Exploring the Skin Benefits of Extremophilic Postbiotics from Exiguobacterium artemiae: A New Frontier in Thermal Protection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Induction of Heat Stress and Sample Treatment
2.3. Sample Preparation for E. artemiae SUPER-T mRNA Extraction
2.4. E. artemiae SUPER-T mRNA Sequencing
2.5. Data Processing and Analysis
2.6. Differential Gene Expression Analysis
2.7. Biocompatibility of SUPER-T and Thermasome
2.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
3.1. Transcriptomic Profiling and Gene Expression Analysis of E. artemiae SUPER-T
3.2. Regenerative Activation of ECM Pathways via SUPER-T Postbiotics
3.3. Sequential Modulation of Heat Shock Protein and Collagen Genes by SUPER-T Under Thermal Stress
3.4. Thermasome, Cosmetic Industrial Application of SUPER-T
3.5. Biocompatibility of SUPER-T and Thermasome
3.6. Effects of SUPER-T and Thermasome on the Gene Expressions of the Basement Membrane Proteins Under Thermal Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| qPCR | Quantitative polymerase chain reaction |
| CFS | Cell-free supernatant |
| SCFAs | Short-chain fatty acids |
| mRNA | Messenger ribonucleic acid |
| miRNA | Micro ribonucleic acid |
| RNA seq | RNA sequencing |
| ATCC | American type culture collection |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| AA | Antibiotic–antimycotic |
| NCBI | National center for biotechnology information |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| TPM | Transcripts per million |
| QC | Quality control |
| PCA | Principal component analysis |
| MDS | Multidimensional scaling |
| FDR | False discovery rate |
| GO | Gene ontology |
| DPBS | Dulbecco’s phosphate-buffered saline |
| CCK-8 | Cell-counting kit-8 |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| TRPV1 | Transient receptor potential vanilloid 1 |
| NID1 | Nidogen 1 |
| COL6A1 | Collagen, type 6, alpha 1 |
| COL1A1 | Collagen, type 1, alpha 1 |
| FBN | Fibrillin |
| COL15A1 | Collagen, type 15, alpha 1 |
| COL24A1 | Collagen, type 24, alpha 1 |
| GC | Guanine–cytosine |
| AT | Adenine–thymine |
| IL-6 | Interleukin-6 |
| PDPN | Podoalanin |
| CTSS | Cathepsin S |
References
- Hylander, B.L.; Repasky, E.A. Temperature as a Modulator of the Gut Microbiome: What Are the Implications and Opportunities for Thermal Medicine? Int. J. Hyperth. 2019, 36, 83–89. [Google Scholar] [CrossRef]
- Shapiro, R.S.; Cowen, L.E. Thermal Control of Microbial Development and Virulence: Molecular Mechanisms of Microbial Temperature Sensing. mBio 2012, 3, e00238-12. [Google Scholar] [CrossRef]
- Berry, E.D.; Foegeding, P.M. Cold Temperature Adaptation and Growth of Microorganisms. J. Food Prot. 1997, 60, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Pikuta, E.V.; Hoover, R.B.; Tang, J. Microbial Extremophiles at the Limits of Life. Crit. Rev. Microbiol. 2007, 33, 183–209. [Google Scholar] [CrossRef]
- Collins, M.D.; Lund, B.M.; Farrow, J.A.E.; Schleifer, K.H. Chemotaxonomic Study of an Alkalophilic Bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. Microbiology 1983, 129, 2037–2042. [Google Scholar] [CrossRef]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An Overview of a Versatile Genus with Potential in Industry and Agriculture. Crit. Rev. Biotechnol. 2018, 38, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Goris, J.; Vishnivetskaya, T.; Gilichinsky, D.; Thomashow, M.F.; Tiedje, J.M. Characterization of Exiguobacterium Isolates from the Siberian Permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles 2006, 10, 285–294. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Shivaji, S. Exiguobacterium Indicum Sp. Nov., a Psychrophilic Bacterium from the Hamta Glacier of the Himalayan Mountain Ranges of India. Int. J. Syst. Evol. Microbiol. 2006, 56, 2765–2770. [Google Scholar] [CrossRef]
- Crapart, S.; Fardeau, M.L.; Cayol, J.L.; Thomas, P.; Sery, C.; Ollivier, B.; Combet-Blanc, Y. Exiguobacterium profundum Sp. Nov., a Moderately Thermophilic, Lactic Acid-Producing Bacterium Isolated from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2007, 57, 287–292. [Google Scholar] [CrossRef]
- Sawle, L.; Ghosh, K. How Do Thermophilic Proteins and Proteomes Withstand High Temperature? Biophys. J. 2011, 101, 217–227. [Google Scholar] [CrossRef]
- Zhou, H.-X. Toward the Physical Basis of Thermophilic Proteins: Linking of Enriched Polar Interactions and Reduced Heat Capacity of Unfolding. Biophys. J. 2002, 83, 3126–3133. [Google Scholar] [CrossRef] [PubMed]
- López-Cortés, A.; Schumann, P.; Pukall, R.; Stackebrandt, E. Exiguobacterium mexicanum Sp. Nov. and Exiguobacterium artemiae Sp. Nov., Isolated from the Brine Shrimp Artemia Franciscana. Syst. Appl. Microbiol. 2006, 29, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.I.; Lee, M.; Jung, Y.; Jeong, M.K.; Ryu, J.H.; Kim, B.J.; Suh, B.F.; Kim, E. Skin Characteristics Following Repeated Exposure to Simulated Outdoor and Indoor Summer Temperatures in South Korea and Southeast Asia. Int. J. Cosmet. Sci. 2021, 43, 352–358. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Z.; Zhang, H.; Arens, E.; Filingeri, D.; Jin, L.; Ghahramani, A.; Chen, W.; He, Y.; Si, B. High-Density Thermal Sensitivity Maps of the Human Body. Build. Environ. 2020, 167, 106435. [Google Scholar] [CrossRef]
- Liu, F.W.; Liu, F.C.; Wang, Y.R.; Tsai, H.I.; Yu, H.P. Aloin Protects Skin Fibroblasts from Heat Stress-Induced Oxidative Stress Damage by Regulating the Oxidative Defense System. PLoS ONE 2015, 10, e0143528. [Google Scholar] [CrossRef]
- Resende, D.I.S.P.; Ferreira, M.S.; Sousa-Lobo, J.M.; Sousa, E.; Almeida, I.F. Usage of Synthetic Peptides in Cosmetics for Sensitive Skin. Pharmaceuticals 2021, 14, 702. [Google Scholar] [CrossRef]
- Chen, Z.; Jin, Y.S.; Yeon, K.K.; Se, R.L.; Kyu, H.K.; Kwang, H.C.; Hee, C.E.; Jin, H.C. Heat Modulation of Tropoelastin, Fibrillin-1, and Matrix Metalloproteinase-12 in Human Skin in Vivo. J. Investig. Dermatol. 2005, 124, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Nischt, R.; Schmidt, C.; Mirancea, N.; Baranowsky, A.; Mokkapati, S.; Smyth, N.; Woenne, E.C.; Stark, H.J.; Boukamp, P.; Breitkreutz, D. Lack of Nidogen-1 and -2 Prevents Basement Membrane Assembly in Skin-Organotypic Coculture. J. Investig. Dermatol. 2007, 127, 545–554. [Google Scholar] [CrossRef]
- Heo, Y.M.; Lee, D.G.; Mun, S.; Kim, M.; Baek, C.; Lee, H.; Yun, S.K.; Kang, S.; Han, K. Skin Benefits of Postbiotics Derived from Micrococcus Luteus Derived from Human Skin: An Untapped Potential for Dermatological Health. Genes Genom. 2024, 46, 13–25. [Google Scholar] [CrossRef]
- Liang, X.; Li, Y.; Zhao, Z.; Ding, R.; Sun, J.; Chi, C. Safety and Efficacy of Adding Postbiotics in Infant Formula: A Systematic Review and Meta-Analysis. Pediatr. Res. 2024, 95, 43–51. [Google Scholar] [CrossRef]
- Liang, B.; Xing, D. The Current and Future Perspectives of Postbiotics. Probiotics Antimicrob. Proteins 2023, 15, 1626–1643. [Google Scholar] [CrossRef] [PubMed]
- Thorakkattu, P.; Khanashyam, A.C.; Shah, K.; Babu, K.S.; Mundanat, A.S.; Deliephan, A.; Deokar, G.S.; Santivarangkna, C.; Nirmal, N.P. Postbiotics: Current Trends in Food and Pharmaceutical Industry. Foods 2022, 11, 3094. [Google Scholar] [CrossRef] [PubMed]
- Safari, B.; Aghazadeh, M.; Davaran, S.; Roshangar, L. Exosome-Loaded Hydrogels: A New Cell-Free Therapeutic Approach for Skin Regeneration. Eur. J. Pharm. Biopharm. 2022, 171, 50–59. [Google Scholar] [CrossRef]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A Novel Nanoparticle Drug Delivery System: The Anti-Inflammatory Activity of Curcumin Is Enhanced When Encapsulated in Exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Chen, S.; Li, H. Heat Stress Regulates the Expression of Genes at Transcriptional and Post-Transcriptional Levels, Revealed by RNA-Seq in Brachypodium Distachyon. Front. Plant Sci. 2017, 7, 2067. [Google Scholar] [CrossRef]
- Liu, X.; Du, F.; Li, N.; Chang, Y.; Yao, D. Gene Expression Profile in the Long-Living Lotus: Insights into the Heat Stress Response Mechanism. PLoS ONE 2016, 11, e0152540. [Google Scholar] [CrossRef]
- Asai, J. The Role of Podoplanin in Skin Diseases. Int. J. Mol. Sci. 2022, 23, 1310. [Google Scholar] [CrossRef] [PubMed]
- Quintanilla, M.; Montero, L.M.; Renart, J.; Villar, E.M. Podoplanin in Inflammation and Cancer. Int. J. Mol. Sci. 2019, 20, 707. [Google Scholar] [CrossRef]
- Bieniasz-Krzywiec, P.; Martín-Pérez, R.; Ehling, M.; García-Caballero, M.; Pinioti, S.; Pretto, S.; Kroes, R.; Aldeni, C.; Di Matteo, M.; Prenen, H.; et al. Podoplanin-Expressing Macrophages Promote Lymphangiogenesis and Lymphoinvasion in Breast Cancer. Cell Metab. 2019, 30, 917–936.e10. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Wang, J.; Xiang, M.X.; Lin, Y.; He, A.; Jin, C.N.; Guan, J.; Sukhova, G.K.; Libby, P.; Wang, J.A.; et al. Cathepsin S-Mediated Fibroblast Trans-Differentiation Contributes to Left Ventricular Remodelling after Myocardial Infarction. Cardiovasc. Res. 2013, 100, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Riau, A.K.; Ong, H.S.; Yam, G.H.F.; Mehta, J.S. Sustained Delivery System for Stem Cell-Derived Exosomes. Front. Pharmacol. 2019, 10, 1368. [Google Scholar] [CrossRef]
- Rosenbaum, T.; Simon, S. TRPV1 Receptors and Signal Transduction. In TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades; CRC Press: Boca Raton, FL, USA, 2007; ISBN 0849340489. [Google Scholar]
- Pfisterer, K.; Shaw, L.E.; Symmank, D.; Weninger, W. The Extracellular Matrix in Skin Inflammation and Infection. Front. Cell Dev. Biol. 2021, 9, 682414. [Google Scholar] [CrossRef]
- Hohenester, E.; Yurchenco, P.D. Laminins in Basement Membrane Assembly. Cell Adh. Migr. 2013, 7, 56–63. [Google Scholar] [CrossRef]
- Theocharidis, G.; Drymoussi, Z.; Kao, A.P.; Barber, A.H.; Lee, D.A.; Braun, K.M.; Connelly, J.T. Type VI Collagen Regulates Dermal Matrix Assembly and Fibroblast Motility. J. Investig. Dermatol. 2016, 136, 74–83. [Google Scholar] [CrossRef]
- Shaw, L.M.; Olsen, B.R. FACIT Collagens: Diverse Molecular Bridges in Extracellular Matrices. Trends Biochem. Sci. 1991, 16, 191–194. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The Capsaicin Receptor: A Heat-Activated Ion Channel in the Pain Pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kang, S.M.; Chung, J.H. The Role of TRPV1 Channel in Aged Human Skin. J. Dermatol. Sci. 2012, 65, 81–85. [Google Scholar] [CrossRef]
- Radtke, C.; Sinis, N.; Sauter, M.; Jahn, S.; Kraushaar, U.; Guenther, E.; Rodemann, H.P.; Rennekampff, H.O. TRPV Channel Expression in Human Skin and Possible Role in Thermally Induced Cell Death. J. Burn Care Res. 2011, 32, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Eming, S.A.; Krieg, T.; Davidson, J.M. Inflammation in Wound Repair: Molecular and Cellular Mechanisms. J. Investig. Dermatol. 2007, 127, 514–525. [Google Scholar] [CrossRef]
- Garbers, C.; Heink, S.; Korn, T.; Rose-John, S. Interleukin-6: Designing Specific Therapeutics for a Complex Cytokine. Nat. Rev. Drug Discov. 2018, 17, 395–412. [Google Scholar] [CrossRef]
- Lin, Z.-Q.; Kondo, T.; Ishida, Y.; Takayasu, T.; Mukaida, N. Essential Involvement of IL-6 in the Skin Wound-Healing Process as Evidenced by Delayed Wound Healing in IL-6-Deficient Mice. J. Leukoc. Biol. 2003, 73, 713–721. [Google Scholar] [CrossRef]
- Asai, J.; Hirakawa, S.; Sakabe, J.I.; Kishida, T.; Wada, M.; Nakamura, N.; Takenaka, H.; Mazda, O.; Urano, T.; Suzuki-Inoue, K.; et al. Platelets Regulate the Migration of Keratinocytes via Podoplanin/CLEC-2 Signaling during Cutaneous Wound Healing in Mice. Am. J. Pathol. 2016, 186, 101–108. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Shi, X.; Shi, H.; Wang, Z.; Peng, C. Review in Isothermal Amplification Technology in Food Microbiological Detection. Food Sci. Biotechnol. 2022, 31, 1501–1511. [Google Scholar] [CrossRef]
- McDowell, S.H.; Gallaher, S.A.; Burden, R.E.; Scott, C.J. Leading the Invasion: The Role of Cathepsin S in the Tumour Microenvironment. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118781. [Google Scholar] [CrossRef]
- Vidak, E.; Javoršek, U.; Vizovišek, M.; Turk, B. Cysteine Cathepsins and Their Extracellular Roles: Shaping the Microenvironment. Cells 2019, 8, 264. [Google Scholar] [CrossRef] [PubMed]
- Dad, H.A.; Gu, T.W.; Zhu, A.Q.; Huang, L.Q.; Peng, L.H. Plant Exosome-like Nanovesicles: Emerging Therapeutics and Drug Delivery Nanoplatforms. Mol. Ther. 2021, 29, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Susa, F.; Limongi, T.; Dumontel, B.; Vighetto, V.; Cauda, V. Engineered Extracellular Vesicles as a Reliable Tool in Cancer Nanomedicine. Cancers 2019, 11, 1979. [Google Scholar] [CrossRef]
- Cescon, M.; Gattazzo, F.; Chen, P.; Bonaldo, P. Collagen VI at a Glance. J. Cell Sci. 2015, 128, 3525–3531. [Google Scholar] [CrossRef] [PubMed]
- Salmivirta, K.; Talts, J.F.; Olsson, M.; Sasaki, T.; Timpl, R.; Ekblom, P. Binding of Mouse Nidogen-2 to Basement Membrane Components and Cells and Its Expression in Embryonic and Adult Tissues Suggest Complementary Functions of the Two Nidogens. Exp. Cell Res. 2002, 279, 188–201. [Google Scholar] [CrossRef]
- Töpfer, U.; Holz, A. Nidogen in Development and Disease. Front. Cell Dev. Biol. 2024, 12, 1380542. [Google Scholar] [CrossRef]

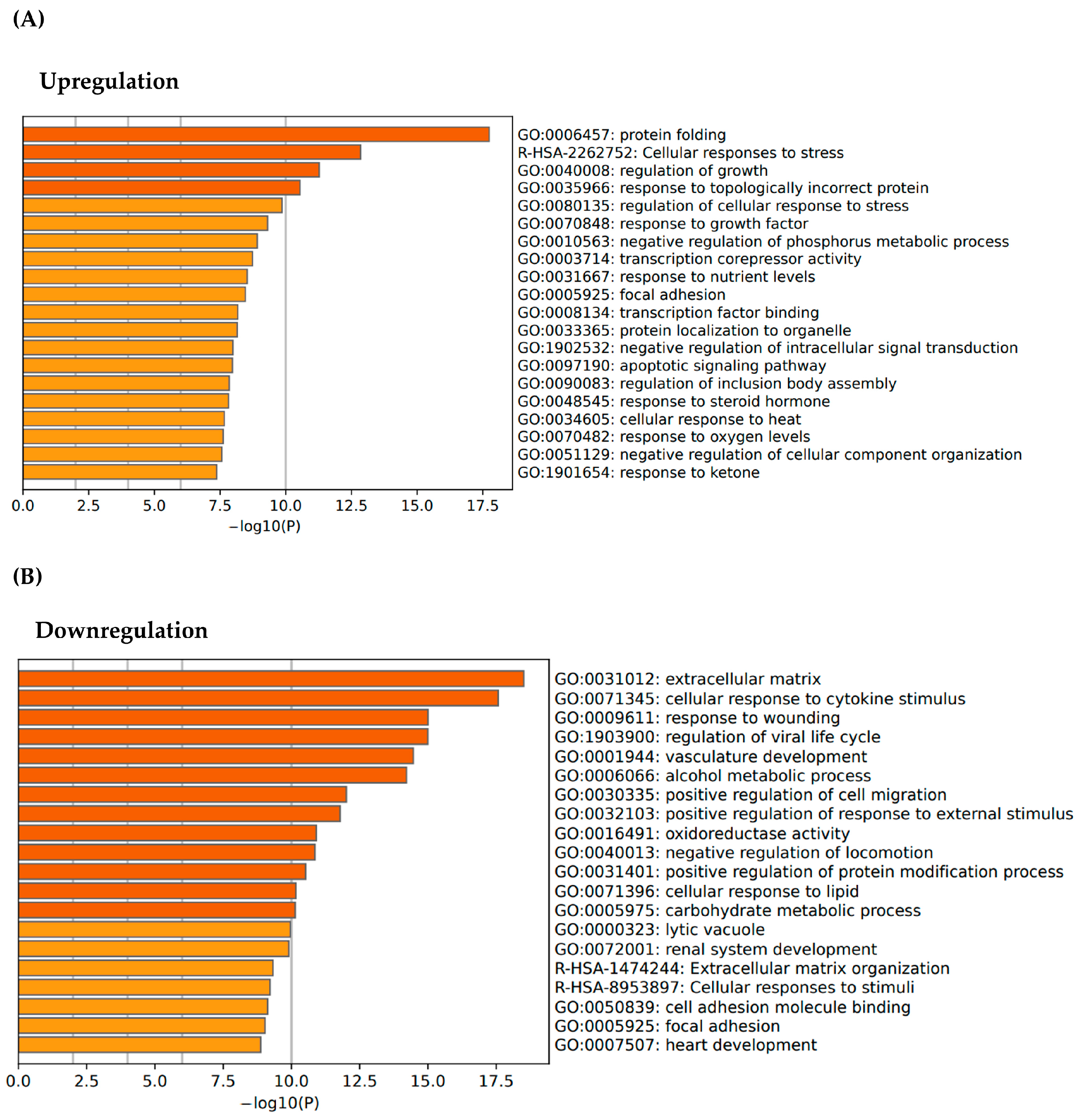

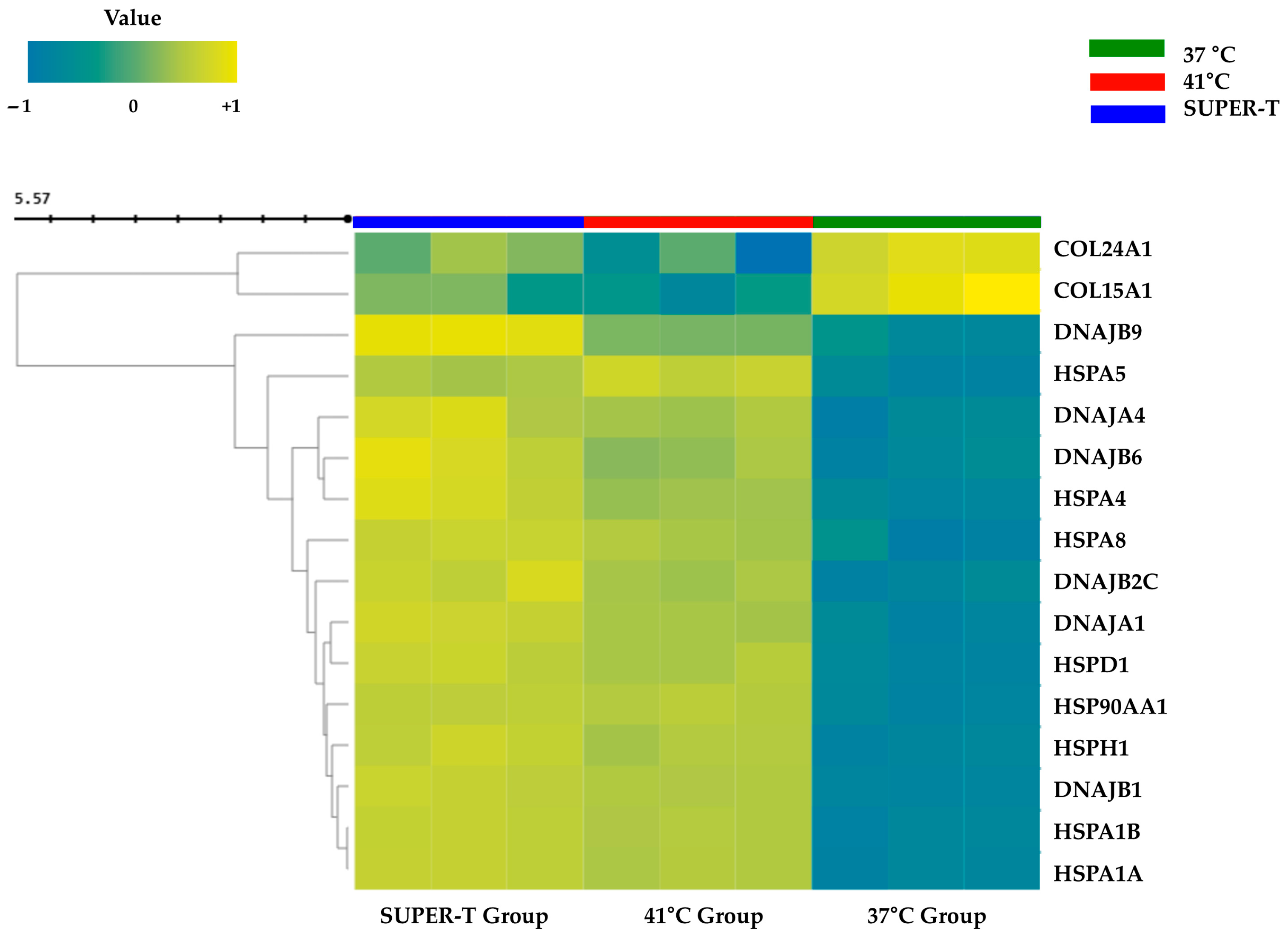

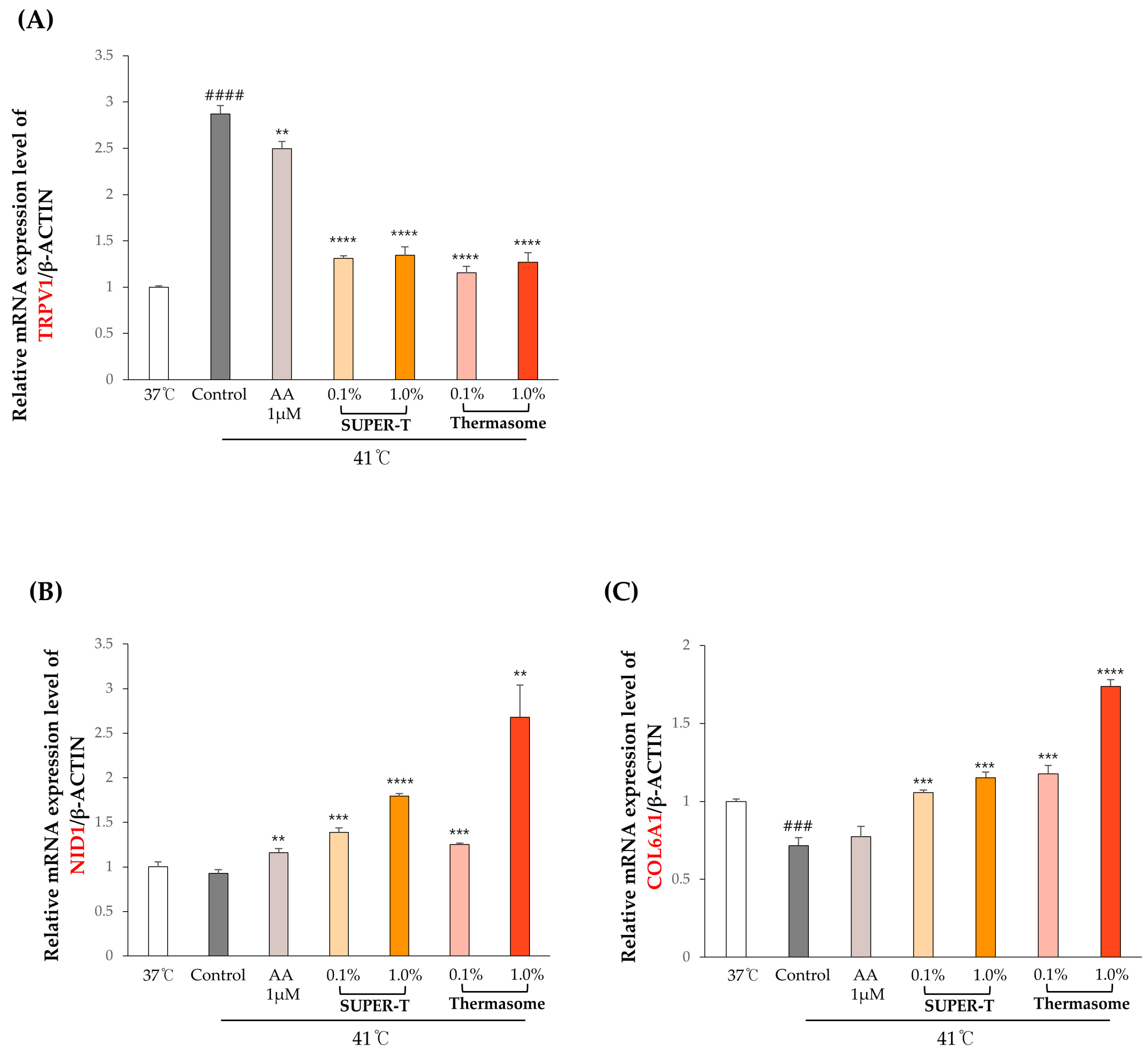

| Sample ID | Total Read Bases (Bp) | Total Reads | GC% | AT% | Q20% | Q30% |
|---|---|---|---|---|---|---|
| 37 °C_1 | 6,301,270,820 | 62,388,820 | 49.1 | 50.9 | 99 | 96.3 |
| 37 °C_2 | 7,270,146,246 | 71,981,646 | 49.4 | 50.6 | 99 | 96.1 |
| 37 °C_3 | 7,509,311,822 | 74,349,622 | 49.2 | 50.8 | 99 | 96.1 |
| 41 °C_1 | 6,868,869,408 | 68,008,608 | 49.7 | 50.3 | 99 | 96 |
| 41 °C_2 | 6,113,338,908 | 60,528,108 | 49.8 | 50.2 | 99 | 96.1 |
| 41 °C_3 | 6,094,026,294 | 60,336,894 | 49.8 | 50.2 | 98.8 | 95.5 |
| SUPER-T_1 | 6,236,462,958 | 61,747,158 | 49.1 | 50.9 | 99 | 96.3 |
| SUPER-T_2 | 7,324,428,696 | 72,519,096 | 49.3 | 50.7 | 98.8 | 95.8 |
| SUPER-T_3 | 7,682,063,030 | 76,060,030 | 49.4 | 50.6 | 99 | 96.2 |
| Diameter/nm | Particles/mL | FWHM/nm | Purity |
|---|---|---|---|
| 117.4 | 82.2 | 89.10% |
| Gene | Forward | Reverse |
|---|---|---|
| hβ-ACTIN | CATGTACGTTGCTATCCAGGC | CTCCTTAATGTCACGCACGAT |
| hTRPV1 | GTGGACAGCTACAGTGAGATGC | GGAAGCCACATACTCCTTGAGG |
| hNID1 | AGGAGCTCTTTCCCTTCGGC | CGGGGGTTCACTCGTAGCAA |
| hCOL6A1 | GCCTTCCTGAAGAATGTCACCG | TCCAGCAGGATGGTGATGTCAG |
| hCOL1A1 | GCTTGGTCCACTTGCTTGAAGA | GAGCATTGCCTTTGATTGCTG |
| hFBN | GCGGAAATCAGTGTATTGTCCC | CAGTGTTGTATGGATCTGGAGC |
| hCOL15A1 | GGACTTGGATTCGAGGATACCG | AATACTGGCTCCATCCATCCC |
| hCOL24A1 | GATACCATGGAGCAGATGGC | CCCTTGTTCACCCTTTGGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Roo, D.; Lee, D.-G.; Kang, S.; Min, J.; Kang, H.; Heo, Y.M.; Lee, K.E. Exploring the Skin Benefits of Extremophilic Postbiotics from Exiguobacterium artemiae: A New Frontier in Thermal Protection. Microorganisms 2025, 13, 1569. https://doi.org/10.3390/microorganisms13071569
Lee H, Roo D, Lee D-G, Kang S, Min J, Kang H, Heo YM, Lee KE. Exploring the Skin Benefits of Extremophilic Postbiotics from Exiguobacterium artemiae: A New Frontier in Thermal Protection. Microorganisms. 2025; 13(7):1569. https://doi.org/10.3390/microorganisms13071569
Chicago/Turabian StyleLee, Haeun, Dayeon Roo, Dong-Geol Lee, Seunghyun Kang, Jinwoo Min, Heecheol Kang, Young Mok Heo, and Kyung Eun Lee. 2025. "Exploring the Skin Benefits of Extremophilic Postbiotics from Exiguobacterium artemiae: A New Frontier in Thermal Protection" Microorganisms 13, no. 7: 1569. https://doi.org/10.3390/microorganisms13071569
APA StyleLee, H., Roo, D., Lee, D.-G., Kang, S., Min, J., Kang, H., Heo, Y. M., & Lee, K. E. (2025). Exploring the Skin Benefits of Extremophilic Postbiotics from Exiguobacterium artemiae: A New Frontier in Thermal Protection. Microorganisms, 13(7), 1569. https://doi.org/10.3390/microorganisms13071569






