Two-Component MprAB System Regulates the Expression of Genes Involved in Cell Envelope Biosynthesis in Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. The MprAB Two-Component System Is Conserved in Corynebacterium and Mycobacterium
3.2. Transcriptome Profile of Altered mprA Expression
3.3. Identification of mprA-Binding Regions
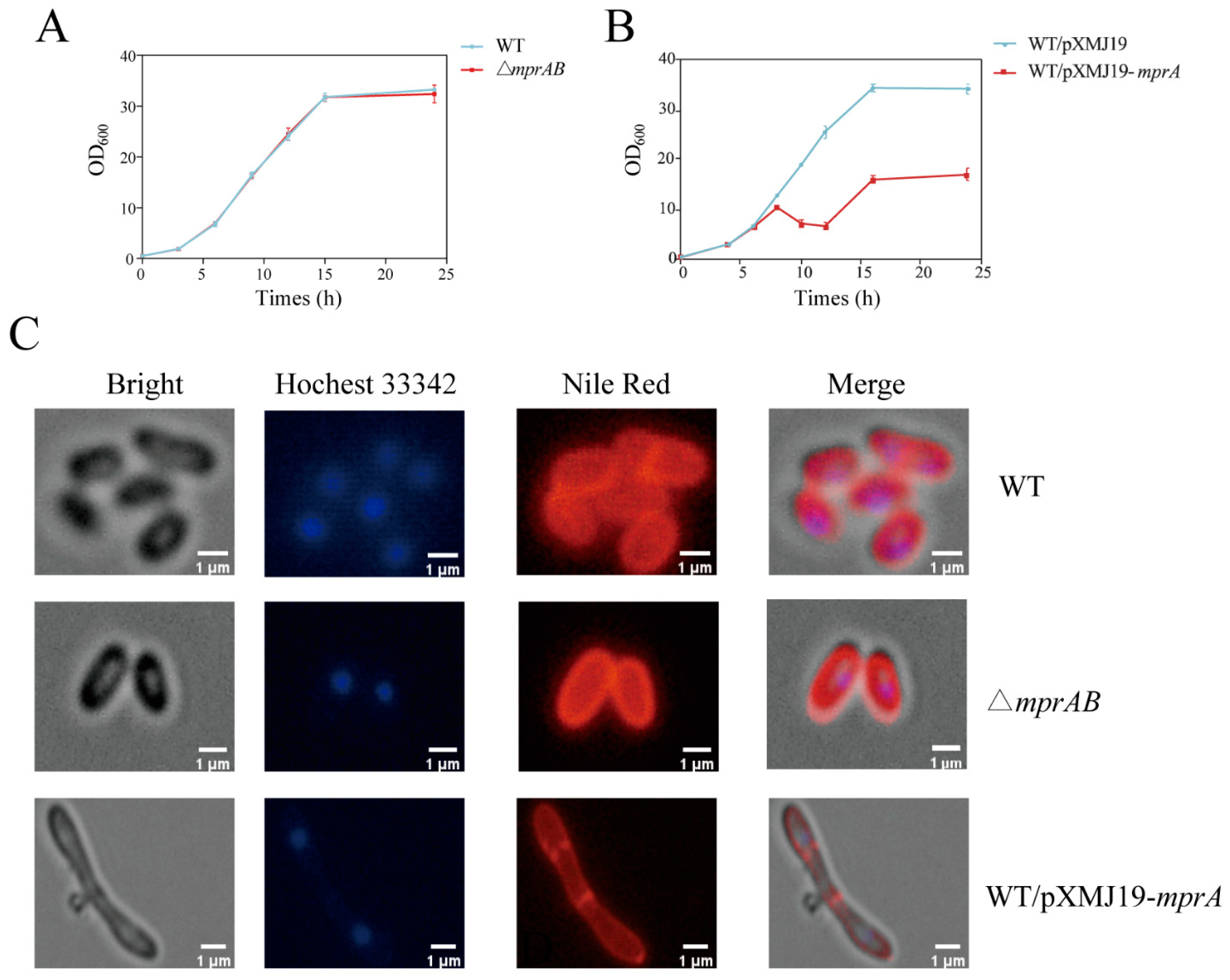
3.4. Overexpression of mprA Caused Cell Envelope Defects and Increased Alanine Titers in C. glutamicum
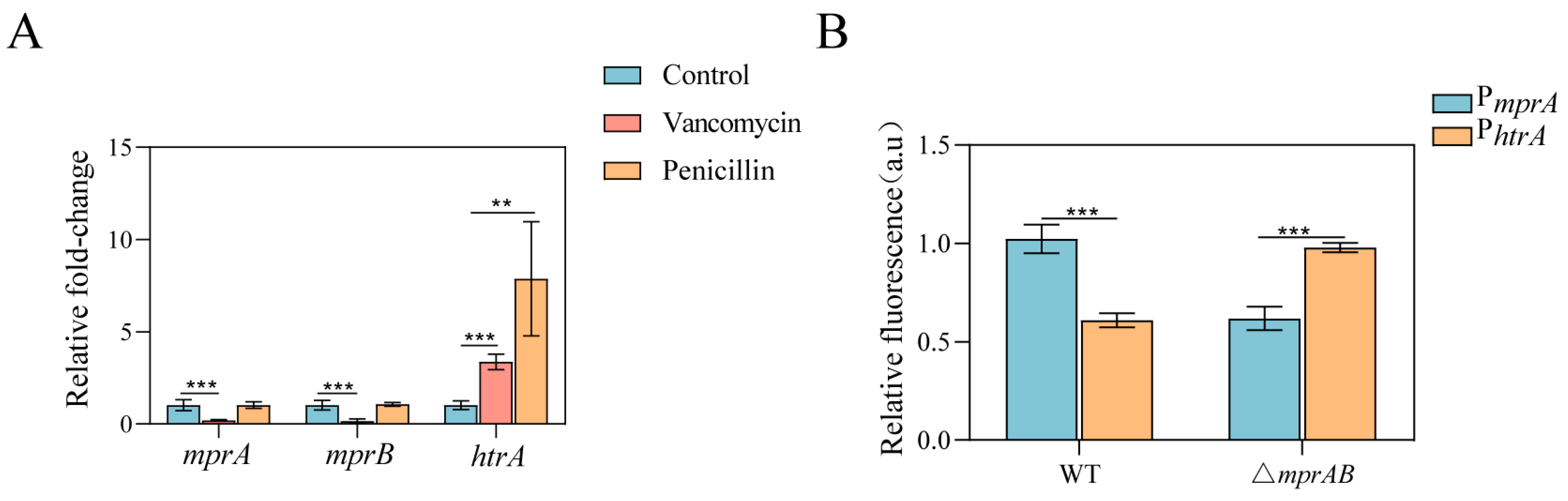
3.5. The MprAB Two-Component System and Its Direct Target Gene htrA Are Induced by Vancomycin
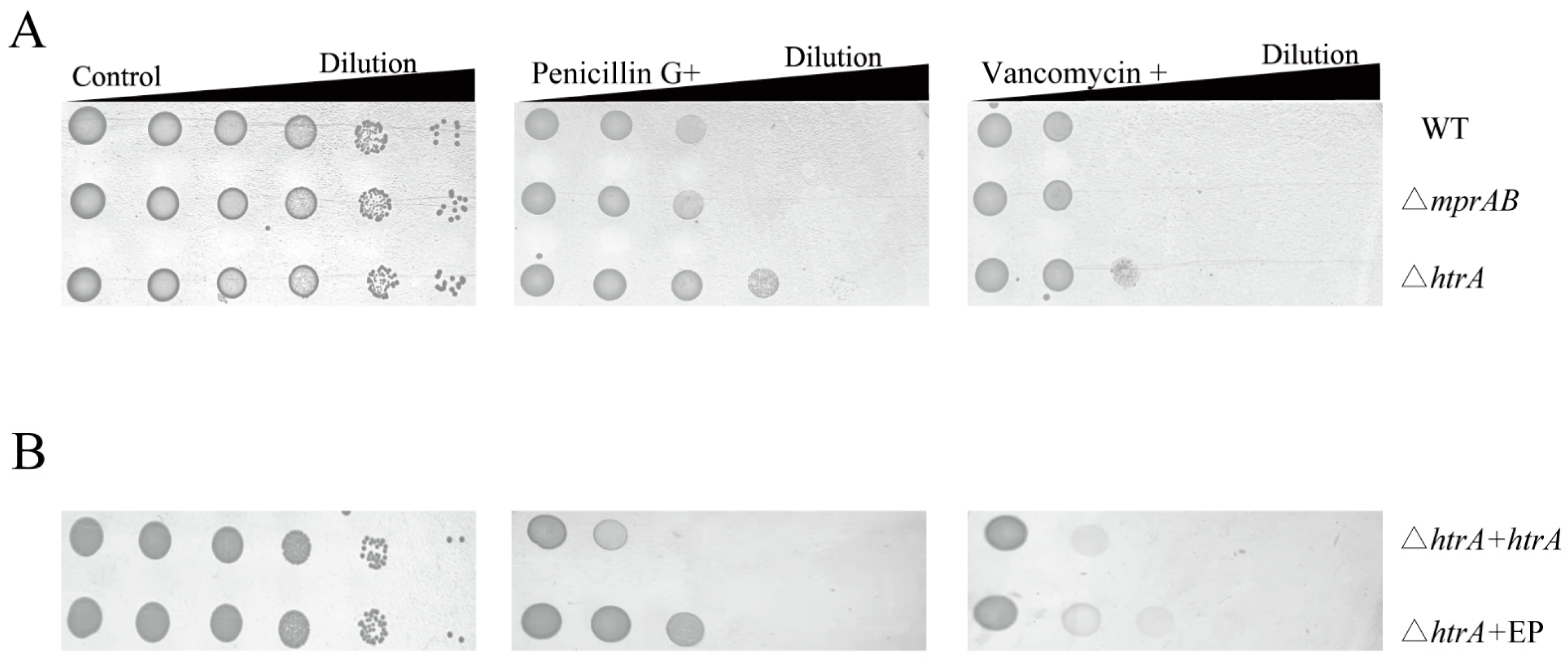
3.6. HtrA Mediates Vancomycin and Penicillin Resistance, but MprAB Does Not
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McKitterick, A.C.; Bernhardt, T.G. Phage resistance profiling identifies new genes required for biogenesis and modification of the corynebacterial cell envelope. eLife 2017, 11, 79981–80005. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Rodriguez-Rivera, F.P.; Lim, H.C.; Bell, J.C.; Bernhardt, T.G.; Bertozzi, C.R.; Theriot, J.A. Sequential assembly of the septal cell envelope prior to V snapping in Corynebacterium glutamicum. Nat. Chem. Biol. 2019, 15, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.C.; Sher, J.W.; Rodriguez-Rivera, F.P.; Fumeaux, C.; Bertozzi, C.R.; Bernhardt, T.G. Identification of new components of the RipC-FtsEX cell separation pathway of Corynebacterineae. PLoS Genet. 2019, 15, e1008284. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Mechaly, A.; Betton, J.M.; Antoine, R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018, 16, 585–593. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Liu, Z.; Gong, G.; Zha, J. Microbial cell surface engineering for high-level synthesis of bio-products. Biotechnol. Adv. 2022, 55, 107912. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, K.; Nie, Y.; Wu, X.L. The roles of the two-component system, MtrAB, in response to diverse cell envelope stresses in Dietzia sp. DQ12-45-1b. Appl. Environ. Microbiol. 2022, 88, e0133722. [Google Scholar] [CrossRef]
- Mike, L.A.; Choby, J.E.; Brinkman, P.R.; Olive, L.Q.; Dutter, B.F.; Ivan, S.J.; Gibbs, C.M.; Sulikowski, G.A.; Stauff, D.L.; Skaar, E.P. Two-component system cross-regulation integrates Bacillus anthracis response to heme and cell envelope stress. PLoS Pathog. 2014, 10, e1004044. [Google Scholar] [CrossRef]
- Komazin, G.; Rizk, A.A.; Armbruster, K.M.; Bonnell, V.A.; Llinás, M.; Meredith, T.C. A copper-responsive two-component system governs lipoprotein remodeling in Listeria monocytogenes. J. Bacteriol. 2003, 205, e0039022. [Google Scholar] [CrossRef]
- Casino, P.; Rubio, V.; Marina, A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 2009, 139, 325–336. [Google Scholar] [CrossRef]
- Trajtenberg, F.; Albanesi, D.; Ruétalo, N.; Botti, H.; Mechaly, A.E.; Nieves, M.; Aguilar, P.S.; Cybulski, L.; Larrieux, N.; de Mendoza, D.; et al. Allosteric activation of bacterial response regulators: The role of the cognate histidine kinase beyond phosphorylation. mBio 2014, 5, e02105. [Google Scholar] [CrossRef]
- Schmidl, S.R.; Ekness, F.; Sofjan, K.; Daeffler, K.N.; Brink, K.R.; Landry, B.P.; Gerhardt, K.P.; Dyulgyarov, N.; Sheth, R.U.; Tabor, J.J. Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat. Chem. Biol. 2019, 15, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Parish, T. Two-component regulatory systems of Mycobacteria. Microbiol. Spectr. 2014, 2, MGM2-0010-2013. [Google Scholar] [CrossRef] [PubMed]
- Möker, N.; Brocker, M.; Schaffer, S.; Krämer, R.; Morbach, S.; Bott, M. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 2004, 54, 420–438. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Hovey, R.; Kane, J.; Singh, V.; Zahrt, T.C. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 2006, 188, 2134–2143. [Google Scholar] [CrossRef]
- Sureka, K.; Dey, S.; Datta, P.; Singh, A.K.; Dasgupta, A.; Rodrigue, S.; Basu, J.; Kundu, M. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 2007, 65, 261–276. [Google Scholar] [CrossRef]
- Zahrt, T.C.; Deretic, V. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc. Natl. Acad. Sci. USA 2001, 98, 12706–12711. [Google Scholar] [CrossRef]
- Datta, P.; Ravi, J.; Guerrini, V.; Chauhan, R.; Neiditch, M.B.; Shell, S.S.; Fortune, S.M.; Hancioglu, B.; Igoshin, O.A.; Gennaro, M.L. The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions. Mol. Microbiol. 2015, 97, 408–422. [Google Scholar] [CrossRef]
- He, H.; Bretl, D.J.; Penoske, R.M.; Anderson, D.M.; Zahrt, T.C. Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J. Bacteriol. 2011, 193, 5105–5118. [Google Scholar] [CrossRef]
- Bretl, D.J.; He, H.; Demetriadou, C.; White, M.J.; Penoske, R.M.; Salzman, N.H.; Zahrt, T.C. MprA and DosR coregulate a Mycobacterium tuberculosis virulence operon encoding Rv1813c and Rv1812c. Infect. Immun. 2012, 80, 3018–3033. [Google Scholar] [CrossRef]
- Pang, X.; Howard, S.T. Regulation of the alpha-crystallin gene acr2 by the MprAB two-component system of Mycobacterium tuberculosis. J. Bacteriol. 2007, 189, 6213–6221. [Google Scholar] [CrossRef]
- Pang, X.; Samten, B.; Cao, G.; Wang, X.; Tvinnereim, A.R.; Chen, X.L.; Howard, S.T. MprAB regulates the espA operon in Mycobacterium tuberculosis and modulates ESX-1 function and host cytokine response. J. Bacteriol. 2013, 195, 66–75. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zahrt, T.C. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J. Bacteriol. 2005, 187, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Sureka, K.; Ghosh, B.; Dasgupta, A.; Basu, J.; Kundu, M.; Bose, I. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS ONE 2008, 3, e1771. [Google Scholar] [CrossRef]
- Pang, X.; Cao, X.; Neuenschwander, P.F.; Haydel, S.E.; Hou, G.; Howard, S.T. The β-propeller gene Rv1057 of Mycobacterium tuberculosis has a complex promoter directly regulated by both the MprAB and TrcRS two-component systems. Tuberculosis 2011, 91, 142–149. [Google Scholar] [CrossRef]
- Pang, X.; Vu, P.; Byrd, T.F.; Ghanny, S.; Soteropoulos, P.; Mukamolova, G.V.; Wu, S.; Samten, B.; Howard, S.T. Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 2007, 153, 1229–1242. [Google Scholar] [CrossRef]
- Bretl, D.J.; Bigley, T.M.; Terhune, S.S.; Zahrt, T.C. The MprB extracytoplasmic domain negatively regulates activation of the Mycobacterium tuberculosis MprAB two-component system. J. Bacteriol. 2014, 196, 391–406. [Google Scholar] [CrossRef]
- Wong, K.W. The Role of ESX-1 in Mycobacterium tuberculosis Pathogenesis. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Zahrt, T.C.; Wozniak, C.; Jones, D.; Trevett, A. Functional analysis of the Mycobacterium tuberculosis MprAB two-component signal transduction system. Infect. Immun. 2003, 71, 6962–6970. [Google Scholar] [CrossRef]
- Pan, L.; Chen, X.; Wang, K.; Mao, Z. Understanding high ε-poly-L-lysine production by Streptomyces albulus using pH shock strategy in the level of transcriptomic. J. Ind. Microbiol. Biotechnol. 2019, 46, 1781–1792. [Google Scholar] [CrossRef]
- Toyoda, K.; Inui, M. Regulons of global transcription factors in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2016, 100, 45–60. [Google Scholar] [CrossRef]
- Kleine, B.; Chattopadhyay, A.; Polen, T.; Pinto, D.; Mascher, T.; Bott, M.; Brocker, M.; Freudl, R. The three-component system EsrISR regulates a cell envelope stress response in Corynebacterium glutamicum. Mol. Microbiol. 2017, 106, 719–741. [Google Scholar] [CrossRef] [PubMed]
- Keilhauer, C.; Eggeling, L.; Sahm, H. Isoleucine synthesis in Corynebacterium glutamicum: Molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993, 175, 5595–5603. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.R.; Wei, S.Y.; Ding, M.Z.; Hou, Z.J.; Wang, D.J.; Xu, Q.M.; Chen, Y.J. Enhancing fengycin production in the co-culture of Bacillus subtilis and Corynebacterium glutamicum by engineering proline transporter. Bioresour. Technol. 2003, 383, 129229. [Google Scholar] [CrossRef] [PubMed]
- SEQC/MAQC-III Consortium. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559–15570. [Google Scholar] [CrossRef]
- Baumgart, M.; Schubert, K.; Bramkamp, M.; Frunzke, J. Impact of LytR-CpsA-Psr proteins on cell wall biosynthesis in Corynebacterium glutamicum. J. Bacteriol. 2016, 198, 3045–3059. [Google Scholar] [CrossRef]
- Toyoda, K.; Inui, M. Extracytoplasmic function sigma factor σD confers resistance to environmental stress by enhancing mycolate synthesis and modifying peptidoglycan structures in Corynebacterium glutamicum. Mol. Microbiol. 2018, 107, 312–329. [Google Scholar] [CrossRef]
- Dietrich, C.; Li de la Sierra-Gallay, I.; Masi, M.; Girard, E.; Dautin, N.; Constantinesco-Becker, N.; Tropis, M.; Daffé, M.; van Tilbeurgh, H.; Bayan, N. The C-terminal domain of Corynebacterium glutamicum mycoloyltransferase A is composed of five repeated motifs involved in cell wall binding and stability. Mol. Microbiol. 2020, 114, 1–16. [Google Scholar] [CrossRef]
- Brand, S.; Niehaus, K.; Pühler, A.; Kalinowski, J. Identification and functional analysis of six mycolyltransferase genes of Corynebacterium glutamicum ATCC 13032: The genes cop1, cmt1, and cmt2 can replace each other in the synthesis of trehalose dicorynomycolate, a component of the mycolic acid layer of the cell envelope. Arch. Microbiol. 2003, 180, 33–44. [Google Scholar] [CrossRef]
- Teramoto, H.; Inui, M.; Yukawa, H. Corynebacterium glutamicum Zur acts as a zinc-sensing transcriptional repressor of both zinc-inducible and zinc-repressible genes involved in zinc homeostasis. FEBS J. 2012, 279, 4385–4397. [Google Scholar] [CrossRef]
- Pfeifer-Sancar, K.; Mentz, A.; Rückert, C.; Kalinowski, J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genom. 2013, 14, 888–911. [Google Scholar] [CrossRef] [PubMed]
- Schröder, J.; Jochmann, N.; Rodionov, D.A.; Tauch, A. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genom. 2010, 7, 12. [Google Scholar] [CrossRef]
- Farkas, A.; Maróti, G.; Kereszt, A.; Kondorosi, É. Comparative Analysis of the Bacterial Membrane Disruption Effect of Two Natural Plant Antimicrobial Peptides. Front. Microbiol. 2017, 23, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, D.; Tan, X.; Huang, D.; Huang, Y.; Zhao, G.; Hu, X.; Wang, X. The role of trehalose biosynthesis on mycolate composition and L-glutamate production in Corynebacterium glutamicum. Microbiol. Res. 2023, 267, 127260. [Google Scholar] [CrossRef] [PubMed]
- Marienhagen, J.; Kennerknecht, N.; Sahm, H.; Eggeling, L. Functional analysis of all aminotransferase proteins inferred from the genome sequence of Corynebacterium glutamicum. J. Bacteriol. 2005, 187, 7639–7646. [Google Scholar] [CrossRef]
- Shimada, T.; Makinoshima, H.; Ogawa, Y.; Miki, T.; Maeda, M.; Ishihama, A. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 2004, 186, 7112–7122. [Google Scholar] [CrossRef]
- Jurischka, S.; Bida, A.; Dohmen-Olma, D.; Kleine, B.; Potzkei, J.; Binder, S.; Schaumann, G.; Bakkes, P.J.; Freudl, R. A secretion biosensor for monitoring Sec-dependent protein export in Corynebacterium glutamicum. Microb. Cell Fact. 2020, 19, 11. [Google Scholar] [CrossRef]
- Bakkes, P.J.; Lenz, P.; Müller, C.; Bida, A.; Dohmen-Olma, D.; Knapp, A.; Oldiges, M.; Jaeger, K.E.; Freudl, R. Biosensor-Based Optimization of Cutinase Secretion by Corynebacterium glutamicum. Front. Microbiol. 2021, 12, 750150–750167. [Google Scholar] [CrossRef]
- White, M.J.; He, H.; Penoske, R.M.; Twining, S.S.; Zahrt, T.C. PepD participates in the mycobacterial stress response mediated through MprAB and SigE. J. Bacteriol. 2010, 192, 1498–1510. [Google Scholar] [CrossRef]


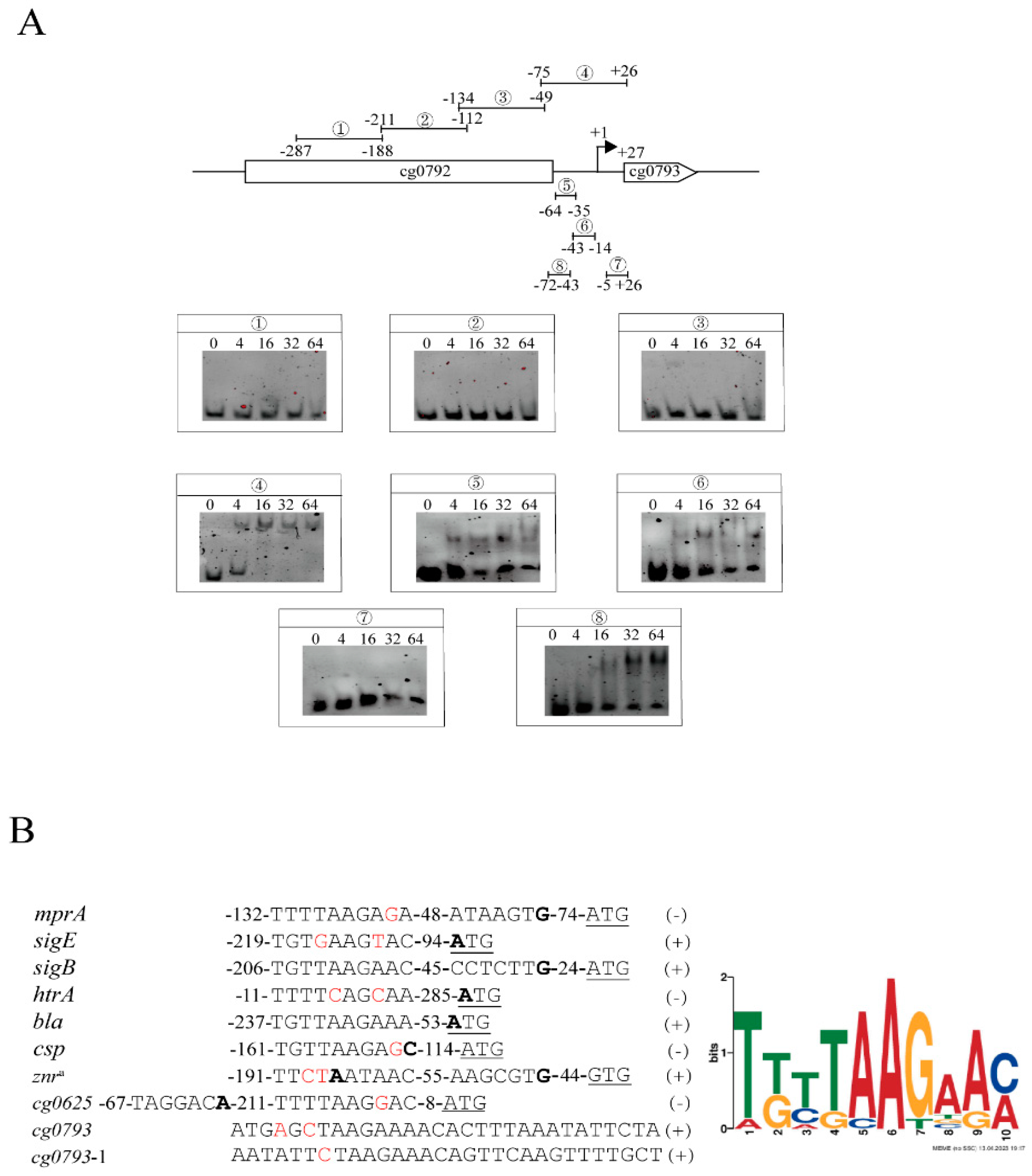

| Gene Locus | Gene | Function | Binding Site a | △mprA b | p-Value c | △mprA/pXMJ19-mprA d | p-Value c |
|---|---|---|---|---|---|---|---|
| cg0996 | mprA | Response regulator of two-component system | TTTTAAGAGA | −3.65 ± 0.17 | 6.29 × 10−101 | 11.11 ± 0.75 | 4.28 × 10−50 |
| cg0997 | mprB | Histidine kinase of two-component system | −4.52 ± 0.17 | 3.91 × 10−148 | −0.82 ± 0.35 | 2.05 × 10−02 | |
| Cell envelope biosynthesis | |||||||
| cg0413 | cmt1 | Trehalose corynomycolyl transferase | −0.03 ± 0.29 | 9.17 × 10−01 | −1.12 ± 0.08 | 3.22 × 10−45 | |
| cg0414 | wzz | Saccharide synthesis | 0.01 ± 0.13 | 9.51 × 10−01 | −1.37 ± 0.08 | 2.64 × 10−74 | |
| cg0650 | L,D-transpeptidases | 0.01 ± 0.13 | 9.51 × 10−01 | −1.62 ± 0.09 | 6.34 × 10−77 | ||
| cg0905 | psp2 | Protein potentially involved in peptidoglycan biosynthesis | 0.21 ± 0.41 | 6.10 × 10−01 | −3.07 ± 0.14 | 4.18 × 10−102 | |
| cg0998 | htrA | HtrA-like serine protease | TTTTCAGCAA | 1.00 ± 0.13 | 1.75 × 10−15 | −0.93 ±0.08 | 9.58 × 10−29 |
| cg1108 | porA | Mycoloyltransferase | 0.08 ± 0.12 | 4.81 × 10−01 | −1.45 ± 0.07 | 5.92 × 10−94 | |
| cg1109 | porB | Mycoloyltransferase | 1.48 ± 0.11 | 9.33 × 10−43 | −0.74 ± 0.04 | 1.13 × 10−71 | |
| cg2069 | Protein of LGFP repeat family | 0.84 ± 0.19 | 8.46 × 10−06 | −2.67 ± 0.15 | 3.69 × 10−67 | ||
| cg2398 | plsC | 1-Acyl-sn-glycerol-3-phosphate acetyltransferase | 0.00 ± 0.12 | 9.85 × 10−01 | −1.16 ± 0.09 | 4.84 × 10−40 | |
| cg2478 | bla | Penicillin-binding protein | TGTTAAGAAA | 0.21 ± 0.14 | 1.20 × 10−01 | −1.31 ± 0.10 | 5.11 × 10−38 |
| cg3197 | csp | Protein potentially involved in peptidoglycan biosynthesis | TGTTAAGAGC | 0.00 ± 0.12 | 9.80 × 10−01 | −2.00 ± 0.08 | 2.76 × 10−144 |
| Regulatory proteins | |||||||
| cg0317 | ArsR family transcriptional regulator | −1.11 ± 0.37 | 2.99 × 10−03 | 1.44 ± 0.49 | 3.54 × 10−03 | ||
| cg1119 | Putative stress-responsive transcriptional regulator | 0.31 ± 0.21 | 1.37 × 10−01 | −1.38 ± 0.39 | 4.16 × 10−04 | ||
| cg1271 | sigE | Sigma factor | TGTGAAGTAC | −0.14 ± 0.15 (0.40 ± 0.10) | 3.50 × 10−01 | −0.10 ± 0.10 (−1.24 ± 0.05) e | 3.17 × 10−01 |
| cg2102 | sigB | Sigma factor | TGTTAAGAAC | −0.29 ± 0.20 (0.35 ± 0.25) | 1.42 × 10−01 | −0.11 ± 0.10 (−1.0 ± 0.07) e | 2.96 × 10−01 |
| cg2115 | sugR | Transcriptional regulators of sugar metabolism | −0.06 ± 0.13 | 6.52 × 10−01 | −1.05 ± 0.11 | 1.74 × 10−21 | |
| cg2500 | znr | Putative transcriptional regulator | −0.92 ± 0.36 | 9.44 × 10−03 | 0.64 ± 0.14 | 3.33 × 10−06 | |
| cg2648 | Bacterial regulatory protein | 3.83 ± 1.32 | 3.78 × 10−03 | −1.02 ± 1.30 | 3.80 × 10−02 | ||
| Secreted | |||||||
| proteins | |||||||
| cg0085 | porH1 | PhoH-like ATPase | 0.12 ± 0.14 | 3.99 × 10−01 | −1.36 ± 0.13 | 1.68 × 10−26 | |
| cg0625 | Secreted protein | TTTTAAGGAC | −0.42 ± 0.20 | 3.27 × 10−02 | 1.10 ± 0.09 | 7.50 × 10−33 | |
| cg0726 | Secreted lipoprotein | 0.22 ± 0.14 | 1.25 × 10−01 | −1.05 ± 0.14 | 1.72 × 10 | ||
| cg0793 | Secreted protein | AGCTAAGAAA ATTCTAAGAA | 3.45 ± 0.19 | 4.35 × 10−76 | −4.15 ± 0.29 | 2.30× 10−45 | |
| cg0918 | Putative secreted protein | −0.03 ± 0.10 | 7.29 × 10−01 | −1.31 ± 0.48 | 6.23 × 10−03 | ||
| cg1247 | Putative secreted protein | 0.10 ± 0.58 | 8.67 × 10−01 | −1.10 ± 0.09 | 4.42 × 10−32 | ||
| cg1514 | Putative secreted protein | −0.89 ± 0.25 | 3.17 × 10−04 | 1.02 ± 0.13 | 6.94 × 10−15 | ||
| cg1936 | Putative secreted protein | −0.09 ± 0.28 | 7.44 × 10−01 | −1.16 ± 0.39 | 2.58 × 10−03 | ||
| cg2061 | Putative secreted protein | −0.25 ± 0.21 | 2.26 × 10−01 | −2.93 ± 0.10 | 6.91 × 10−177 | ||
| cg2518 | Putative secreted protein | 0.12 ± 0.17 | 4.69 × 10−01 | −2.60 ± 0.12 | 7.07 × 10−113 | ||
| cg2566 | Putative secreted protein | −0.06 ± 0.21 | 7.80 × 10−01 | −2.07 ± 0.12 | 3.88 × 10−68 | ||
| cg3197 | Putative secreted protein | 0.00 ± 0.12 | 9.80 × 10−01 | −2.00 ± 0.08 | 2.76 × 10−144 | ||
| cg3343 | Putative secreted protein | 0.42 ± 0.25 | 9.70 × 10−02 | −2.90 ± 0.10 | 1.96 × 10−180 | ||
| cg3394 | Putative secreted protein | 0.35 ± 0.27 | 1.98 × 10−01 | −1.70 ± 0.20 | 2.73 × 10−17 | ||
| Other metabolism | |||||||
| cg0010 | Hypothetical protein | 0.06 ± 0.30 | 8.54 × 10−01 | −1.19 ± 0.19 | 2.70 × 10−10 | ||
| cg0088 | Citrate transporter | −40 ± 0.32 | 2.15 × 10−01 | 1.00 ± 0.40 | 1.19 × 10−02 | ||
| cg0096 | Hypothetical protein | −0.44 ± 0.47 | 3.56 × 10−01 | 1.16 ± 0.46 | 7.20 × 10−124 | ||
| cg0107 | Putative integral membrane transport protein | −0.25 ± 0.60 | 6.78 × 10−01 | −5.07 ± 0.23 | 1.08 × 10−02 | ||
| cg0108 | Sirtuin-type KDAC homologues | −0.08 ± 0.15 | 5.98 × 10−01 | −1.48 ± 0.27 | 4.42 × 10−08 | ||
| cg0133 | abgT | Secondary transporter of the AbgT family | −0.08 ± 0.14 | 5.94 × 10−01 | −2.03 ± 0.08 | 1.43 × 10−137 | |
| cg0134 | abgB | Peptidase | −0.05 ± 0.16 | 7.75 × 10−01 | −2.01 ± 0.09 | 4.58 × 10−122 | |
| cg0135 | Putative inner membrane protein | 0.07 ± 0.20 | 7.28 × 10−01 | 1.13 ± 0.16 | 8.57 × 10−13 | ||
| cg0182 | tagA2 | DNA-3-methyladenine glycosylase I | −0.28 ± 0.30 | 3.63 × 10−01 | −0.65 ± 0.05 | 1.40 × 10−108 | |
| cg0253 | Flavodoxin reductase | 0.28 ± 0.0.24 | 2.45 × 10−01 | −1.04 ± 0.17 | 1.63 × 10−09 | ||
| cg0254 | Alanine symporter | −0.98 ± 0.15 | 1.42 × 10−10 | −1.06 ± 0.08 | 5.51 × 10−37 | ||
| cg0291 | 3,4-dioxygenase beta subunit | 0.15 ± 0.10 | 1.10 × 10−01 | 1.16 ± 0.10 | 1.07 × 10−22 | ||
| cg0391 | UDP-glucose 4-epimerase | −0.07 ± 0.12 | 5.75 × 10−01 | −1.97 ± 0.08 | 4.70 × 10−123 | ||
| cg0415 | ptpA2 | Low molecular weight protein-tyrosine phosphatase | 0.05 ± 0.12 | 6.65 × 10−01 | −1.11 ± 0.09 | 1.62 × 10−35 | |
| cg0437 | Membrane protein | 0.22 ± 0.14 | 1.03 × 10−01 | −1.19 ± 0.12 | 5.66 × 10−24 | ||
| cg0441 | lpd | Dihydrolipoyl dehydrogenase | 0.04 ± 0.14 | 7.70 × 10−01 | −1.31 ± 0.09 | 6.20 × 10−48 | |
| cg0623 | Cobalt transport system | −0.12 ± 0.19 | 5.28 × 10−01 | 1.10 ± 0.08 | 7.30 × 10−43 | ||
| cg0646 | IclR family proteins | 0.12 ± 0.19 | 5.09 × 10−01 | −1.93 ± 0.15 | 7.33 × 10−38 | ||
| cg0692 | Transposase | 1.37 ± 0.58 | 1.77 × 10−02 | 0.51 ± 0.41 | 2.13 × 10−01 | ||
| cg0703 | guaA | GMP synthase | 0.21 ± 0.41 | 6.10 × 10−01 | −1.42 ± 0.63 | 2.49 × 10−02 | |
| cg0721 | crtB2 | Hytoene synthetase | 0.2 ± 0.20 | 9.26 × 10−01 | −1.11 ± 0.21 | 1.82 × 10−07 | |
| cg0723 | crtE | Geranylgeranyl diphosphate synthase | 0.03 ± 0.18 | 8.69 × 10−01 | −1.06 ± 016 | 7.74 × 10−11 | |
| cg0727 | Nucleoside-diphosphate-sugar epimerase | 0.16 ± 0.12 | 1.81 × 10−01 | −1.17 ± 0.11 | 1.76 × 10−24 | ||
| cg0739 | Putative integral membrane protein | 0.06 ± 0.15 | 6.81 × 10−01 | −1.42 ± 0.16 | 1.51 × 10−18 | ||
| cg0740 | Membrane protein | 0.13 ± 0.18 | 4.85 × 10−01 | −1.23 ± 0.13 | 1.53 × 10−21 | ||
| cg0755 | metY | O-Acetylhomoserine-lyase | 0.03 ± 0.12 | 7.96 × 10−01 | −1.05 ± 0.13 | 1.53 × 10−21 | |
| cg0770 | Fe3+-siderophores transport system | −1.18 ± 0.12 | 1.32 × 10−22 | −1.57 ± 0.14 | 6.58 × 10−31 | ||
| cg0794 | ycic | Cobalamin synthesis protein | 3.27 ± 0.14 | 7.50 × 10−123 | −3.42 ± 0.11 | 3.91 × 10−195 | |
| cg0830 | Membrane protein | 0.58 ± 0.27 | 3.23 × 10−02 | −1.33 ± 0.45 | 3.05 × 10−03 | ||
| cg0844 | Type II restriction enzyme | −0.59 ± 0.17 | 4.15 × 10−04 | −1.09 ± 0.09 | 9.28 × 10−35 | ||
| cg0845 | Superfamily II DNA/RNA helicase | −0.17 ± 0.22 | 4.26 × 10−01 | −1.27 ± 0.13 | 2.73 × 10−21 | ||
| cg0923 | Membrane protein | 0.15 ± 0.15 | 6.02 × 10−01 | −1.14 ± 0.22 | 2.44 × 10−07 | ||
| cg0924 | Fe3+-siderophores transport system | −0.09 ± 0.09 | 7.50 × 10−01 | −1.04 ± 0.16 | 4.79 × 10−11 | ||
| cg1016 | betP | Glycine betaine transporter | 0.09 ± 0.21 | 6.72 × 10−01 | −1.05 ± 0.11 | 1.62 × 10−11 | |
| cg1018 | recQ | ATP-dependent DNA helicase | −0.28 ± 0.16 | 8.29 × 10−02 | 1.20 ± 0.08 | 2.81 × 10−51 | |
| cg1019 | Metal-dependent hydrolase | 0.06 ± 0.19 | 7.77 × 10−01 | 1.21 ± 0.36 | 8.24 × 10−04 | ||
| cg1055 | menG | 2-demethylmenaquinone methyltransferase | 0.08 ± 0.19 | 6.71 × 10−01 | 1.31 ± 0.09 | 1.54 × 10−47 | |
| cg1061 | urtA | Urea transport system substrate-binding protein | 1.09 ± 0.47 | 1.96 × 10−02 | −0.52 ± 0.27 | 5.50 × 10−02 | |
| cg1077 | Permease of the major facilitator superfamily | 0.18 ± 0.15 | 2.30 × 10−01 | −1.22 ± 0.19 | 5.56 × 10−11 | ||
| cg1088 | ABC-type multidrug/protein/lipid transport system | 0.22 ± 0.17 | 1.83 × 10−01 | −1.35 ± 0.08 | 1.67 × 10−60 | ||
| cg1178 | Transposase | −2.43 ± 0.27 | 1.69 × 10−19 | −0.61 ± 0.21 | 4.00 × 10−03 | ||
| cg1182 | Putative membrane protein | −1.97 ± 0.19 | 3.55 × 10−24 | −0.20 ± 0.16 | 2.20 × 10−01 | ||
| cg1183 | Predicted dinucleotide-utilizing enzyme | −2.15 ± 0.21 | 1.78 × 10−25 | −0.13 ± 0.15 | 3.89 × 10−01 | ||
| cg1184 | Transposase | −1.80 ± 0.20 | 8.97 × 10−20 | −0.13 ± 0.19 | 3.89 × 10−01 | ||
| cg1244 | Arsenate reductase or related protein | 0.17 ± 0.39 | 6.55 × 10−01 | −1.37 ± 0.50 | 6.53 × 10−03 | ||
| cg1305 | Amino acid permease | −0.09 ± 0.19 | 6.20 × 10−01 | −1.15 ± 0.21 | 6.84 × 10−08 | ||
| cg1427 | Extracellular deoxyribonuclease | 0.43 ± 0.25 | 8.64 × 10−02 | 1.45 ± 0.12 | 3.40 × 10−35 | ||
| cg1438 | ATPase component | 1.83 ± 0.93 | 4.98 × 10−02 | −0.27 ± 0.24 | 1.54 × 10−01 | ||
| cg1493 | D-alanine--d-alanine ligase A | 0.00 ± 0.13 | 8.67 × 10−01 | −1.07 ± 0.10 | 2.12 × 10−29 | ||
| cg1551 | uspA1 | Universal stress protein | 0.12 ± 0.11 | 2.71 × 10−01 | −1.13 ± 0.08 | 1.18 × 10−50 | |
| cg1642 | Siderophore-interacting protein | −1.02 ± 0.49 | 3.80 × 10−02 | 1.33 ± 0.40 | 8.43 × 10−04 | ||
| cg2043 | Hypothetical protein | −0.02 ± 0.13 | 8.98 × 10−01 | −1.01 ± 0.08 | 5.60 × 10−37 | ||
| cg2181 | ABC-type peptide transport system | −0.59 ± 0.96 | 5.40 × 10−01 | −1.62 ± 0.26 | 5.46 × 10−10 | ||
| cg2343 | Decarboxylase | 0.03 ± 0.27 | 9.24 × 10−01 | −1.05 ± 0.08 | 5.25 × 10−40 | ||
| cg2358 | Hypothetical protein | −0.19 ± 0.40 | 6.25 × 10−01 | −1.33 ± 0.31 | 1.47 × 10−05 | ||
| cg2359 | Isoleucine-tRNA ligase-like protein | −0.16 ± 0.13 | 1.96 × 10−01 | −1.21 ± 0.09 | 1.92 × 10−39 | ||
| cg2386 | Hypothetical protein | −0.27 ± 0.28 | 3.36 × 10−01 | −1.01 ± 0.14 | 1.05 × 10−12 | ||
| cg2397 | Putative membrane protein | 0.04 ± 0.11 | 7.07 × 10−01 | −1.49 ± 0.10 | 9.83 × 10−02 | ||
| cg2565 | Hypothetical protein | −0.14 ± 0.13 | 3.03 × 10−01 | −2.45 ± 0.11 | 1.09 × 10−103 | ||
| cg2651 | Hypothetical protein | 1.37 ± 0.26 | 1.96 × 10−07 | −3.71 ± 0.13 | 1.29 × 10−187 | ||
| cg2662 | pepN | Aminopeptidase | −0.19 ± 0.14 | 1.65 × 10−01 | −1.44 ± 0.09 | 8.29 × 10−53 | |
| cg2807 | tnp11a | Transposase | 0.31 ± 0.21 | 1.31 × 10−01 | −1.04 ± 0.15 | 2.78 × 10−12 | |
| cg2844 | pstA | ABC-type phosphate transport system | −0.11 ± 0.26 | 6.65 × 10−01 | −1.12 ± 0.16 | 9.03 × 10−12 | |
| cg2845 | pstC | ABC-type phosphate transport system | −0.28 ± 0.24 | 2.39 × 10−01 | −1.21 ± 0.12 | 2.31 × 10−19 | |
| cg2846 | pstS | ABC-type phosphate transport system | −0.26 ± 0.24 | 2.80 × 10−01 | −1.36 ± 0.12 | 1.97 × 10−29 | |
| cg2870 | dctA | Na+/H+-dicarboxylate symporter | 0.50 ± 0.20 | 1.16 × 10−02 | −1.47 ± 0.13 | 1.46 × 10−28 | |
| cg2895 | Permease of the major facilitator superfamily | −0.00 ± 0.13 | 2.80 × 10−01 | −2.43 ± 0.12 | 2.59 × 10−92 | ||
| cg2896 | Endoglucanase | −0.05 ± 0.14 | 7.52 × 10−01 | −1.36 ± 0.12 | 1.97 × 10−29 | ||
| cg3086 | L,L-Cystathionine gamma-Lyase | −0.21 ± 0.18 | 2.36 × 10−01 | −1.02 ± 0.20 | 1.93 × 10−07 | ||
| cg3105 | Hypothetical protein | −0.30 ± 0.17 | 7.70 × 10−02 | 1.04 ± 0.13 | 1.97 × 10−29 | ||
| cg3106 | Hypothetical protein | −0.13 ± 0.14 | 3.66 × 10−01 | −1.14 ± 0.09 | 6.64 × 10−37 | ||
| cg3270 | Hypothetical protein | 0.34 ± 0.30 | 2.53 × 10−01 | −2.63 ± 0.40 | 7.68 × 10−05 | ||
| cg3292 | Copper chaperone | −0.07 ± 0.16 | 6.73 × 10−01 | −1.19 ± 0.18 | 3.97 × 10−11 | ||
| cg3395 | proP | Proline/betaine transporter | 0.34 ± 0.19 | 7.13 × 10−02 | −2.37 ± 0.10 | 1.25 × 10−122 | |
| cg3403 | Permease of the major facilitator superfamily | 0.28 ± 0.30 | 3.60 × 10−01 | −1.32 ± 0.44 | 2.75 × 10−03 | ||
| cg3404 | ABC-type transport system | −0.19 ± 0.30 | 5.26 × 10−01 | −1.15 ± 0.21 | 2.89 × 10−08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, Y.; Huang, D.; Liu, X.; Yang, Y.; Liu, C.; Li, Y.; Bai, Z. Two-Component MprAB System Regulates the Expression of Genes Involved in Cell Envelope Biosynthesis in Corynebacterium glutamicum. Microorganisms 2025, 13, 1120. https://doi.org/10.3390/microorganisms13051120
Zou Y, Huang D, Liu X, Yang Y, Liu C, Li Y, Bai Z. Two-Component MprAB System Regulates the Expression of Genes Involved in Cell Envelope Biosynthesis in Corynebacterium glutamicum. Microorganisms. 2025; 13(5):1120. https://doi.org/10.3390/microorganisms13051120
Chicago/Turabian StyleZou, Yu, Danni Huang, Xiuxia Liu, Yankun Yang, Chunli Liu, Ye Li, and Zhonghu Bai. 2025. "Two-Component MprAB System Regulates the Expression of Genes Involved in Cell Envelope Biosynthesis in Corynebacterium glutamicum" Microorganisms 13, no. 5: 1120. https://doi.org/10.3390/microorganisms13051120
APA StyleZou, Y., Huang, D., Liu, X., Yang, Y., Liu, C., Li, Y., & Bai, Z. (2025). Two-Component MprAB System Regulates the Expression of Genes Involved in Cell Envelope Biosynthesis in Corynebacterium glutamicum. Microorganisms, 13(5), 1120. https://doi.org/10.3390/microorganisms13051120






