Cell Factories 2020, 19, 39. [Google Scholar] [CrossRef]
- Marienhagen, J. Engineering of Corynebacterium glutamicum for the Synthesis of Aromatic Compounds. Appl. Microbiol. Biotechnol. 2025, 109, 132. [Google Scholar] [CrossRef]
- Ikeda, M.; Nakagawa, S. The Corynebacterium glutamicum Genome: Features and Impacts on Biotechnological Processes. Appl. Microbiol. Biotechnol. 2003, 62, 99–109. [Google Scholar] [CrossRef]
- Kalinowski, J.; Bathe, B.; Bartels, D.; Bischoff, N.; Bott, M.; Burkovski, A.; Dusch, N.; Eggeling, L.; Eikmanns, B.J.; Gaigalat, L.; et al. The Complete Corynebacterium glutamicum ATCC 13032 Genome Sequence and Its Impact on the Production of L-Aspartate-Derived Amino Acids and Vitamins. J. Biotechnol. 2003, 104, 5–25. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Al Makishah, N.H.; Sun, X.; Wen, Z.; Jiang, Y.; Yang, S. Advances and Perspectives for Genome Editing Tools of Corynebacterium glutamicum. Front. Microbiol. 2021, 12, 654058. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Vertès, A.A.; Okino, S.; Inui, M.; Yukawa, H. Engineering of a Xylose Metabolic Pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2006, 72, 3418–3428. [Google Scholar] [CrossRef]
- Meiswinkel, T.M.; Gopinath, V.; Lindner, S.N.; Nampoothiri, K.M.; Wendisch, V.F. Accelerated Pentose Utilization by Corynebacterium glutamicum for Accelerated Production of Lysine, Glutamate, Ornithine and Putrescine. Microb. Biotechnol. 2013, 6, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Li, G.; Chang, Z.; Tao, R.; Cui, Z.; Wang, Z.; Tang, Y.; Chen, T.; Zhao, X. Metabolic Engineering of Corynebacterium glutamicum for Efficient Production of Succinate from Lignocellulosic Hydrolysate. Biotechnol. Biofuels 2018, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Huang, J.; Wu, Y.; Wu, W.; Zhang, Y.; Liu, D. Metabolic Engineering of Corynebacterium glutamicum for the Production of 3-Hydroxypropionic Acid from Glucose and Xylose. Metab. Eng. 2017, 39, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Mao, Y.; Luo, J.; Liu, P.; Jiang, M.; He, G.; Zhang, Z.; Cao, Q.; Shen, J.; Ma, H.; et al. Global Cellular Metabolic Rewiring Adapts Corynebacterium glutamicum to Efficient Nonnatural Xylose Utilization. Appl. Environ. Microbiol. 2022, 88, e01518–e01522. [Google Scholar] [CrossRef]
- Shi, X.; Chang, J.; Kim, M.; Lee, M.-E.; Shin, H.-Y.; Ok Han, S. Isopropanol Production Using Engineered Corynebacterium glutamicum from Waste Rice Straw Biomass. Bioresour. Technol. 2024, 396, 130416. [Google Scholar] [CrossRef]
- Sasaki, M.; Jojima, T.; Inui, M.; Yukawa, H. Xylitol Production by Recombinant Corynebacterium glutamicum under Oxygen Deprivation. Appl. Microbiol. Biotechnol. 2009, 86, 1057–1066. [Google Scholar] [CrossRef]
- Zhong, Z.; Ma, Z.; Cao, Y.; Zhang, H.; Qi, Y.; Wei, L.; Jiang, J.; Xu, N.; Liu, J. Metabolic Engineering of Corynebacterium glutamicum for Efficient L-Homoserine Production from Lignocellulose-Derived Sugars. ACS Sustain. Chem. Eng. 2025, 13, 3441–3451. [Google Scholar] [CrossRef]
- Jo, S.; Yoon, J.; Lee, S.-M.; Um, Y.; Han, S.O.; Woo, H.M. Modular Pathway Engineering of Corynebacterium glutamicum to Improve Xylose Utilization and Succinate Production. J. Biotechnol. 2017, 258, 69–78. [Google Scholar] [CrossRef]
- Tenhaef, N.; Brüsseler, C.; Radek, A.; Hilmes, R.; Unrean, P.; Marienhagen, J.; Noack, S. Production of D-Xylonic Acid Using a Non-Recombinant Corynebacterium glutamicum Strain. Bioresour. Technol. 2018, 268, 332–339. [Google Scholar] [CrossRef]
- Werner, F.; Schwardmann, L.S.; Siebert, D.; Rückert-Reed, C.; Kalinowski, J.; Wirth, M.-T.; Hofer, K.; Takors, R.; Wendisch, V.F.; Blombach, B. Metabolic Engineering of Corynebacterium glutamicum for Fatty Alcohol Production from Glucose and Wheat Straw Hydrolysate. Biotechnol. Biofuels Bioprod. 2023, 16, 116. [Google Scholar] [CrossRef]
- Mutz, M.; Brüning, V.; Brüsseler, C.; Müller, M.-F.; Noack, S.; Marienhagen, J. Metabolic Engineering of Corynebacterium glutamicum for the Production of Anthranilate from Glucose and Xylose. Microb. Biotechnol. 2024, 17, e14388. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, V.; Meiswinkel, T.M.; Wendisch, V.F.; Nampoothiri, K.M. Amino Acid Production from Rice Straw and Wheat Bran Hydrolysates by Recombinant Pentose-Utilizing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2011, 92, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.N.; Seibold, G.M.; Henrich, A.; Krämer, R.; Wendisch, V.F. Phosphotransferase System-Independent Glucose Utilization in Corynebacterium glutamicum by Inositol Permeases and Glucokinases. Appl. Environ. Microbiol. 2011, 77, 3571–3581. [Google Scholar] [CrossRef] [PubMed]
- Brüsseler, C.; Radek, A.; Tenhaef, N.; Krumbach, K.; Noack, S.; Marienhagen, J. The Myo-Inositol/Proton Symporter IolT1 Contributes to d-Xylose Uptake in Corynebacterium glutamicum. Bioresour. Technol. 2018, 249, 953–961. [Google Scholar] [CrossRef]
- Dragosits, M.; Mattanovich, D. Adaptive Laboratory Evolution—Principles and Applications for Biotechnology. Microb. Cell Factories 2013, 12, 64. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on Transformation of Escherichia Coli with Plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Kinoshita, S.; Nakayama, K.; Akita, S. Taxonomical Study of Glutamic Acid Accumulating Bacteria, Micrococcus glutamicus Nov. sp. Bull. Agric. Chem. Soc. Jpn. 1958, 22, 176–185. [Google Scholar] [CrossRef]
- Abe, S.; Takayama, K.-I.; Kinoshita, S. Taxonomical Studies on Glutamic Acid-Producing Bacteria. J. Gen. Appl. Microbiol. 1967, 13, 279–301. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1989; ISBN 978-0-87969-309-1. [Google Scholar]
- Eggeling, L.; Bott, M. (Eds.) Handbook of Corynebacterium glutamicum; Taylor & Francis: Boca Raton, FL, USA, 2005; ISBN 978-0-8493-1821-4. [Google Scholar]
- Eikmanns, B.J.; Thum-Schmitz, N.; Eggeling, L.; Lüdtke, K.-U.; Sahm, H. Nucleotide Sequence, Expression and Transcriptional Analysis of the Corynebacterium glutamicum gltA Gene Encoding Citrate Synthase. Microbiology 1994, 140, 1817–1828. [Google Scholar] [CrossRef]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small Mobilizable Multi-Purpose Cloning Vectors Derived from the Escherichia coli Plasmids pK18 and pK19: Selection of Defined Deletions in the Chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G. Enzymatic Assembly of Overlapping DNA Fragments. Methods Enzymol. 2011, 498, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Haas, T.; Müller, F.; Buchmann, A.; Harm-Bekbenbetova, J.; Freund, A.; Nieß, A.; Persicke, M.; Kalinowski, J.; Blombach, B.; et al. Continuous Adaptive Evolution of a Fast-Growing Corynebacterium glutamicum Strain Independent of Protocatechuate. Front. Microbiol. 2019, 10, 1648. [Google Scholar] [CrossRef] [PubMed]
- Veit, A.; Rittmann, D.; Georgi, T.; Youn, J.-W.; Eikmanns, B.J.; Wendisch, V.F. Pathway Identification Combining Metabolic Flux and Functional Genomics Analyses: Acetate and Propionate Activation by Corynebacterium glutamicum. J. Biotechnol. 2009, 140, 75–83. [Google Scholar] [CrossRef]
- Teramoto, H.; Inui, M.; Yukawa, H. Transcriptional Regulators of Multiple Genes Involved in Carbon Metabolism in Corynebacterium glutamicum. J. Biotechnol. 2011, 154, 114–125. [Google Scholar] [CrossRef]
- Klaffl, S.; Brocker, M.; Kalinowski, J.; Eikmanns, B.J.; Bott, M. Complex Regulation of the Phosphoenolpyruvate Carboxykinase Gene pck and Characterization of Its GntR-Type Regulator IolR as a Repressor of myo-Inositol Utilization Genes in Corynebacterium glutamicum. J. Bacteriol. 2013, 195, 4283–4296. [Google Scholar] [CrossRef]
- Pfeifer, E.; Gätgens, C.; Polen, T.; Frunzke, J. Adaptive Laboratory Evolution of Corynebacterium glutamicum towards Higher Growth Rates on Glucose Minimal Medium. Sci. Rep. 2017, 7, 16780. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, L.; Tuyishime, P.; Liu, J.; Zhang, K.; Gao, N.; Zhang, Z.; Ni, X.; Feng, J.; Yuan, Q.; et al. Adaptive Laboratory Evolution Enhances Methanol Tolerance and Conversion in Engineered Corynebacterium glutamicum. Commun. Biol. 2020, 3, 217. [Google Scholar] [CrossRef]
- Halle, L.; Hollmann, N.; Tenhaef, N.; Mbengi, L.; Glitz, C.; Wiechert, W.; Polen, T.; Baumgart, M.; Bott, M.; Noack, S. Robotic Workflows for Automated Long-Term Adaptive Laboratory Evolution: Improving Ethanol Utilization by Corynebacterium glutamicum. Microb. Cell Factories 2023, 22, 175. [Google Scholar] [CrossRef]
- Weikert, C.; Sauer, U.; Bailey, J.E. Use of a Glycerol-Limited, Long-Term Chemostat for Isolation of Escherichia coli Mutants with Improved Physiological Properties. Microbiology 1997, 143, 1567–1574. [Google Scholar] [CrossRef]
- Zambrano, M.M.; Siegele, D.A.; Almirón, M.; Tormo, A.; Kolter, R. Microbial Competition: Escherichia coli Mutants That Take Over Stationary Phase Cultures. Science 1993, 259, 1757–1760. [Google Scholar] [CrossRef] [PubMed]
- Liebl, W.; Klamer, R.; Schleifer, K.-H. Requirement of Chelating Compounds for the Growth of Corynebacterium glutamicum in Synthetic Media. Appl. Microbiol. Biotechnol. 1989, 32, 205–210. [Google Scholar] [CrossRef]
- Graf, M.; Zieringer, J.; Haas, T.; Nieß, A.; Blombach, B.; Takors, R. Physiological Response of Corynebacterium glutamicum to Increasingly Nutrient-Rich Growth Conditions. Front. Microbiol. 2018, 9, 2058. [Google Scholar] [CrossRef] [PubMed]
- Radek, A.; Krumbach, K.; Gätgens, J.; Wendisch, V.F.; Wiechert, W.; Bott, M.; Noack, S.; Marienhagen, J. Engineering of Corynebacterium glutamicum for Minimized Carbon Loss during Utilization of D-Xylose Containing Substrates. J. Biotechnol. 2014, 192, 156–160. [Google Scholar] [CrossRef]
- Martín, J.F.; Barreiro, C.; González-Lavado, E.; Barriuso, M. Ribosomal RNA and Ribosomal Proteins in Corynebacteria. J. Biotechnol. 2003, 104, 41–53. [Google Scholar] [CrossRef]
- Xu, Z.; Culver, G.M. Differential Assembly of 16S rRNA Domains during 30S Subunit Formation. RNA 2010, 16, 1990–2001. [Google Scholar] [CrossRef]
- Wennerhold, J.; Bott, M. The DtxR Regulon of Corynebacterium glutamicum. J. Bacteriol. 2006, 188, 2907–2918. [Google Scholar] [CrossRef]
- Rolfe, M.D.; Rice, C.J.; Lucchini, S.; Pin, C.; Thompson, A.; Cameron, A.D.S.; Alston, M.; Stringer, M.F.; Betts, R.P.; Baranyi, J.; et al. Lag Phase Is a Distinct Growth Phase That Prepares Bacteria for Exponential Growth and Involves Transient Metal Accumulation. J. Bacteriol. 2012, 194, 686–701. [Google Scholar] [CrossRef]
- Thoma, F.; Appel, C.; Russ, D.; Huber, J.; Werner, F.; Blombach, B. Improving Growth Properties of Corynebacterium glutamicum by Implementing an Iron-responsive Protocatechuate Biosynthesis. Microb. Biotechnol. 2023, 16, 1041–1053. [Google Scholar] [CrossRef]
- Brune, I.; Werner, H.; Hüser, A.T.; Kalinowski, J.; Pühler, A.; Tauch, A. The DtxR Protein Acting as Dual Transcriptional Regulator Directs a Global Regulatory Network Involved in Iron Metabolism of Corynebacterium glutamicum. BMC Genom. 2006, 7, 21. [Google Scholar] [CrossRef]
- Walter, T.; Veldmann, K.H.; Götker, S.; Busche, T.; Rückert, C.; Kashkooli, A.B.; Paulus, J.; Cankar, K.; Wendisch, V.F. Physiological Response of Corynebacterium glutamicum to Indole. Microorganisms 2020, 8, 1945. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, M.; Luder, K.; Grover, S.; Gätgens, C.; Besra, G.S.; Frunzke, J. IpsA, a Novel LacI-Type Regulator, Is Required for Inositol-Derived Lipid Formation in Corynebacteria and Mycobacteria. BMC Biol. 2013, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Strehlow, K.G.; Robertson, A.D.; Baldwin, R.L. Proline for Alanine Substitutions in the C-Peptide Helix of Ribonuclease A. Biochemistry 1991, 30, 5810–5814. [Google Scholar] [CrossRef] [PubMed]


| Strain | Relevant Characteristics | References |
|---|---|---|
| E. coli DH5α | F-Φ80lacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR17 (rK−, mK+) supE44 thi-1 gyrA96 relA1 phoA | [26] |
| E. coli S17-1 | recA pro hsdR RP4-2-Tc::Mu-Km::Tn7 | [27] |
| C. glutamicum WT | C. glutamicum wild type (WT), ATCC13032 | [28,29] |
| C. glutamicum gX | C. glutamicum ΔactA::xylAB (C. glutamicum WT derivate with the synthetic operon, consisting of xylA of Xanthomonas campestris and xylB of C. glutamicum WT, integrated into the gene locus actA (cg2840) with evolved sequence upstream of xylA) | [20] |
| C. glutamicum gXP | C. glutamicum gX with the deletion of gene ptsG encoding PTS system glucose-specific EIICBA component | This study |
| C. glutamicum gXPΔiolR | C. glutamicum gXP with the deletion of gene iolR encoding the transcriptional regulator IolR | This study |
| C. glutamicum ALE_1 | Population of evolved C. glutamicum gXPΔiolR after stage 1 (300 h of continuous adaptation) | This study |
| C. glutamicum ALE_2 | Population of evolved C. glutamicum gXPΔiolR after stage 2 (600 h of continuous adaptation) | This study |
| C. glutamicum gXPΔiolRΔiolT1 | C. glutamicum gXPΔiolR with the deletion of gene iolT1, encoding myo-inositol/proton symporter IolT1 | This study |
| C. glutamicum gXPΔiolRΔiolT1::mut iolT1 | C. glutamicum gXPΔiolRΔiolT1 with integrated mutiolT1 (iolT1A452P), encoding myo-inositol/proton symporter IolT1 | This study |
| Gene | Mutation | Frequency [%] |
|---|---|---|
| iolT1 (cg0223) | C190823G → A452P | 99.86 |
| 16s rRNA copy E | C2689275T | 100.00 |
| Gene | Mutation | Frequency [%] |
|---|---|---|
| iolT1 (cg0223) | C190823G → A452P | 99.86 |
| 16s rRNA copy E | C2689275T | 90.91 |
| 16s rRNA copy E | G2689278A | 90.91 |
| dtxR (cg2103) | G1992621A → R103H | 97.66 |
| ipsA (cg2910) | C2769644T → D141N | 97.46 |
| C. glutamicum gXPΔiolR Strain | µmax [h−1] | qglc [g g−1 h−1] | qxyl [g g−1 h−1] |
|---|---|---|---|
| parental | 0.20 ± 0.00 *** | 0.36 ± 0.02 *** | 0.15 ± 0.01 * |
| ALE_1 | 0.27 ± 0.01 | 0.68 ± 0.03 | 0.18 ± 0.01 |
| mutiolT1 | 0.27 ± 0.01 | 0.66 ± 0.04 | 0.18 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hofer, K.; Schwardmann, L.S.; Youn, J.-W.; Wendisch, V.F.; Takors, R. Single Mutation in iolT1 in ptsG-Deficient Corynebacterium glutamicum Enables Growth Boost in Xylose-Containing Media. Microorganisms 2025, 13, 1606. https://doi.org/10.3390/microorganisms13071606
Hofer K, Schwardmann LS, Youn J-W, Wendisch VF, Takors R. Single Mutation in iolT1 in ptsG-Deficient Corynebacterium glutamicum Enables Growth Boost in Xylose-Containing Media. Microorganisms. 2025; 13(7):1606. https://doi.org/10.3390/microorganisms13071606
Chicago/Turabian StyleHofer, Katharina, Lynn S. Schwardmann, Jung-Won Youn, Volker F. Wendisch, and Ralf Takors. 2025. "Single Mutation in iolT1 in ptsG-Deficient Corynebacterium glutamicum Enables Growth Boost in Xylose-Containing Media" Microorganisms 13, no. 7: 1606. https://doi.org/10.3390/microorganisms13071606
APA StyleHofer, K., Schwardmann, L. S., Youn, J.-W., Wendisch, V. F., & Takors, R. (2025). Single Mutation in iolT1 in ptsG-Deficient Corynebacterium glutamicum Enables Growth Boost in Xylose-Containing Media. Microorganisms, 13(7), 1606. https://doi.org/10.3390/microorganisms13071606








