Comparison and Analysis of the Genomes of Three Strains of Botrytis cinerea Isolated from Pomegranate
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Evaluation of B. cinerea Development in Various Culture Media
2.3. Production of Enzymes Involved in the Degradation of Plant Cell Wall Polymers
2.4. Evaluation of Disease Progression Caused by B. cinerea Strains in Pomegranate Fruits
2.5. Statistical Analyses
2.6. Data Quality Control and Genome Assembly
2.7. Phylogenetic Analysis
2.8. Functional Annotation
2.9. Comparative Analysis of Coding Sequences
2.10. Search for Genes Related to Plant Cell Wall Degradation and Pathogenicity
3. Results
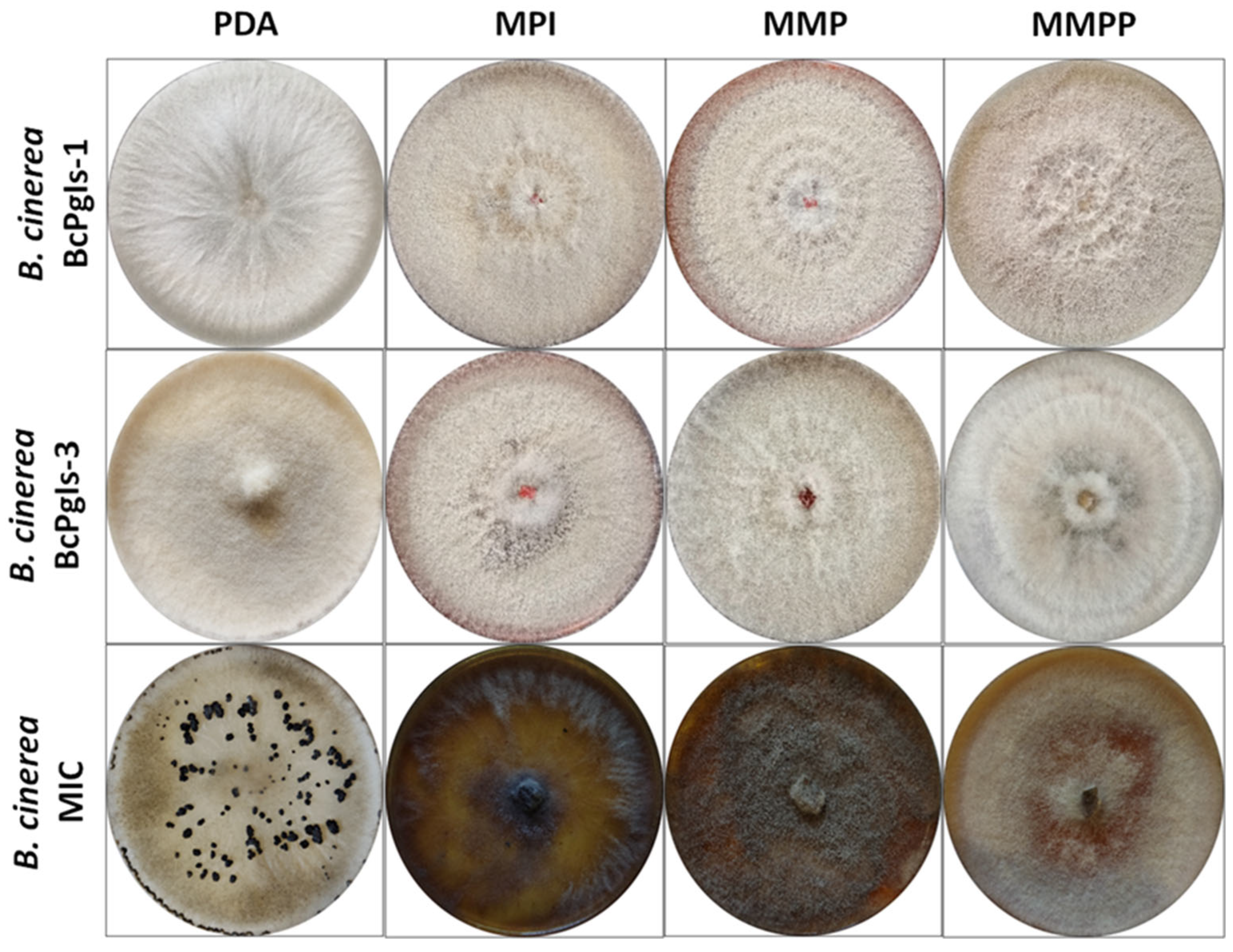
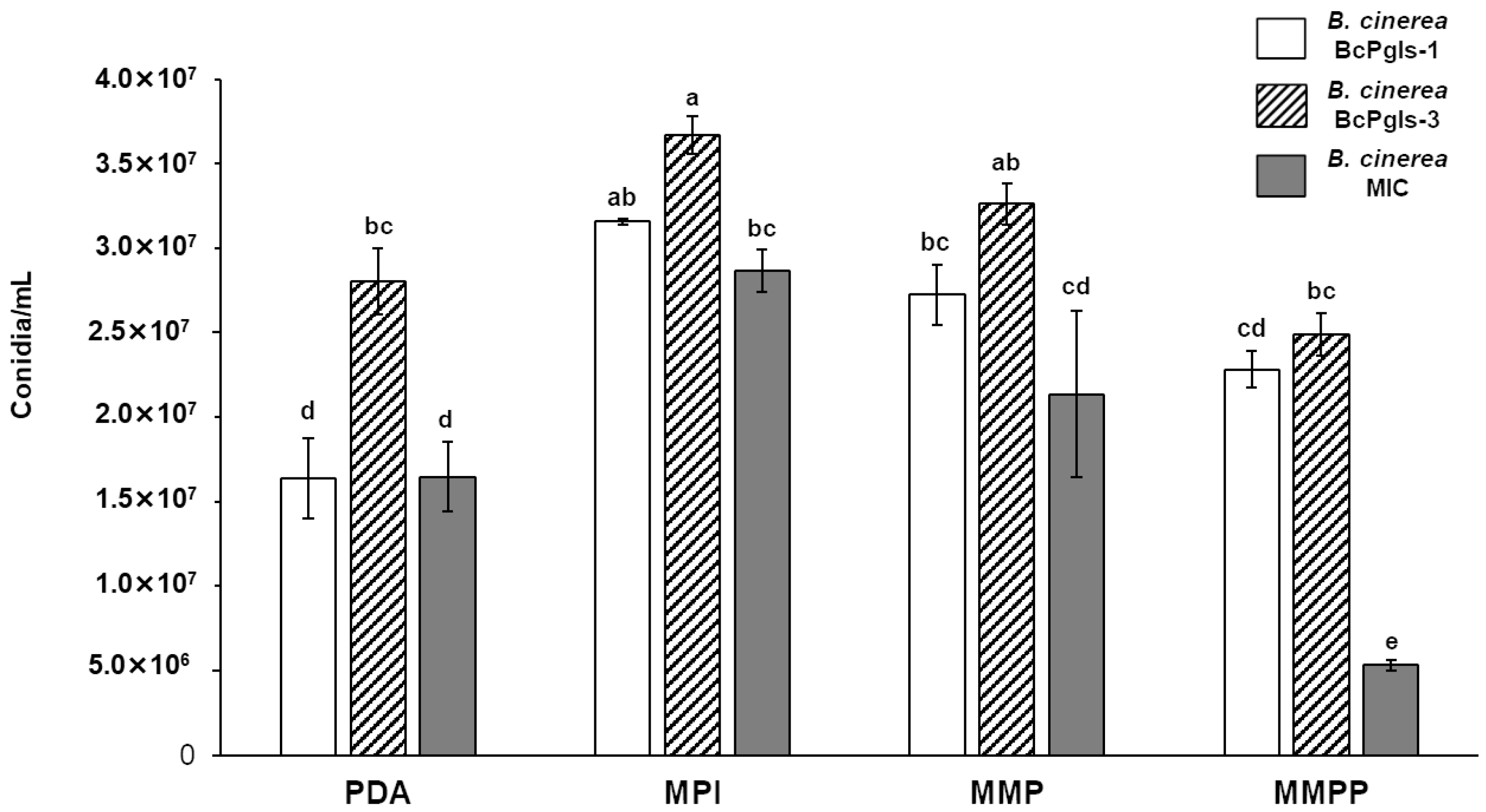
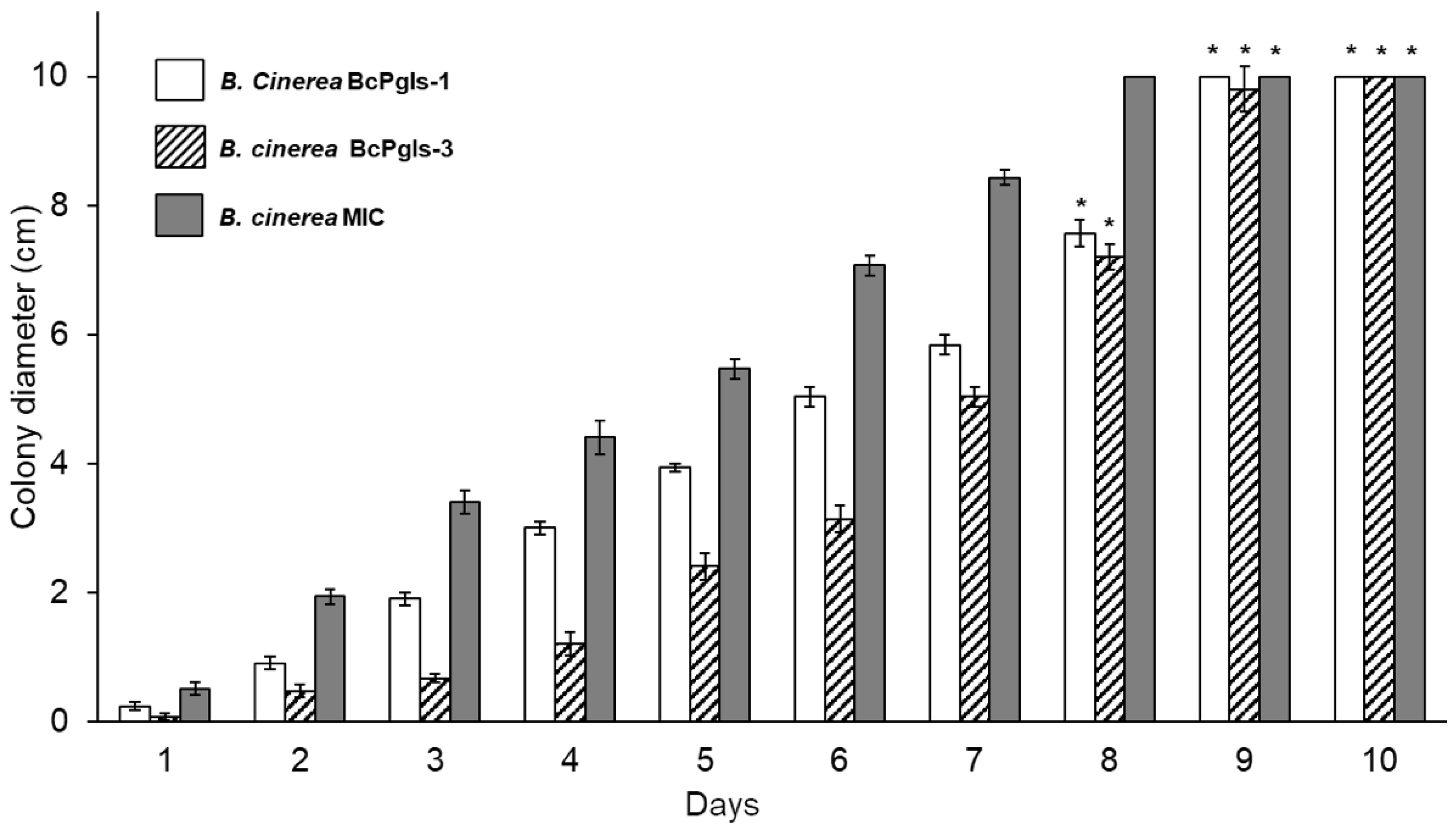
3.1. Evaluation of the Development of B. cinerea in Distinct Culture Media
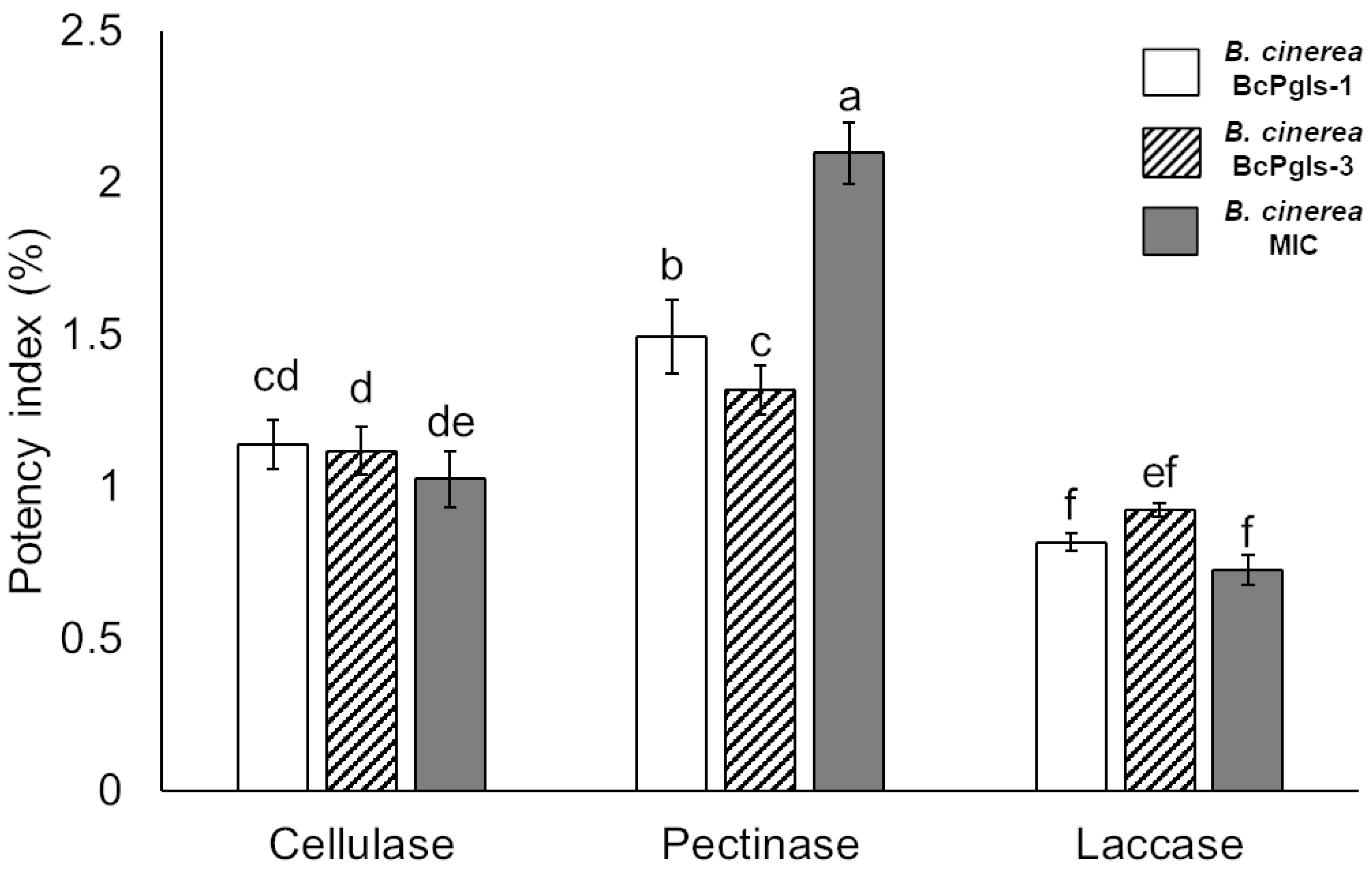
3.2. Enzyme Production Associated with the Degradation of Plant Cell Wall Polymers
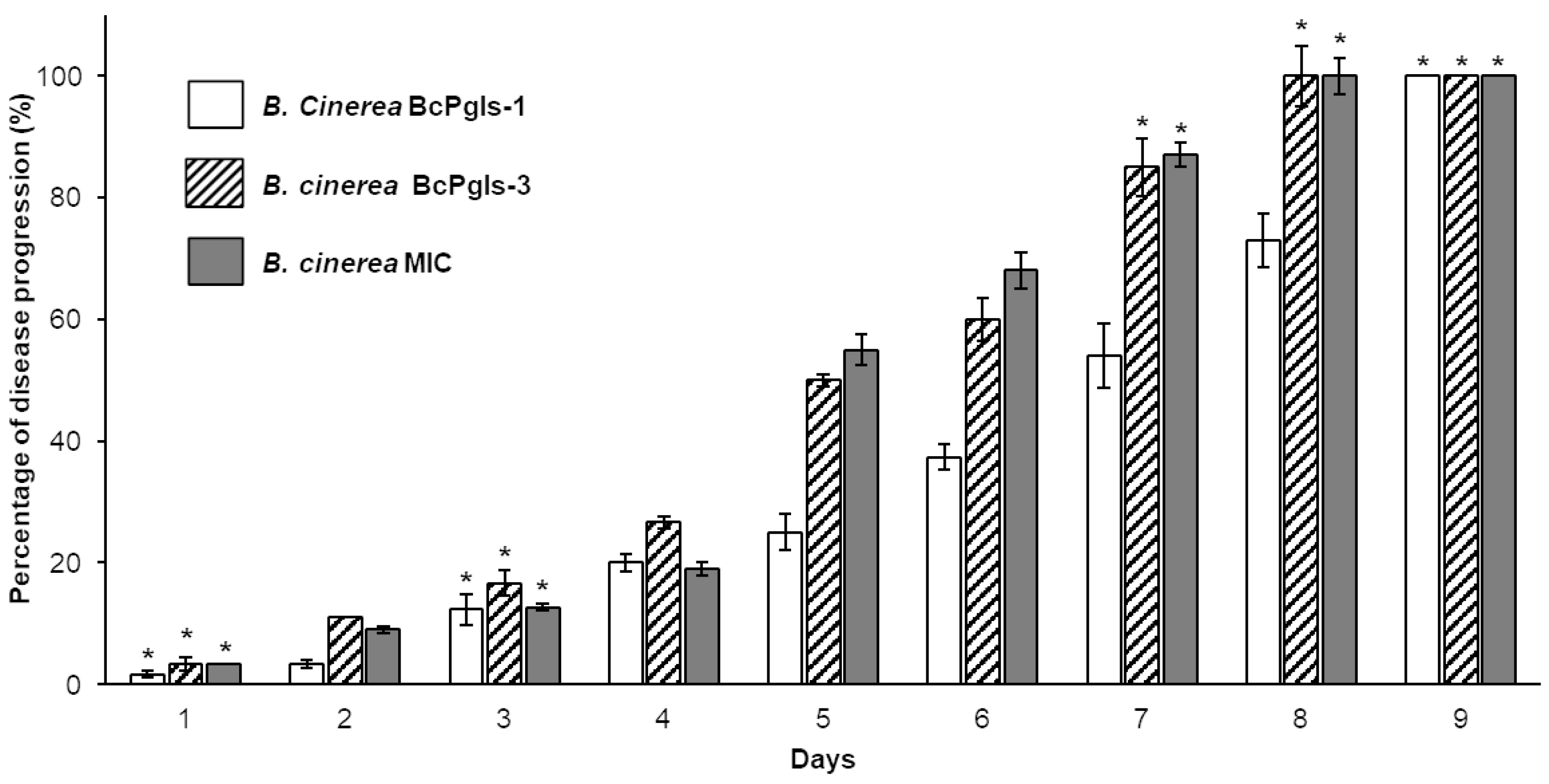
3.3. Tests of the Advance of Infection on Pomegranate Fruits
3.4. Genome Sequencing and Assembly
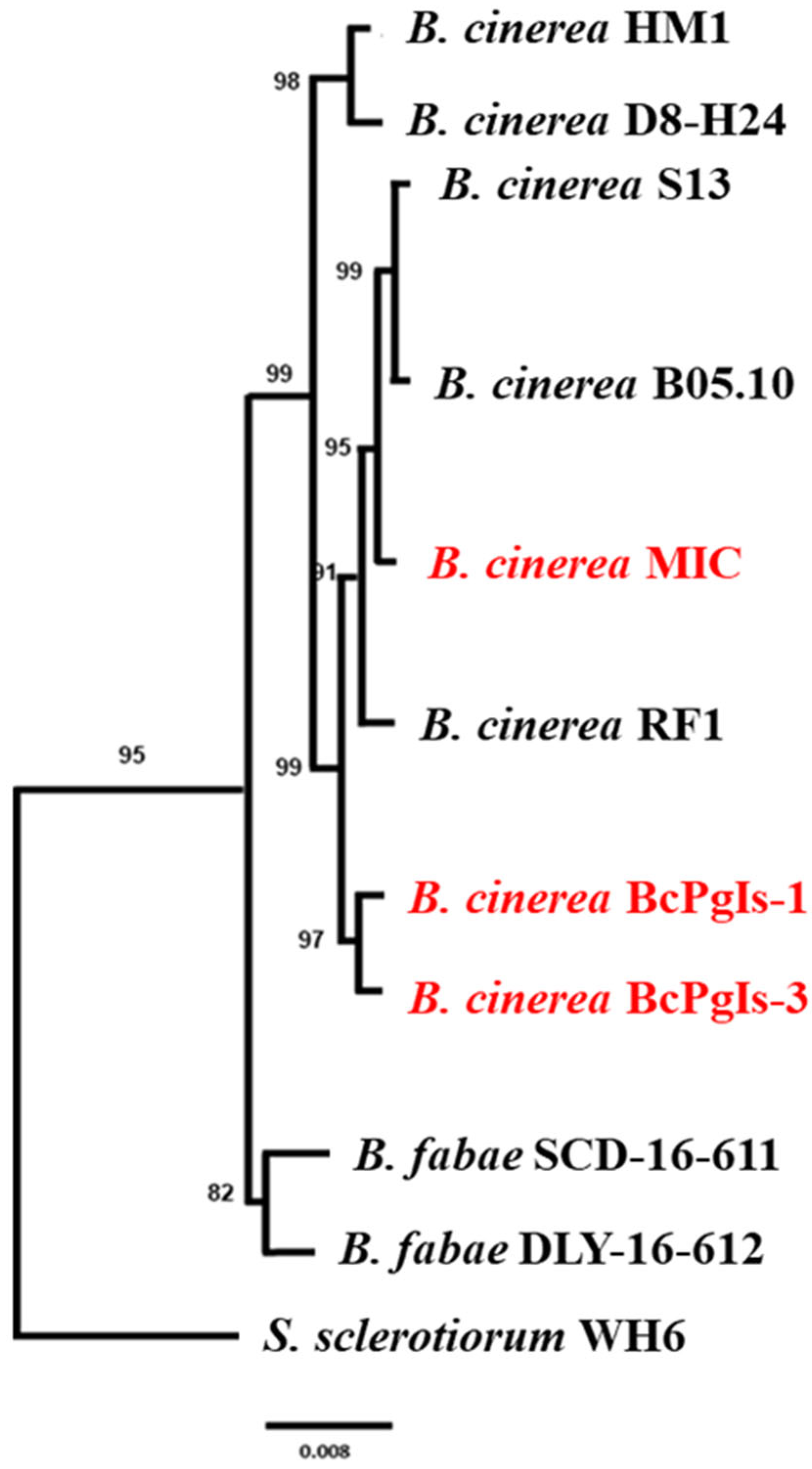
3.5. Phylogenomic Analysis
3.6. Functional Annotation
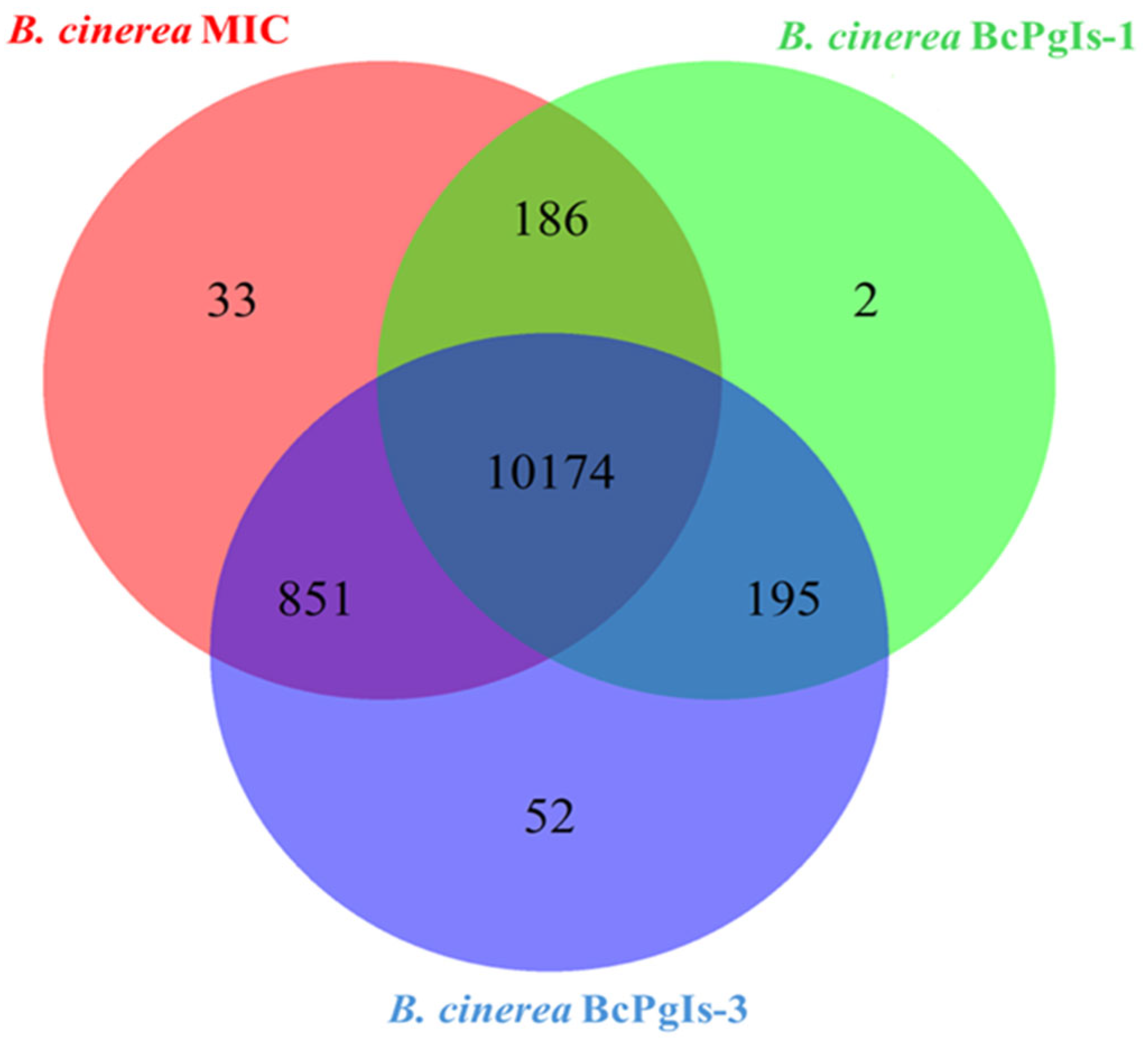
3.7. Comparative Analysis of Coding Sequences
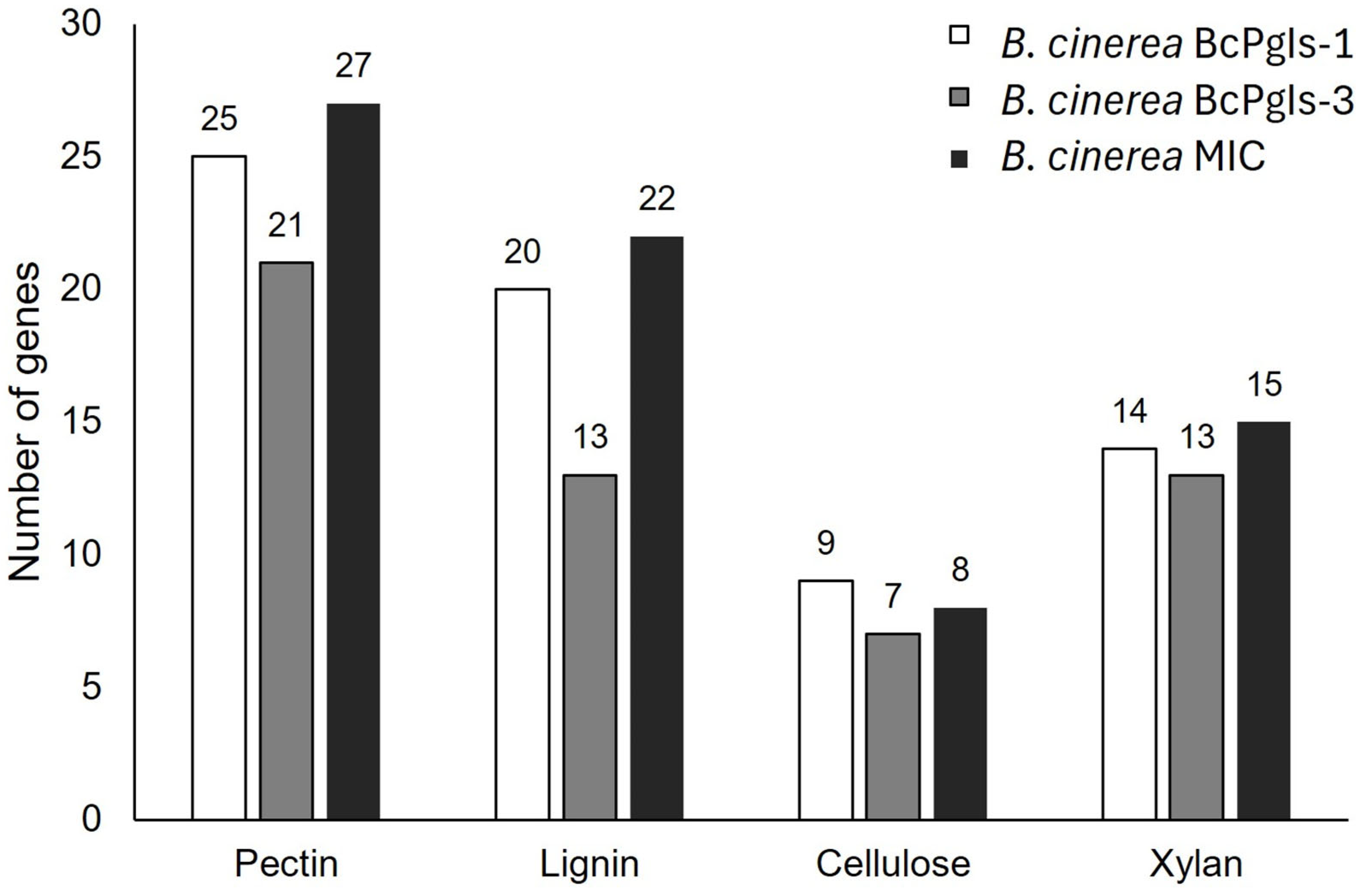
3.8. Search for Genes Related to Plant Cell Wall Degradation and Pathogenicity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohammadi, M.; Boghrati, Z.; Emami, S.A.; Akaberi, M. Pomegranate: A review of the heavenly healer’s past, present, and future. Iran. J. Basic Med. Sci. 2023, 26, 1245–1264. [Google Scholar] [PubMed]
- Magangana, T.P.; Makunga, N.P.; Fawole, O.A.; Opara, U.L. Processing factors affecting the phytochemical and nutritional properties of pomegranate (Punica granatum L.) peel waste: A review. Molecules 2020, 25, 4690. [Google Scholar] [CrossRef]
- Manjunatha, N.; Sharma, J.; Pokhare, S.S.; Agarrwal, R.; Patil, P.G.; Sirsat, J.D.; Chakranarayan, M.G.; Bicchal, A.; Ukale, A.S.; Marathe, R.A. Characterization of Alternaria and Colletotrichum species associated with pomegranate (Punica granatum L.) in Maharashtra State of India. J. Fungi 2022, 8, 1040. [Google Scholar] [CrossRef]
- Bardas, G.A.; Tzelepis, G.D.; Lotos, L.; Karaoglanidis, G.S. First report of Botrytis cinerea causing gray mold of pomegranate (Punica granatum) in Greece. Plant Dis. 2009, 93, 1346. [Google Scholar] [CrossRef] [PubMed]
- Patricio-Hernández, A.; Moreno-Velázquez, M.; Mercado-Flores, Y.; Quezada-Salinas, A. First report of Botrytis cinerea causing gray mold of pomegranate (Punica granatum L.) in Mexico. J. Plant Dis. Prot. 2023, 130, 663–667. [Google Scholar] [CrossRef]
- Cheung, N.; Tian, L.; Liu, X.; Li, X. The destructive fungal pathogen Botrytis cinerea-insights from genes studied with mutant analysis. Pathogens 2020, 9, 923. [Google Scholar] [CrossRef]
- Rhouma, A.; Hajji-Hedfi, L.; Kouadri, M.; Atallaoui, K.; Matrood, A.; Khrieba, M. Botrytis cinerea: The cause of tomatoes gray mold. Egypt. J. Phytopathol. 2023, 51, 68–75. [Google Scholar] [CrossRef]
- Singh, R.; Caseys, C.; Kliebenstein, D.J. Genetic and molecular landscapes of the generalist phytopathogen Botrytis cinerea. Mol. Plant Pathol. 2024, 25, e13404. [Google Scholar] [CrossRef]
- Petrasch, S.; Knapp, S.J.; van Kan, J.A.L.; Blanco-Ulate, B. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Mol. Plant Pathol. 2019, 20, 877–892. [Google Scholar] [CrossRef]
- Choquer, M.; Rascle, C.; Gonçalves, I.R.; de Vallée, A.; Ribot, C.; Loisel, E.; Smilevski, P.; Ferria, J.; Savadogo, M.; Souibgui, E.; et al. The infection cushion of Botrytis cinerea: A fungal ‘weapon’ of plant-biomass destruction. Environ. Microbiol. 2021, 23, 2293–2314. [Google Scholar] [CrossRef]
- Van-Kan, J.A.L.; Stassen, J.H.M.; Mosbach, A.; Van Der Lee, T.A.J.; Faino, L.; Farmer, A.D.; Papasotiriou, D.G.; Zhou, S.; Seidl, M.F.; Cottam, E.; et al. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 2017, 18, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Caseys, C.; Shi, G.; Soltis, N.; Gwinner, R.; Corwin, J.; Atwell, S.; Kliebenstein, D.J. Quantitative interactions: The disease outcome of Botrytis cinerea across the plant kingdom. G3 2021, 11, jkab175. [Google Scholar] [CrossRef] [PubMed]
- Patricio-Hernandez, A.; Quezada-Salinas, A.Q.-S.; Anducho-Reyes, M.A.; Mercado-Flores, Y. Application of Bacillus velezensis 160 for the biocontrol of Botrytis cinerea in pomegranate. Plant Dis. 2025. Advance online publication. [Google Scholar] [CrossRef]
- Saranya, G.; Ravinder, A.; Swaminathan, T.; Ravikrishna, R. Comparison and evaluation of enumeration methods for measurement of fungal spore emission. J. Aerosol. Sci. 2022, 165, 106033. [Google Scholar]
- Andro, T.; Chambost, J.P.; Kotoujansky, A.; Catteneo, J.; Bertheau, Y.; Barras, F.; van Gijsegem, F.; Colleno, A. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 1984, 160, 1199–1203. [Google Scholar] [CrossRef]
- Parveen, S.; Wani, A.H.; Bhat, M.Y.; Koka, J.A.; Fazili, M.A. Variability in production of extracellular enzymes by different fungi isolated from rotten pear, peach and grape fruits. Braz. J. Biol. Sci. 2017, 4, 259–264. [Google Scholar] [CrossRef]
- Chairattanamanokorn, P.; Imai, T.; Kondo, R.; Ukita, M.; Prasertsan, P. Screening thermotolerant white-rot fungi for decolorization of wastewaters. Appl. Biochem. Biotechnol. 2006, 128, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Patricio-Hernández, A.; Mercado-Flores, Y.; Téllez-Jurado, A.; Ramírez-Vargas, M.R.; Quezada-Salinas, A. Diagrammatic scale to evaluate the severity of gray mold (Botrytis cinerea) in pomegranate. Rev. Mex. De Fitopatol. 2023, 41, 524–530. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Armstrong, J.; Raney, B.J.; Streeter, I.; Dunn, M.; Yang, F.; Odom, D.; Flicek, P.; Keane, T.M.; Thybert, D.; et al. Chromosome assembly of large and complex genomes using multiple references. Genome Res. 2018, 28, 1720–1732. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation com-pleteness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4–10. [Google Scholar] [CrossRef]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef]
- Korf, I. Gene finding in novel genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Testa, A.C.; Hane, J.K.; Ellwood, S.R.; Oliver, R.P. CodingQuarry: Highly accurate hidden Markov model gene prediction in fungal genomes using RNA-seq transcripts. BMC Genom. 2015, 16, 170. [Google Scholar] [CrossRef]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.O.; Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [PubMed]
- Törönen, P.; Holm, L. PANNZER-A practical tool for protein function prediction. Protein Sci. 2022, 31, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Morton, F.R.; Kok, C.Y.; Kong, J.; Barrett, A.J. MEROPS: The peptidase database. Nucleic Acids Res. 2008, 36, D320–D325. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Nonavinakere, N.; Sahoo, J.; Sahu, P.; Iyer, V.L.; Khamari, L.; Martinez, M.C.; Hammond-Kosack, K.E. PHI-base-the multi-species pathogen-host interaction database in 2025. Nucleic Acids Res. 2025, 53, D826–D838. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; de Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. antiSMASH 8.0: Extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Ren, W.; Liu, N.; Sang, C.; Shi, D.; Zhou, M.; Chen, C.; Qin, Q.; Chen, W. The autophagy gene BcATG8 regulates the vegetative differentiation and pathogenicity of Botrytis cinerea. Appl. Environ. Microbiol. 2018, 84, e02455-17. [Google Scholar] [CrossRef]
- Minz-Dub, A.; Sharon, A. The Botrytis cinerea PAK kinase BcCla4 mediates morphogenesis, growth and cell cycle regulating processes downstream of BcRac. Mol. Microbiol. 2017, 104, 487–498. [Google Scholar] [CrossRef]
- Gourgues, M.; Brunet-Simon, A.; Lebrun, M.H.; Levis, C. The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol. Microbiol. 2004, 51, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Schamber, A.; Leroch, M.; Diwo, J.; Mendgen, K.; Hahn, M. The role of mitogen-activated protein (MAP) kinase signalling components and the Ste12 transcription factor in germination and pathogenicity of Botrytis cinerea. Mol. Plant Pathol. 2010, 11, 105–119. [Google Scholar] [CrossRef]
- Zheng, L.; Campbell, M.; Murphy, J.; Lam, S.; Xu, J.R. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. MPMI 2000, 13, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Sui, Z.; Li, R.; Xu, Y.; Xiang, L.; Fu, S.; Wei, J.; Cai, X.; Wu, M.; Zhang, J.; et al. The Gβ-like protein Bcgbl1 regulates development and pathogenicity of the gray mold Botrytis cinerea via modulating two MAP kinase signaling pathways. PLoS Pathog. 2023, 19, e1011839. [Google Scholar] [CrossRef]
- Acosta-Morel, W.; Anta-Fernández, F.; Baroncelli, R.; Becerra, S.; Thon, M.R.; van-Kan, J.A.L.; Díaz-Mínguez, J.M.; Benito, E.P. A major effect gene controlling development and pathogenicity in Botrytis cinerea identified through genetic analysis of natural mycelial non-pathogenic isolates. Front. Plant Sci. 2021, 12, 663870. [Google Scholar] [CrossRef] [PubMed]
- Spada, M.; Pugliesi, C.; Fambrini, M.; Pecchia, S. Challenges and opportunities arising from host-Botrytis cinerea interactions to outline novel and sustainable control strategies: The key role of RNA interference. Int. J. Mol. Sci. 2024, 25, 6798. [Google Scholar] [CrossRef]
- Mobasher-Amini, M.; Mirzaei, S.; Heidari, A. A growing threat: Investigating the high incidence of benzimidazole fungicides resistance in Iranian Botrytis cinerea isolates. PLoS ONE 2023, 18, e0294530. [Google Scholar] [CrossRef]
- Liñeiro, E.; Macias-Sánchez, A.J.; Espinazo, M.; Cantoral, J.M.; Moraga, J.; Collado, I.G.; Fernández-Acero, F.J. Phenotypic effects and inhibition of botrydial biosynthesis induced by different plant-based elicitors in Botrytis cinerea. Curr. Microbiol. 2018, 75, 431–440. [Google Scholar] [CrossRef]
- Westfall, A.K.; Telemeco, R.S.; Grizante, M.B.; Waits, D.S.; Clark, A.D.; Simpson, D.Y.; Klabacka, R.L.; Sullivan, A.P.; Perry, G.H.; Sears, M.W.; et al. A chromosome-level genome assembly for the eastern fence lizard (Sceloporus undulatus), a reptile model for physiological and evolutionary ecology. GigaScience 2021, 10, giab066. [Google Scholar] [CrossRef]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef]
- Simon, A.; Mercier, A.; Gladieux, P.; Poinssot, B.; Walker, A.S.; Viaud, M. Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs. Peer Community J. 2022, 2, e83. [Google Scholar] [CrossRef]
- Plesken, C.; Pattar, P.; Reiss, B.; Noor, Z.N.; Zhang, L.; Klug, K.; Huettel, B.; Hahn, M. Genetic diversity of Botrytis cinerea revealed by multilocus sequencing, and identification of B. cinerea populations showing genetic isolation and distinct host adaptation. Front. Plant Sci. 2021, 12, 663027. [Google Scholar] [CrossRef] [PubMed]
- Makris, G.; Nikoloudakis, N.; Samaras, A.; Karaoglanidis, G.S.; Kanetis, L.I. Under pressure: A comparative study of Botrytis cinerea populations from conventional and organic farms in Cyprus and Greece. Phytopathology 2022, 112, 2236–2247. [Google Scholar] [CrossRef]
- Mousavi-Derazmahalleh, M.; Chang, S.; Thomas, G.; Derbyshire, M.; Bayer, P.E.; Edwards, D.; Nelson, M.N.; Erskine, W.; Lopez-Ruiz, F.J.; Clements, J.; et al. Prediction of pathogenicity genes involved in adaptation to a lupin host in the fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum via comparative genomics. BMC Genom. 2019, 20, 385. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.; Zheng, J.; Liu, G.; Chen, L. Importance of cell wall permeability and cell wall degrading enzymes during infection of Botrytis cinerea in Hazelnut. Forests 2023, 14, 565. [Google Scholar] [CrossRef]
- Bi, K.; Liang, Y.; Mengiste, T.; Sharon, A. Killing softly: A roadmap of Botrytis cinerea pathogenicity. Trends Plant Sci. 2023, 28, 211–222. [Google Scholar] [CrossRef]
- Zhang, Z.; He, C.; Chen, Y.; Li, B.; Tian, S. DNA Methyltransferases regulate pathogenicity of Botrytis cinerea to horticultural crops. J. Fungi 2021, 7, 659. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, J.; Wang, Y.; Du, J.; Zhang, J.; Luisi, B.F.; Liang, W. Broad-spectrum chemicals block ROS detoxification to prevent plant fungal invasion. Curr. Biol. 2022, 32, 3886–3897. [Google Scholar] [CrossRef]
- Boissinot, S. On the base composition of transposable elements. Int. J. Mol. Sci. 2022, 23, 4755. [Google Scholar] [CrossRef]
- Raffaele, S.; Kamoun, S. Genome evolution in filamentous plant pathogens: Why bigger can be better. Nat. Rev. Microbiol. 2012, 10, 417–430. [Google Scholar] [CrossRef]
- Zhong, R.; Zhou, D.; Chen, L.; Rose, J.P.; Wang, B.C.; Ye, Z.H. Plant cell wall polysaccharide O-acetyltransferases. Plants 2024, 13, 2304. [Google Scholar] [CrossRef]
- Tundo, S.; Paccanaro, M.C.; Elmaghraby, I.; Moscetti, I.; D’Ovidio, R.; Favaron, F.; Sella, L. The xylanase inhibitor TAXI-I increases plant resistance to Botrytis cinerea by inhibiting the BcXyn11a xylanase necrotizing activity. Plants 2020, 9, 601. [Google Scholar] [CrossRef] [PubMed]
- Janusz, G.; Pawlik, A.; Sulej, J.; Swiderska-Burek, U.; Jarosz-Wilkolazka, A.; Paszczynski, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [PubMed]
- John, E.; Singh, K.B.; Oliver, R.P.; Tan, K.C. Transcription factor control of virulence in phytopathogenic fungi. Mol. Plant Pathol. 2021, 22, 858–881. [Google Scholar] [CrossRef]
- Tao, Z.; Yan, F.; Hahn, M.; Ma, Z. Regulatory roles of epigenetic modifications in plant-phytopathogen interactions. Crop Health 2023, 1, 6. [Google Scholar] [CrossRef]


| Feature | B. cinerea Strain | ||
|---|---|---|---|
| BcPgIs-1 | BcPgIs-3 | MIC | |
| Genome size (pb) | 41,776,945 | 41,672,452 | 41,417,038 |
| Number of scaffolds | 17 | 17 | 17 |
| GC content (%) | 42.45 | 42.22 | 42.48 |
| Shortest scaffolding (pb) | 189,773 | 230,524 | 208,218 |
| Longest scaffold (pb) | 4,215,149 | 4,076,546 | 4,062,.245 |
| N50 (pb) | 2,607,131 | 2,575,794 | 2,589,848 |
| N90 (pb) | 2,054,873 | 2,025,540 | 2,015,948 |
| L50 | 7 | 7 | 7 |
| L90 | 14 | 14 | 14 |
| BUSCO (%) | 93.2 | 97.0 | 96.9 |
| Feature | BcPgIs-1 | BcPgIs-3 | MIC |
|---|---|---|---|
| Number of coding sequences | 10,708 | 10,787 | 10,668 |
| mRNA number | 10,449 | 10,578 | 10,459 |
| tRNA number | 209 | 209 | 209 |
| Interproscan (number of genes) | 7943 | 8323 | 8312 |
| PFam (number of genes) | 6726 | 7129 | 7098 |
| CAZymes (number of genes) | 419 | 435 | 445 |
| MEROPS (number of genes) | 326 | 330 | 334 |
| Stain of B. cinerea | Gen | Known Function | Reference | |||
|---|---|---|---|---|---|---|
| B05.10 (Reference Strain) | BcPgIs-1 | BcPgIs-3 | MIC | |||
| BCIN_02g02570 | FUN_003004-T1 | FUN_007008-T1 | FUN_008163-T1 | BcATG8 | Autophagy, mycelial development, conidiation, sclerotia formation, and virulence | [39] |
| BCIN_03g02930 | FUN_000057-T1 | FUN_002146-T1 | FUN_000245-T1 | BcCLA4 | Growth, morphogenesis, conidia production, and pathogenicity | [40] |
| BCIN_09g06130 | FUN_011469-T1 | FUN_010843-T1 | FUN_010711-T1 | Bcpls1 | Necessary for proper performance of the appressorium | [41] |
| BCIN_10g05560 | FUN_000702-T1 | FUN_004250-T1 | FUN_002577-T1 | Bcste12 | Transcription factor that controls penetration efficiency | [8,42] |
| BCIN_02g08170 | FUN_001816-T1 | FUN_001878-T1 | FUN_001831-T1 | Bmp1 | Protein kinases required for surface recognition and host penetration | [43,44] |
| BCIN_09g02390 | FUN_006454-T1 | FUN_006459-T1 | FUN_006454-T1 | Bmp3 | Protein kinases required for surface recognition and host penetration | [43,44] |
| BCIN_14g03010 | FUN_009511-T1 | FUN_009579-T1 | FUN_009472-T1 | Bcgbl1 | Regulates protein kinase signaling pathways (Bmp1 and Bmp3) | [44] |
| BCIN_04g03490 | FUN_003110-T1 | FUN_003142-T1 | FUN_003113-T1 | Hypotetical protein | Protein with GAL4-like DNA-binding domain and acetyltransferase activity, which controls key processes such as pathogenicity | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patricio-Hernández, A.; Anducho-Reyes, M.A.; Téllez-Jurado, A.; Ramírez-Vargas, R.; Quezada-Salinas, A.; Mercado-Flores, Y. Comparison and Analysis of the Genomes of Three Strains of Botrytis cinerea Isolated from Pomegranate. Microorganisms 2025, 13, 1605. https://doi.org/10.3390/microorganisms13071605
Patricio-Hernández A, Anducho-Reyes MA, Téllez-Jurado A, Ramírez-Vargas R, Quezada-Salinas A, Mercado-Flores Y. Comparison and Analysis of the Genomes of Three Strains of Botrytis cinerea Isolated from Pomegranate. Microorganisms. 2025; 13(7):1605. https://doi.org/10.3390/microorganisms13071605
Chicago/Turabian StylePatricio-Hernández, Alberto, Miguel Angel Anducho-Reyes, Alejandro Téllez-Jurado, Rocío Ramírez-Vargas, Andrés Quezada-Salinas, and Yuridia Mercado-Flores. 2025. "Comparison and Analysis of the Genomes of Three Strains of Botrytis cinerea Isolated from Pomegranate" Microorganisms 13, no. 7: 1605. https://doi.org/10.3390/microorganisms13071605
APA StylePatricio-Hernández, A., Anducho-Reyes, M. A., Téllez-Jurado, A., Ramírez-Vargas, R., Quezada-Salinas, A., & Mercado-Flores, Y. (2025). Comparison and Analysis of the Genomes of Three Strains of Botrytis cinerea Isolated from Pomegranate. Microorganisms, 13(7), 1605. https://doi.org/10.3390/microorganisms13071605







