Development of a Strain-Specific Detection and Quantification Method for Bifidobacterium animalis subsp. lactis HN019 Using WGS-SNP Analysis and qPCR
Abstract
1. Introduction
2. Materials and Methods
2.1. SNP Analysis
2.2. Primer and Probe Design
2.3. DNA Extraction
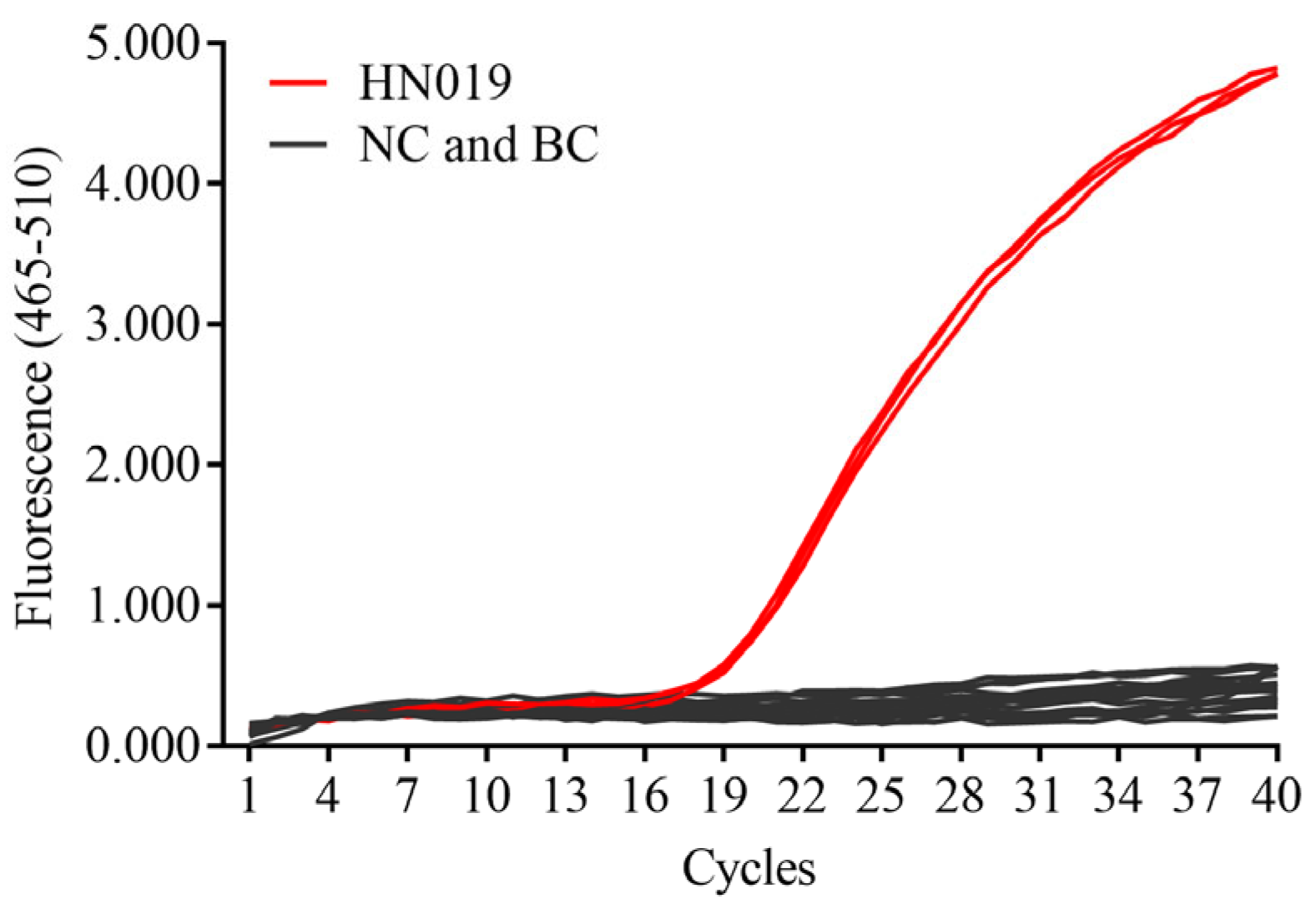
2.4. Specificity
2.5. Stability During Passage
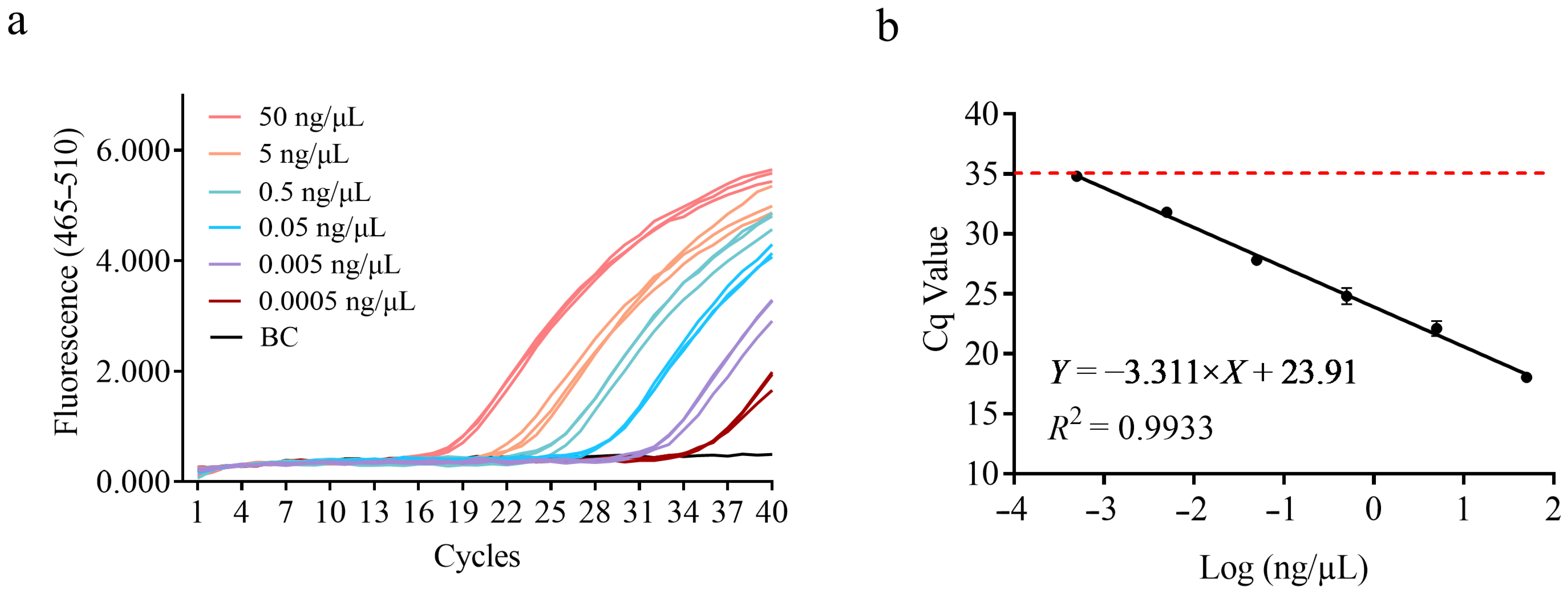
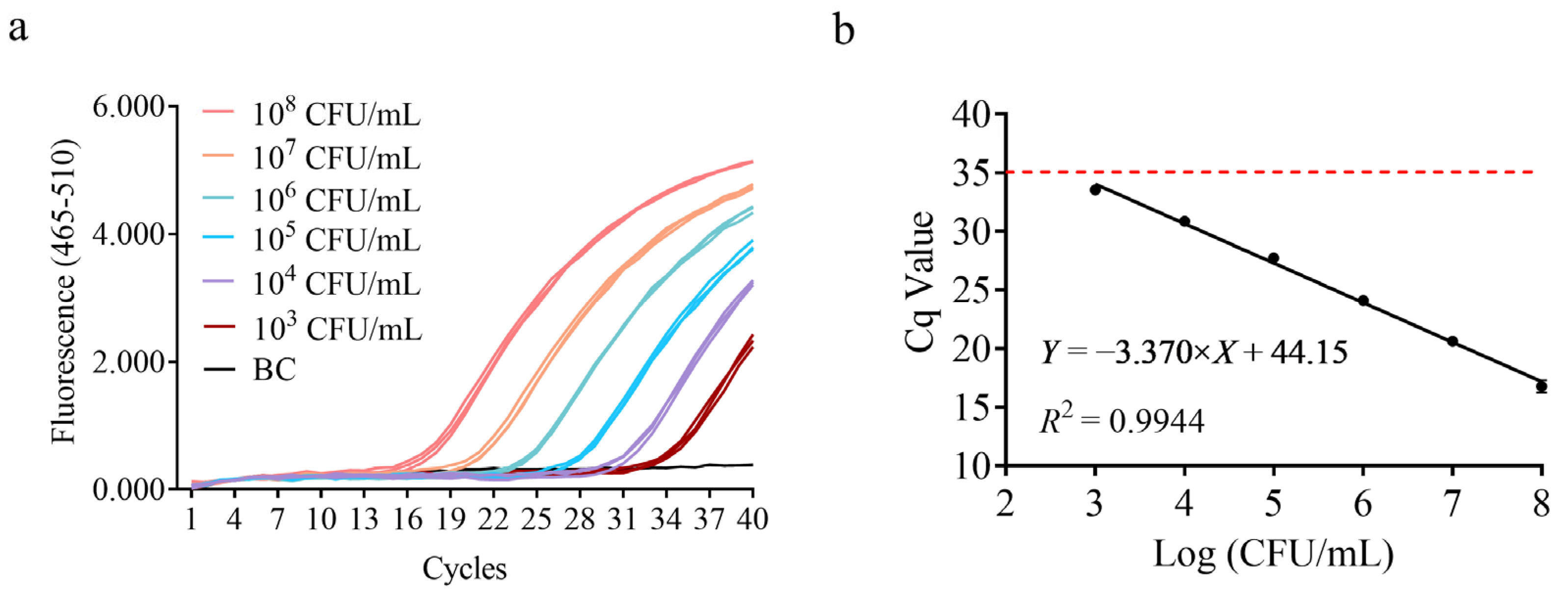
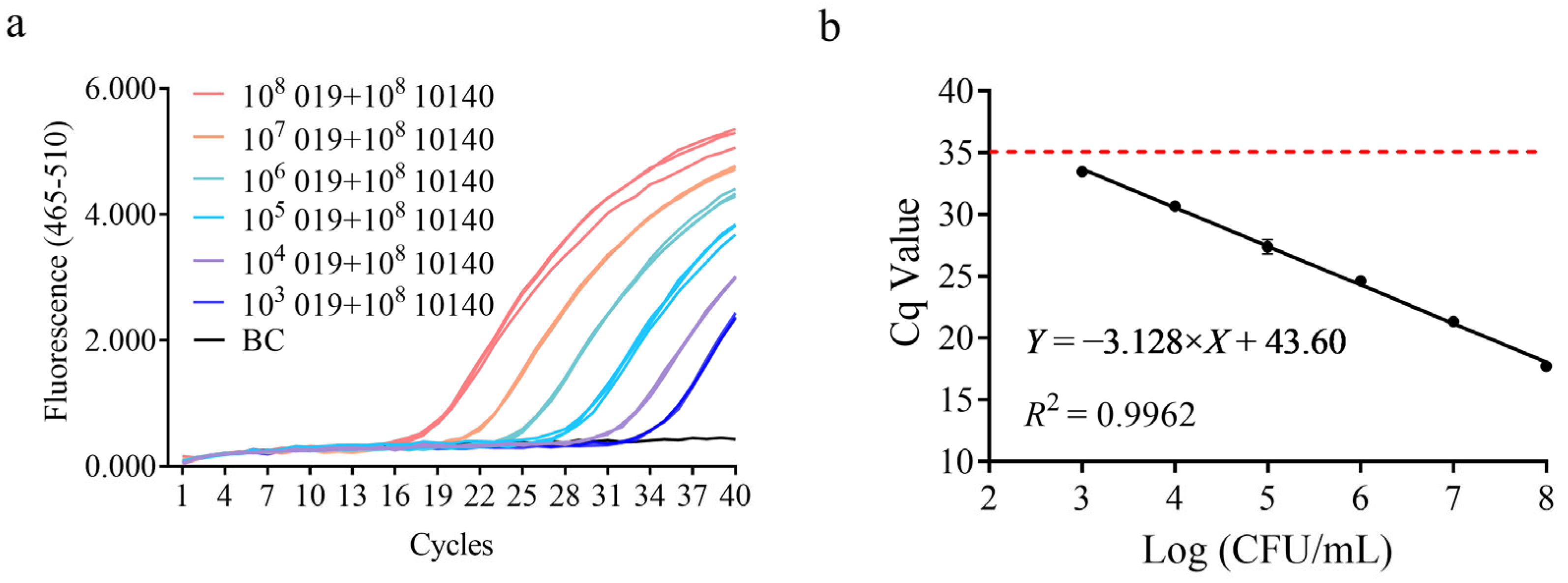
2.6. Sensitivity and Efficiency
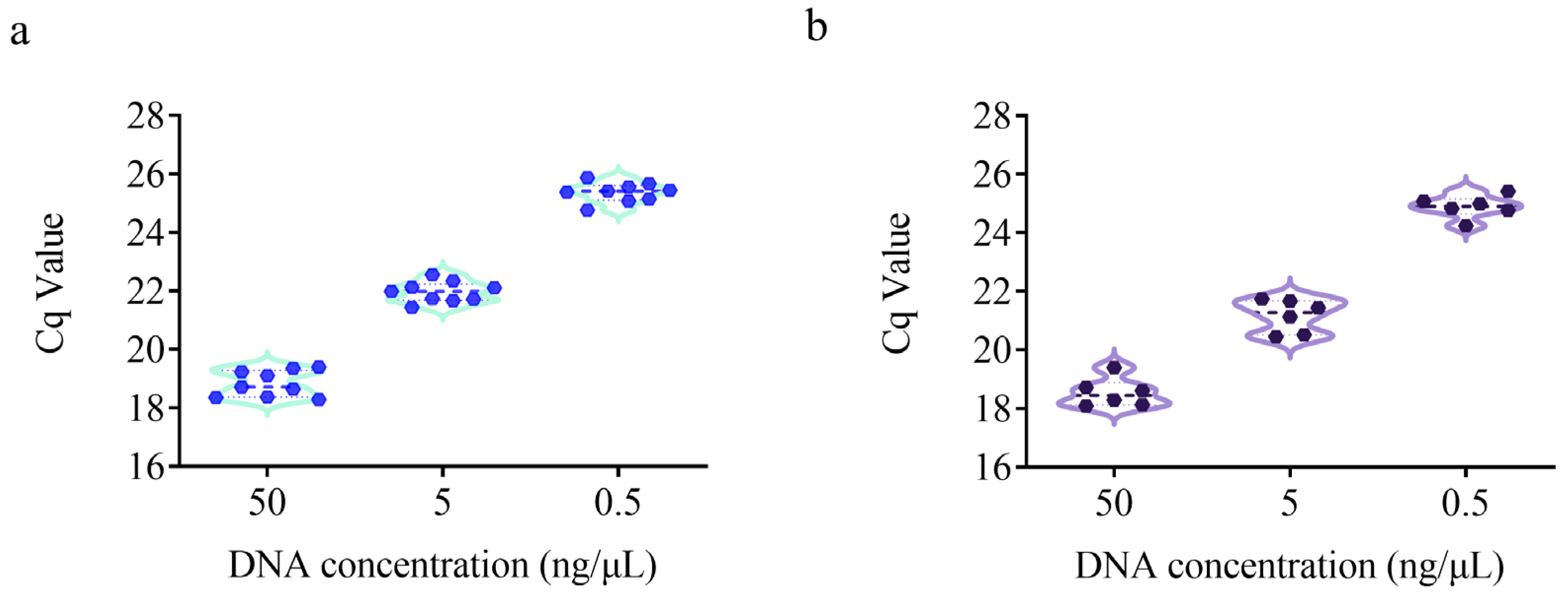
2.7. Repeatability and Reproducibility
2.8. Interference Resistance
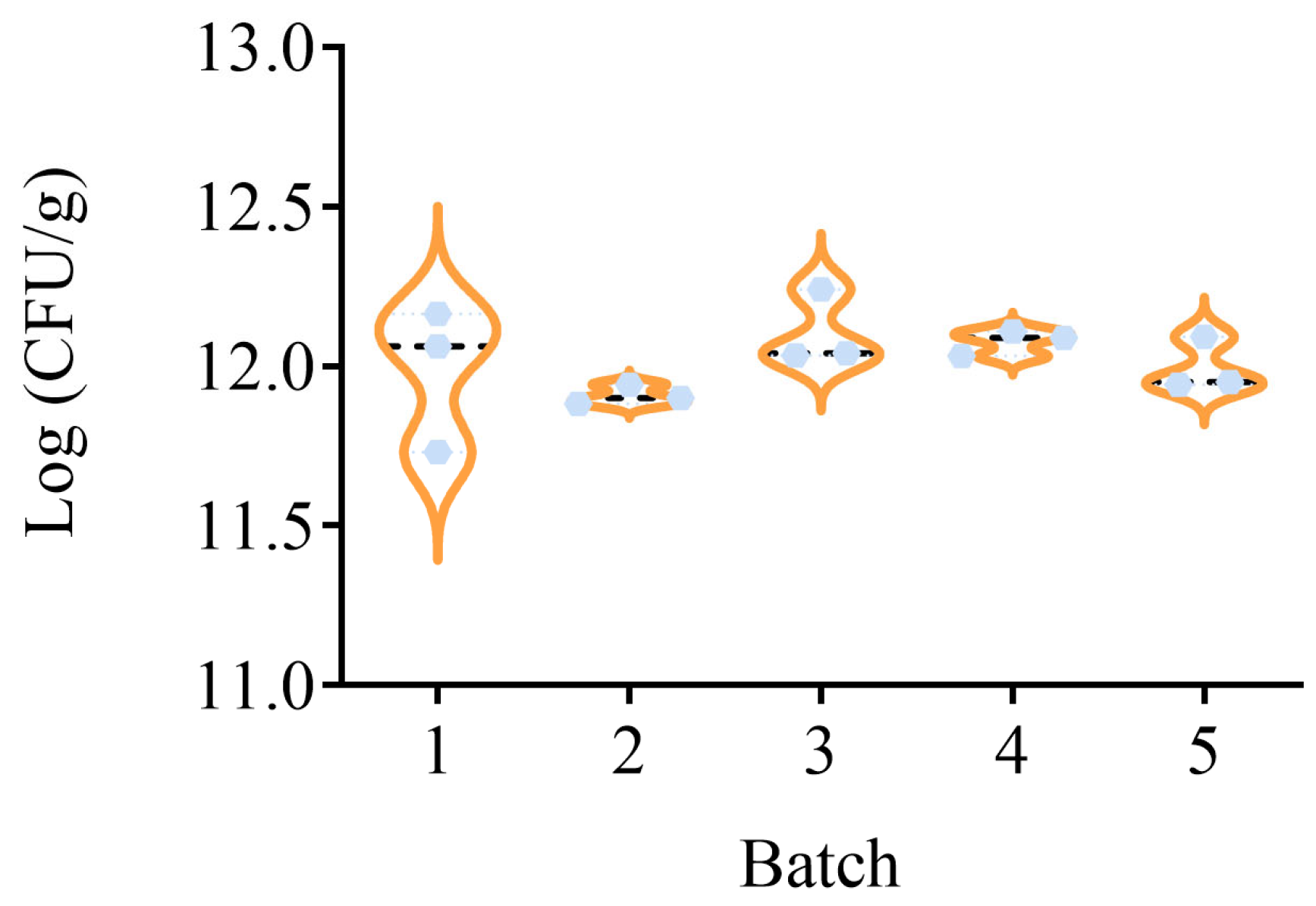
2.9. Actual Sample Test
2.10. Statistical Analysis
3. Results
3.1. SNP Analysis and Specificity Test
3.2. Passage Stability Test
3.3. Sensitivity Test
3.4. Repeatability and Reproducibility Test
3.5. Interference Resistance Test
3.6. Quantification of HN019 Strain in Probiotic Powder Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, T.K.; Pradhan, S.; Chakrabarti, S.; Mondal, K.C.; Ghosh, K. Current status of probiotic and related health benefits. Appl. Food Res. 2022, 2, 100185. [Google Scholar] [CrossRef]
- Yan, M.; Man, S.; Sun, B.; Ma, L.; Guo, L.; Huang, L.; Gao, W. Gut liver brain axis in diseases: The implications for therapeutic interventions. Signal Transduct. Target. Ther. 2023, 8, 443. [Google Scholar] [CrossRef]
- Loh, J.S.; Mak, W.Q.; Tan, L.K.S.; Ng, C.X.; Chan, H.H.; Yeow, S.H.; Foo, J.B.; Ong, Y.S.; How, C.W.; Khaw, K.Y. Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct. Target. Ther. 2024, 9, 37. [Google Scholar]
- Kearns, R. Gut–brain axis and neuroinflammation: The role of gut permeability and the kynurenine pathway in neurological disorders. Cell. Mol. Neurobiol. 2024, 44, 64. [Google Scholar] [CrossRef] [PubMed]
- Lugli, G.A.; Argentini, C.; Tarracchini, C.; Mancabelli, L.; Viappiani, A.; Anzalone, R.; Angelini, L.; Alessandri, G.; Longhi, G.; Bianchi, M.G.; et al. Characterization of a Bifidobacterium animalis subsp. lactis reference strain based on ecology and transcriptomics. Appl. Environ. Microbiol. 2024, 90, e01080-24. [Google Scholar] [CrossRef]
- D’Ambrosio, S.; Dabous, A.; Sadiq, S.; Casillo, A.; Schiraldi, C.; Cassese, E.; Bedini, E.; Corsaro, M.M.; Cimini, D. Bifidobacterium animalis subsp. lactis HN019 live probiotics and postbiotics: Production strategies and bioactivity evaluation for potential therapeutic properties. Front. Bioeng. Biotechnol. 2024, 12, 1379574. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Laitila, A.; Ouwehand, A.C. Bifidobacterium animalis subsp. lactis HN019 effects on gut health: A review. Front. Nutr. 2021, 8, 790561. [Google Scholar] [CrossRef]
- Sharma, M.; Wasan, A.; Sharma, R.K. Recent developments in probiotics: An emphasis on Bifidobacterium. Food Biosci. 2021, 41, 100993. [Google Scholar] [CrossRef]
- Putaala, H.; Salusjärvi, T.; Nordström, M.; Saarinen, M.; Ouwehand, A.C.; Bech Hansen, E.; Rautonen, N. Effect of four probiotic strains and Escherichia coli O157:H7 on tight junction integrity and cyclo-oxygenase expression. Res. Microbiol. 2008, 159, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Bernini, L.J.; Simão, A.N.C.; de Souza, C.H.B.; Alfieri, D.F.; Segura, L.G.; Costa, G.N.; Dichi, I. Effect of Bifidobacterium lactis HN019 on inflammatory markers and oxidative stress in subjects with and without the metabolic syndrome. Br. J. Nutr. 2018, 120, 645–652. [Google Scholar] [CrossRef]
- Invernici, M.M.; Furlaneto, F.A.C.; Salvador, S.L.; Ouwehand, A.C.; Salminen, S.; Mantziari, A.; Vinderola, G.; Ervolino, E.; Santana, S.I.; Silva, P.H.F.; et al. Bifidobacterium animalis subsp lactis HN019 presents antimicrobial potential against periodontopathogens and modulates the immunological response of oral mucosa in periodontitis patients. PLoS ONE 2020, 15, e0238425. [Google Scholar] [CrossRef] [PubMed]
- Bettocchi, S.; Comotti, A.; Elli, M.; De Cosmi, V.; Berti, C.; Alberti, I.; Mazzocchi, A.; Rosazza, C.; Agostoni, C.; Milani, G.P. Probiotics and Fever Duration in Children with Upper Respiratory Tract Infections: A Randomized Clinical Trial. JAMA Netw. Open 2025, 8, e250669. [Google Scholar] [CrossRef]
- Ala-Jaakkola, R.; Forssten, S.D.; Cheng, J.; Griffon, F.; Metreau, I.; Sturm, Y.; Lecerf, J.M.; Donazzolo, Y.; Junnila, J.; Nordlund, A.; et al. Effect of an 8-week Bifidobacterium lactis HN019 supplementation on functional constipation: A multi-center, triple-blind, randomized, placebo-controlled trial. Mol. Nutr. Food Res. 2025, e70081. [Google Scholar] [CrossRef]
- Jena, R.; Choudhury, P.K. Bifidobacteria in fermented dairy foods: A health beneficial outlook. Probiotics Antimicrob. Prot. 2025, 17, 1–22. [Google Scholar] [CrossRef]
- Waller, P.A.; Gopal, P.K.; Leyer, G.J.; Ouwehand, A.C.; Reifer, C.; Stewart, M.E.; Miller, L.E. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand. J. Gastroenterol. 2011, 46, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of probiotic Bifidobacteria for treating human diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar] [CrossRef]
- Huang, C.H.; Li, S.W.; Huang, L.; Watanabe, K. Identification and classification for the Lactobacillus casei group. Front. Microbiol. 2018, 9, 1974. [Google Scholar] [CrossRef] [PubMed]
- Li, M.N.; Han, Q.; Wang, N.; Wang, T.; You, X.M.; Zhang, S.; Zhang, C.C.; Shi, Y.Q.; Qiao, P.Z.; Man, C.L.; et al. 16S rRNA gene sequencing for bacterial identification and infectious disease diagnosis. Biochem. Biophys. Res. Commun. 2024, 739, 150974. [Google Scholar] [CrossRef]
- Ruiz-Moyano, S.; Tao, N.; Underwood, M.A.; Mills, D.A. Rapid discrimination of Bifidobacterium animalis subspecies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Food Microbiol. 2012, 30, 432–437. [Google Scholar] [CrossRef]
- Huang, C.H.; Liou, J.S.; Huang, L.; Watanabe, K. Developing novel species-specific DNA markers for PCR-based species identification of the Lactobacillus sakei group. Lett. Appl. Microbiol. 2018, 66, 138–144. [Google Scholar] [CrossRef]
- Schürch, A.C.; Arredondo-Alonso, S.; Willems, R.J.L.; Goering, R.V. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene by gene—Based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, S.; Zhang, Y.; Llaca, V.; Ye, L.; Sanyal, A.; King, M.; May, G.; Lin, H. A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. Nat. Commun. 2018, 9, 4844. [Google Scholar] [CrossRef]
- Lo, K.J.; Lin, S.S.; Lu, C.W.; Kuo, C.H.; Liu, C.T. Whole-genome sequencing and comparative analysis of two plant-associated strains of Rhodopseudomonas palustris (PS3 and YSC3). Sci. Rep. 2018, 8, 12769. [Google Scholar] [CrossRef] [PubMed]
- Leekitcharoensophon, P.; Kaas, R.S.; Thomsen, M.C.; Friis, C.; Rasmussen, S.; Aarestrup, F.M. snpTree a web server to identify and construct SNP trees from whole genome sequence data. BMC Genom. 2012, 13, S6. [Google Scholar]
- Shao, N.; Li, F.; Nie, K.; Fu, S.H.; Zhang, W.J.; He, Y.; Lei, W.W.; Wang, Q.Y.; Liang, G.D.; Cao, Y.X.; et al. TaqMan real-time rt-pcr assay for detecting and differentiating Japanese encephalitis virus. Biomed. Environ. Sci. 2018, 31, 208–214. [Google Scholar]
- Huang, S.; Wang, X.; Chen, X.; Liu, X.; Xu, Q.; Zhang, L.; Huang, G.; Wu, J. Rapid and sensitive detection of Pseudomonas aeruginosa by isothermal amplification combined with Cas12a-mediated detection. Sci. Rep. 2023, 13, 19199. [Google Scholar] [CrossRef] [PubMed]
- Sound, J.K.; Peters, A.; Bellamy-Carter, J.; Rad-Menéndez, C.; MacKechnie, K.; Green, D.H.; Leney, A.C. Rapid cyanobacteria species identification with high sensitivity using native mass spectrometry. Anal. Chem. 2021, 93, 14293–14299. [Google Scholar] [CrossRef]
- Hsieh, A.Y.Y.; Saberi, S.; Ajaykumar, A.; Hukezalie, K.; Gadawski, I.; Sattha, B.; Côté, H.C.F. Optimization of a relative telomere length assay by monochromatic multiplex real-time quantitative pcr on the LightCycler 480: Sources of variability and quality control considerations. J. Mol. Diagn. 2016, 18, 425–437. [Google Scholar] [CrossRef]
- GB 4789.34–2016; National Food Safety Standard–Food Microbiological Examination: Examination of Bifidobacteria. China Standards Press: Beijing, China, 2016.
- Yao, Y.T.; Zhang, X.; Wang, C.Y.; Zhang, Y.H.; Li, D.W.; Yang, W.D.; Li, H.Y.; Zou, L.G. Optimizing longifolene production in Yarrowia lipolytica via metabolic and protein engineering. Synth. Syst. Biotechnol. 2025, in press. [Google Scholar] [CrossRef]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef]
- Von Hertwig, A.M.; Sant’Ana, A.S.; Sartori, D.; da Silva, J.J.; Nascimento, M.S.; Iamanaka, B.T.; Pelegrinelli Fungaro, M.H.; Taniwaki, M.H. Real-time PCR-based method for rapid detection of Aspergillus niger and Aspergillus welwitschiae isolated from coffee. J. Microbiol. Methods 2018, 148, 87–92. [Google Scholar] [CrossRef] [PubMed]
- SN/T 5642.3-2023; Professional Standard of the People’s Republic of China for Entry-Exit Inspection and Quarantine–Detection of lactic acid bacteria in dairy products for export–Enumeration method of digital PCR–Part3: Bifidobacterium animalis. China Standards Press: Beijing, China, 2023.
- Wang, R.; Zhang, W.; Ye, R.; Pan, Z.; Li, G.; Su, S. One-step multiplex TaqMan probe-based method for real-time PCR detection of four canine diarrhea viruses. Mol. Cell. Probes 2020, 53, 101618. [Google Scholar] [CrossRef]
- Herbel, S.R.; Lauzat, B.; Von Nickisch-Rosenegk, M.; Kuhn, M.; Murugaiyan, J.; Wieler, L.H.; Guenther, S. Species-specific quantification of probiotic lactobacilli in yoghurt by quantitative real-time PCR. J. Appl. Microbiol. 2013, 115, 1402–1410. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berben, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- Dekker, J.W.; Wickens, K.; Black, P.N.; Stanley, T.V.; Mitchell, E.A.; Fitzharris, P. Safety aspects of probiotic bacterial strains Lactobacillus rhamnosus HN001 and Bifidobacterium animalis subsp. lactis HN019 in human infants aged 0–2 years. Int. Dairy J. 2009, 19, 149–154. [Google Scholar]
- Cheng, J.; Gao, C.; Ala-Jaakkola, R. Eight-Week Supplementation with Bifidobacterium lactis HN019 and Functional Constipation: A Randomized Clinical Trial. JAMA Netw. Open 2024, 7, e2436888. [Google Scholar] [CrossRef] [PubMed]
- Solano-Aguilar, G.; Dawson, H.; Restrepo, M.; Andrews, K.; Vinyard, B.; Urban, J.F., Jr. Detection of Bifidobacterium animalis subsp. lactis (Bb12) in the intestine after feeding of sows and their piglets. Appl. Environ. Microbiol. 2008, 74, 6338–6347. [Google Scholar]
- Taipale, T.; Pieniähäkkänen, K.; Isolauri, E.; Larsen, C.; Brockmann, E.; Alanen, P.; Jokela, J.; Söderling, E. Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in infancy. Br. J. Nutr. 2011, 105, 409–416. [Google Scholar]
- Requena, T.; Burton, J.; Matsuki, T.; Munro, K.; Simon, M.A.; Tanaka, R.; Watanabe, K.; Tannock, G.W. Identification, detection, and enumeration of human bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 2002, 68, 2420–2427. [Google Scholar] [CrossRef]
| Species | Strain | Assembly Number | Scaffold Number | Genome Size (Mb) |
|---|---|---|---|---|
| B. animalis subsp. lactis | HN019 * | GCA_003606305.1 | 1 | 1.935423 |
| 19-D-1 | GCA_020309945.1 | 1 | 1.963057 | |
| AD011 | GCA_000021425.1 | 1 | 1.933695 | |
| ATCC 27673 | GCA_000471945.1 | 1 | 1.963012 | |
| B420 | GCA_000277325.1 | 1 | 1.938595 | |
| BAMA-B06 | GCA_028463865.1 | 1 | 1.944252 | |
| BB-12 | GCA_000025245.2 | 1 | 1.944152 | |
| BF052 | GCA_000818055.1 | 1 | 1.938624 | |
| BGI-N3 | GCA_040084745.1 | 1 | 1.944238 | |
| BI040 | GCA_038086955.1 | 1 | 1.944141 | |
| Bi-07 | GCA_000277345.1 | 1 | 1.938822 | |
| Bl-04 | GCA_000022705.1 | 1 | 1.938709 | |
| Bl12 | GCA_000414215.2 | 1 | 1.944036 | |
| BLC1 | GCA_000224965.2 | 1 | 1.938583 | |
| CGMCC1.15623 | GCA_044866785.1 | 1 | 1.944145 | |
| CNCM I-2494 | GCA_000220885.1 | 1 | 1.943113 | |
| DSM 10140 | GCA_000022965.1 | 1 | 1.938483 | |
| DSM 15954 | GCA_021018785.1 | 1 | 1.944152 | |
| GOLDGUT-BB18 | GCA_034561975.1 | 1 | 1.944144 | |
| H1 | GCA_016835115.1 | 1 | 1.944352 | |
| H3 | GCA_016835135.1 | 1 | 1.944351 | |
| HOM2120 | GCA_044502405.1 | 1 | 1.946900 | |
| i797 | GCA_019576095.1 | 1 | 1.943538 | |
| IDCC4301 | GCA_003428375.1 | 1 | 1.944141 | |
| KLDS 2.0603 | GCA_000816205.1 | 1 | 1.946899 | |
| LPL-RH | GCA_030284625.1 | 1 | 1.944223 | |
| MH-02 | GCA_029167605.1 | 1 | 1.944290 | |
| S7 | GCA_003390755.1 | 1 | 1.944072 | |
| SF | GCA_029542645.1 | 1 | 1.944374 | |
| TCI604 | GCA_036923305.1 | 1 | 1.944134 | |
| V9 | GCA_000092765.1 | 1 | 1.944050 |
| Gene | The Sequences of Primers and Probe | Product Length (bp) |
|---|---|---|
| galK | TGATATGGCGAATGCTTGCA | 61 |
| CGGCTTGTGTGTCGTCATG | ||
| FAM-ACCCTTTTCATTTCCCTGC-MGB |
| No. | Species | Strain |
|---|---|---|
| 1 | B. animalis subsp. lactis | HN019 |
| 2 | Bi-07 | |
| 3 | Bl-04 | |
| 4 | BB12 | |
| 5 | DSMZ 10140 | |
| 6 | CICC 21718 | |
| 7 | ATCC 25527 | |
| 8 | ATCC 27673 | |
| 9 | B. animalis | ATCC 27672 |
| 10 | CICC 6165 | |
| 11 | CICC 6250 | |
| 12 | B. adolescentis | ATCC 15703 |
| 13 | B. longum subsp. infantis | ATCC 15697 |
| 14 | B. breve | ATCC 15700 |
| 15 | CICC 6185 | |
| 16 | CICC 6181 | |
| 17 | B. bifidum | CICC 6173 |
| 18 | B. longum | CICC 6207 |
| 19 | CICC 6199 | |
| 20 | Bl05 |
| Number of Generations | Cq Value | Mean Cq Value | SD | RSD (%) | ||
|---|---|---|---|---|---|---|
| 5 | 18.88 | 18.78 | 18.98 | 18.88 | 0.24 | 1.31 |
| 15 | 18.92 | 18.86 | 18.27 | 18.68 | ||
| 25 | 18.73 | 18.38 | 18.89 | 18.67 | ||
| 35 | 17.98 | 18.4 | 18.5 | 18.29 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, D.; Zhao, L.; Zhao, B.; Xu, H.; Zhang, Q. Development of a Strain-Specific Detection and Quantification Method for Bifidobacterium animalis subsp. lactis HN019 Using WGS-SNP Analysis and qPCR. Microorganisms 2025, 13, 1596. https://doi.org/10.3390/microorganisms13071596
Mao D, Zhao L, Zhao B, Xu H, Zhang Q. Development of a Strain-Specific Detection and Quantification Method for Bifidobacterium animalis subsp. lactis HN019 Using WGS-SNP Analysis and qPCR. Microorganisms. 2025; 13(7):1596. https://doi.org/10.3390/microorganisms13071596
Chicago/Turabian StyleMao, Da, Lei Zhao, Bo Zhao, Hongbin Xu, and Qinghe Zhang. 2025. "Development of a Strain-Specific Detection and Quantification Method for Bifidobacterium animalis subsp. lactis HN019 Using WGS-SNP Analysis and qPCR" Microorganisms 13, no. 7: 1596. https://doi.org/10.3390/microorganisms13071596
APA StyleMao, D., Zhao, L., Zhao, B., Xu, H., & Zhang, Q. (2025). Development of a Strain-Specific Detection and Quantification Method for Bifidobacterium animalis subsp. lactis HN019 Using WGS-SNP Analysis and qPCR. Microorganisms, 13(7), 1596. https://doi.org/10.3390/microorganisms13071596






