Antibiotic Resistance and Characteristics of Vibrio parahaemolyticus Isolated from Seafood Distributed in South Korea from 2021 to 2022
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Procurement
2.2. Enrichment and Isolation
2.3. Antibiotic Susceptibility Testing
2.4. PCR Detection of Toxins and Related Genes
2.5. WGS
2.6. Gene Analysis and MLST
2.7. Virulence Factor Analysis Using WGS Data
3. Results
3.1. Isolation of V. parahaemolyticus from Fish Samples
3.2. Antibiotic Susceptibility Test
3.3. PCR Detection of Toxin and Related Genes
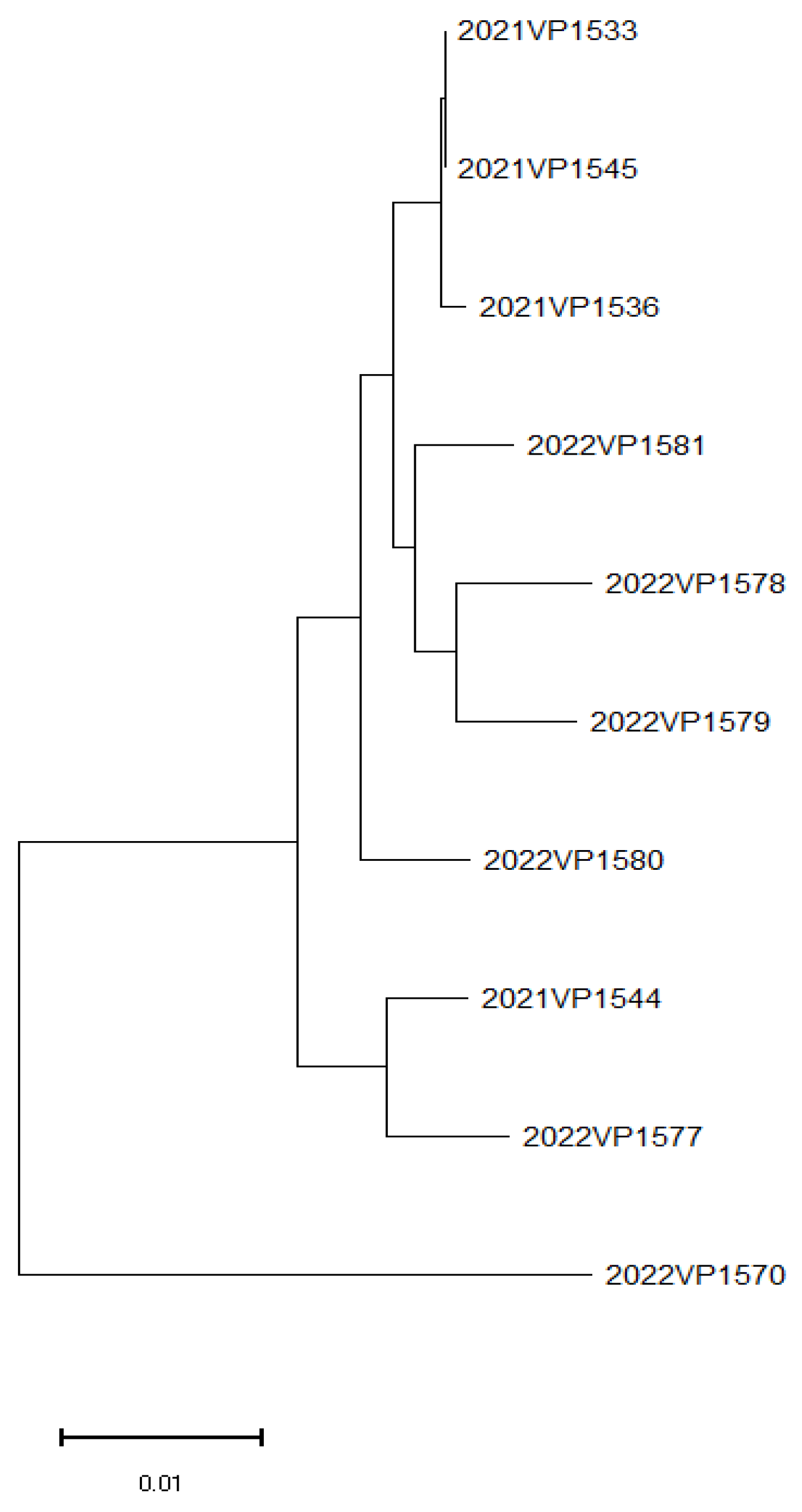
3.4. WGS and MLST
3.5. Virulence Factor Analysis
3.5.1. Commonly Identified Virulence Factors
3.5.2. Individually Identified Virulence Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martinez-Urtaza, J.; Baker-Austin, C. Vibrio parahaemolyticus . Trends Microbiol. 2020, 28, 867–868. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-cholera vibrios: The microbial barometer of climate change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, K.; Zhang, L.; Zhang, X.; Zhu, B.; Lv, N.; Mi, K. Genetic adaptation of Vibrio parahaemolyticus in response to global warming. Microbiol. Spectr. 2023, 11, 01502–01523. [Google Scholar]
- Wang, Y.; Li, X.; Chen, Z.; Liu, M.; Zhang, H.; Zhou, Q.; Xu, J. Environmental changes driving virulence evolution in Vibrio parahaemolyticus. BMC Microbiol. 2024, 24, 3303. [Google Scholar]
- Jang, W.; Cho, J.H.; Lee, D.; Kim, Y. Trends in seafood consumption and factors influencing the consumption of seafood among the old adults based on the Korea National Health and Nutrition Examination survey 2009–2019. J. Korean Soc. Food Sci. Nutr. 2022, 51, 651–659. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, J.; Park, S.; Choi, H.; Jung, S.; Lim, D.; Han, K.; Seo, M.; Kang, T. The impact of antibiotic overuse in aquaculture on antimicrobial resistance in marine bacteria. Korean J. Fish. Aquat. Sci. 2019, 52, 198–210. Available online: https://www.koreascience.kr/article/JAKO201918454912838.pdf (accessed on 28 June 2025).
- Jeong, H.-W.; Kim, J.-A.; Jeon, S.-J.; Choi, S.-S.; Kim, M.-K.; Yi, H.-J.; Cho, S.-J.; Kim, I.-Y.; Chon, J.-W.; Kim, D.-H.; et al. Prevalence, antibiotic-resistance, and virulence characteristics of Vibrio parahaemolyticus in restaurant fish tanks in Seoul, South Korea. Foodborne Pathog. Dis. 2019, 16, 482–488. [Google Scholar] [CrossRef]
- Ko, J.-M.; Oh, S.-W.; Hong, J.-H. Sensory drivers of sliced raw fish in Korea: Case study on flounder (Paralichthys olivaceus) and rockfish (Sebastes schlegeli). J. Korean Soc. Food Sci. Nutr. 2016, 45, 1192–1201. [Google Scholar] [CrossRef]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef]
- Gonzalez-Escalona, N.; Martinez-Urtaza, J.; Romero, J.; Espejo, R.T.; Jaykus, L.A.; DePaola, A. MLST analysis of Vibrio parahaemolyticus from clinical and environmental sources in Thailand and its implications for seafood safety. Appl. Environ. Microbiol. 2008, 74, 7451–7453. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- ISO 6887-3:2017; Microbiology of the Food Chain–Preparation of Test Samples, Initial Suspension and Decimal Dilutions for Microbiological Examination–Part 3: Specific Rules for the Preparation of Fish and Fishery Products. International Organization for Standardization: Geneva, Switzerland, 2017.
- Ministry of Food and Drug Safety (MFDS). Food Standards and Specifications (Food Code). Notification No. 2024–71; Ministry of Food and Drug Safety: Cheongju, Republic of Korea, 2024. Available online: https://www.mfds.go.kr (accessed on 20 May 2025).
- ISO 21872-1:2017; Microbiology of the Food Chain–Horizontal Method for the Determination of Vibrio spp.–Part 1: Detection of Potentially Enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. International Organization for Standardization: Geneva, Switzerland, 2017.
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: M45, 3rd ed.; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.; Jones, D.D.; Kaysner, C.A. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Casselli, T.; Lynch, T.; Southward, C.M.; Jones, B.W.; DeVinney, R. Vibrio parahaemolyticus inhibition of Rho family GTPase activation requires a functional chromosome I type III secretion system. Infect. Immun. 2008, 76, 2202–2211. [Google Scholar] [CrossRef]
- Dong, X.; Li, Z.; Wang, X.; Zhou, M.; Lin, L.; Zhou, Y.; Li, J. Characteristics of Vibrio parahaemolyticus isolates obtained from crayfish (Procambarus clarkii) in freshwater. Int. J. Food Microbiol. 2016, 238, 132–138. [Google Scholar] [CrossRef]
- Okada, N.; Iida, T.; Park, K.S.; Goto, N.; Yasunaga, T.; Hida, T.; Honda, T. Identification and characterization of a novel type III secretion system in Vibrio parahaemolyticus. Infect. Immun. 2009, 77, 1473–1483. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, H.; Li, J.; Zhang, P.; Wu, B.; Zhu, B.; Zhang, Y.; Fang, W. Putative type VI secretion systems of Vibrio parahaemolyticus contribute to adhesion to cultured cell monolayers. Arch. Microbiol. 2012, 194, 827–835. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef]
- Newton, A.; Kendall, M.; Vugia, D.J.; Henao, O.L.; Mahon, B.E. Increasing rates of vibriosis in the United States, 1996–2010: Review of surveillance data from 2 systems. Clin. Infect. Dis. 2012, 54 (Suppl. S5), S391–S395. [Google Scholar] [CrossRef]
- Daniels, N.A.; MacKinnon, L.; Bishop, R.; Altekruse, S.; Ray, B.; Hammond, R.M.; Thompson, S.; Wilson, S.; Bean, N.H.; Griffin, P.M.; et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J. Infect. Dis. 2000, 181, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Hara-Kudo, Y.; Sugiyama, K.; Nishibuchi, M.; Chowdhury, A.; Yatsuyanagi, J.; Ohtomo, Y.; Saito, A.; Nagano, H.; Nishina, T.; Nakagawa, H.; et al. Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 2003, 69, 3883–3891. [Google Scholar] [CrossRef] [PubMed]
- Broberg, C.A.; Calder, T.J.; Orth, K. Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 2011, 13, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G. Influence of climatic factors on the occurrence of Vibrio parahaemolyticus food poisoning in the Republic of Korea. Climate 2024, 12, 25. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.A.; Taylor, N.G.H.; Hartnell, R.; Siitonen, A.; Martinez-Urtaza, J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Change 2013, 3, 73–77. [Google Scholar] [CrossRef]
- Liu, M.; Yang, D.; Jin, Q.; Wang, C.; Shen, X. Prevalence of Vibrio parahaemolyticus in seafood from South China and its genetic diversity. J. Food Prot. 2014, 77, 611–618. [Google Scholar] [CrossRef]
- Yano, Y.; Hamano, K.; Satomi, M.; Tsukamoto, T.; Narahara, K. The effect of seafood distribution and storage conditions on the prevalence of Vibrio parahaemolyticus in seafood. Int. J. Food Microbiol. 2010, 137, 79–85. [Google Scholar] [CrossRef]
- Zhang, L.; Li, N.; Ling, J.; Gu, T.; Tang, Y. High prevalence and genetic diversity of Vibrio parahaemolyticus in seafood from Southeast Asia. Food Control 2019, 98, 133–140. [Google Scholar]
- Caburlotto, G.; Gennari, M.; Ghidini, V.; Tafi, M.; Lleo, M.M. Effect of seafood storage and processing on Vibrio parahaemolyticus contamination levels in different regions. Int. J. Food Microbiol. 2010, 141, 87–92. [Google Scholar] [CrossRef]
- Yamamoto, S.; Nakayama, T.; Takahashi, H.; Kanki, M. Antibiotic resistance in Vibrio parahaemolyticus isolated from seafood. Int. J. Food Microbiol. 2015, 203, 23–28. [Google Scholar] [CrossRef]
- Joseph, S.W.; Colwell, R.R.; Kaper, J.B. In Vitro response to chloramphenicol, tetracycline, ampicillin, gentamicin, and beta-lactamase production by halophilic Vibrios from human and environmental source. Antimicrob. Agents Chemother. 1979, 16, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, H.; Sun, J. Influence of β-lactam antibiotics on antimicrobial resistance in Vibrio parahaemolyticus from marine environments. Mar. Pollut. Bull. 2019, 149, 110631. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Baker-Austin, C.; Jones, J.L. Horizontal gene transfer and antimicrobial resistance in Vibrio parahaemolyticus. Environ. Microbiol. 2018, 20, 21–30. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, E. Antimicrobial resistance and resistance transfer of Vibrio parahaemolyticus and Morganella morganii from commercial fisheries products. Fish Pathol. 2019, 32, 97–104. [Google Scholar]
- Miranda, C.D.; Zemelman, R. Bacterial resistance to oxytetracycline in Chilean salmon farming. Aquaculture 2002, 212, 31–47. [Google Scholar] [CrossRef]
- Leal, J.F.; Santos, E.B.H.; Esteves, V.I. Oxytetracycline in intensive aquaculture: Water quality during and after its administration, environmental fate, toxicity and bacterial resistance. Rev. Aquac. 2019, 11, 1176–1194. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, L.; Zhou, J.L.; Zhao, H.; Ying, G.G.; Liu, Y.S. A risk-based approach for managing aquaculture-used oxytetracycline and its ecological risks in China. Front. Pharmacol. 2021, 12, 803499. [Google Scholar] [CrossRef]
- Paranjpye, R.N.; Hamel, O.S.; Stojanovski, A.; Liermann, M. Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 2012, 78, 8631–8638. [Google Scholar] [CrossRef]
- Cui, Y.; Yang, X.; Wang, H.; Sun, Q.; Wang, R.; Yuan, S.; Wang, L. Genomic diversity and phylogenetic relationships of Vibrio parahaemolyticus isolates from global sources. Front. Microbiol. 2015, 6, 1251. [Google Scholar] [CrossRef]
- Letchumanan, V.; Chan, K.G.; Lee, L.H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2015, 6, 703. [Google Scholar] [CrossRef]
- Xu, F.; Ilyas, S.; Hall, J.A.; Jones, S.H.; Cooper, V.S.; Whistler, C.A. Genetic characterization of clinical and environmental Vibrio parahaemolyticus from the Northeast USA reveals emerging resident and non-indigenous pathogen lineages. Front. Microbiol. 2015, 6, 272. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Urtaza, J.; Bowers, J.C.; Trinanes, J.; DePaola, A. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res. Int. 2010, 43, 1780–1790. [Google Scholar] [CrossRef]
- Vezzulli, L.; Grande, C.; Reid, P.C.; Hélaouët, P.; Edwards, M.; Höfle, M.G.; Brettar, I.; Colwell, R.R.; Pruzzo, C. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl Acad. Sci. USA 2016, 113, E5062–E5071. [Google Scholar] [CrossRef]
- Letchumanan, V.; Chan, K.G.; Lee, L.H. Vibrio parahaemolyticus: A review on antimicrobial resistance and virulence mechanisms. FEMS Microbiol. Lett. 2015, 362, fnv067. [Google Scholar] [CrossRef]
- Parveen, S.; Hettiarachchi, K.A.; Wong, E.; Koonse, R.; Sheth, A. Microbial risk assessment and whole genome sequencing in seafood safety. Front. Microbiol. 2020, 11, 1815. [Google Scholar] [CrossRef]
- FDA (US Food and Drug Administration). Fish and Fishery Products Hazards and Controls Guidance; United States Department of Health and Human Services: Washington, DC, USA, 2021. Available online: https://www.fda.gov/food/seafood-guidance-documents-regulatory-information/fish-and-fishery-products-hazards-and-controls (accessed on 28 June 2025).
- FDA (US Food and Drug Administration). Selecting and Serving Fresh and Frozen Seafood Safely; United States Department of Health and Human Services: Washington, DC, USA, 2023. Available online: https://www.fda.gov/food/buy-store-serve-safe-food/selecting-and-serving-fresh-and-frozen-seafood-safely (accessed on 28 June 2025).
| Region | |||||
|---|---|---|---|---|---|
| Year | Jeonla | Gangwon | Chungcheong | Seoul–Gyeonggi | Gyeongsang |
| 2021 | 40 | 6 | 63 | 71 | 70 |
| 2022 | 40 | 10 | 50 | 80 | 70 |
| Total | 80 | 16 | 113 | 151 | 140 |
| Antimicrobial Subclass | Antimicrobial Agent (Abbreviation) | Range Tested | Breakpoint |
|---|---|---|---|
| Aminoglycosides | Gentamicin (GEN) | 1–64 | ≥16 (1) |
| Streptomycin (STR) | 16–128 | ND * | |
| Aminopenicillin | Ampicillin (AMP) | 2–64 | ≥32 (1) |
| β-lactam/β-lactamase inhibitor combinations | Amoxicillin/clavulanic acid (AMC) | 2/1–32/16 | ≥32/16 (1) |
| Cephamycin | Cefoxitin (FOX) | 1–32 | ≥32 (1) |
| Cephalosporin III | Cefotaxime (CTX) | 0.5–8 | ≥4 (1) |
| Ceftazidime (CAZ) | 1–16 | ≥16 (1) | |
| Cephalosporin IV | Cefepime (FEP) | 0.25–16 | ≥16 (1) |
| Carbapenem | Meropenem (MEM) | 0.25–4 | ≥4 (1) |
| Fluoroquinolone | Ciprofloxacin (CIP) | 0.12–16 | ≥4 (1) |
| Folate pathway inhibitors | Trimethoprim/Sulfamethoxazole (SXT) | 0.12/2.38–4/76 | ≥4/76 (1) |
| Sulfisoxazole (FIS) | 16–256 | ≥512 (1) | |
| Phenicols | Chloramphenicol (CHL) | 2–64 | ≥32 (1) |
| Polymyxins | Colistin (COL) | 2–16 | ND |
| Quinolone | Nalidixic acid (NAL) | 2–128 | ND |
| Tetracyclines | Tetracycline (TET) | 2–128 | ≥16 (1) |
| Target Gene | Primer | Sequence (5′→3′) | Target Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| tlh | tl-F | AAAGCGGATTATGCAGAAGCACTG | 405 | [16] |
| tl-R | GCTACTTTCTAGCATTTTCTCTGC | |||
| tdh | tdh-F | GTAAAGGTCTCTGACTTTTGGAC | 259 | [16] |
| tdh-R | TGGAATAGAACCTTCATCTTCACC | |||
| trh | trh-F | TTGGCTTCGATATTTTCAGTATCT | 500 | [16] |
| rth-R | CATAACAAACATATGCCCATTTCCG | |||
| T3SS1 | vscN1-F | GGGGCTGTGGTGCCGGGCGTA | 1325 | [17] |
| vscN1-R | GGGGCGATGCCTTTCAGTTGAGC | |||
| T3SS2α | vscN2-F | AAACGTACTCACCGACTCGAATG | 1120 | [17] |
| vscN2-R | TGAAATCGTTAAGGTGACAGGC | |||
| T3SS2β | vcrD2-F | GGTAACACTGCCTGGTGTGGTCATCG | 1594 | [19] |
| vcrD2-R | GTCTCTCAAAGTCTTCAAACTCACCTGC | |||
| T6SS1 | icmF1-F | AGTACCGCCTGCCAATAAGACAAC | 411 | [20] |
| icmF1-R | GACGCATCGGCAAACTCAACAG | |||
| T6SS2 | icmF2-F | AATGGATTGGGACTAGGGAGGTTG | 452 | [20] |
| icmF2-R | TACGCGTTATTTGCTGCTTGAGA |
| Year | Origin | Category | No. of Samples | No. of Isolates (%) |
|---|---|---|---|---|
| 2021 | Domestic | Fish | 250 | 8(3.2) |
| 2022 | Domestic | Fish | 250 | 9(3.6) |
| Total | 500 | 3.4% | ||
| Antimicrobial Subclass | Antimicrobial Agent (Abbreviation) | No. of Isolates (%) |
|---|---|---|
| Resistant | ||
| Aminoglycosides | Gentamicin (GEN) | 0 (0) |
| Streptomycin (STR) | ND * | |
| Aminopenicillin | Ampicillin (AMP) | 10 (58.8) |
| β-lactam/β-lactamase inhibitor combinations | Amoxicillin/clavulanic acid (AMC) | 0 (0) |
| Cephamycin | Cefoxitin (FOX) | 0 (0) |
| Cephalosporin III | Cefotaxime (CTX) | 0 (0) |
| Ceftazidime (CAZ) | 0 (0) | |
| Cephalosporin IV | Cefepime (FEP) | 0 (0) |
| Carbapenem | Meropenem (MEM) | 0 (0) |
| Fluoroquinolone | Ciprofloxacin (CIP) | 0 (0) |
| Folate pathway inhibitors | Trimethoprim/Sulfamethoxazole (SXT) | 0 (0) |
| Sulfisoxazole (FIS) | 0 (0) | |
| Phenicols | Chloramphenicol (CHL) | 0 (0) |
| Polymyxins | Colistin (COL) | ND * |
| Quinolone | Nalidixic acid (NAL) | ND * |
| Tetracyclines | Tetracycline (TET) | 0 (0) |
| Sample | tlh | tdh | trh | T3SS1 | T3SS2α | T3SS2β | T6SS1 | T6SS2 |
|---|---|---|---|---|---|---|---|---|
| 21_VP_1530 | + | − | + | + | − | − | + | + |
| 21_VP_1531 | + | − | + | + | − | − | − | + |
| 21_VP_1533 | + | − | + | + | − | − | + | + |
| 21_VP_1536 | + | − | + | + | − | − | + | + |
| 21_VP_1542 | + | − | + | + | − | − | − | + |
| 21_VP_1544 | + | − | + | + | − | − | − | + |
| 21_VP_1545 | + | − | + | + | − | − | + | + |
| 21_VP_1774 | + | − | + | + | − | − | + | + |
| 22_VP_1570 | + | − | + | + | − | − | − | + |
| 22_VP_1572 | + | − | + | + | − | − | + | + |
| 22_VP_1574 | + | − | + | + | − | − | − | + |
| 22_VP_1576 | + | − | − | + | − | − | − | + |
| 22_VP_1577 | + | − | + | + | − | − | + | + |
| 22_VP_1578 | + | − | − | + | − | − | − | + |
| 22_VP_1579 | + | − | − | + | − | − | + | + |
| 22_VP_1580 | + | − | − | + | − | − | + | + |
| 22_VP_1581 | + | − | − | + | − | − | − | + |
| Total | 17/17(100) | 0/17(0) | 12/17(70.6) | 17/17(100) | 0/17(0) | 0/17(0) | 9/17(52.9) | 17/17(100) |
| Sample ID | Genetic Background | Antimicrobial | Class | WGS-Predicted Phenotype | ST | Nearest STs |
|---|---|---|---|---|---|---|
| 21_VP_1533 | blaCARB-40 | AMX, AMP, PIP | Beta-lactam | Resistant | 114 | |
| 21_VP_1536 | blaCARB-21 | AMX, AMP, PIP | Beta-lactam | Resistant | Unknown | 2902, 1989, 114, 2170 |
| 21_VP_1544 | blaCARB-26 | AMX, AMP, PIP | Beta-lactam | Resistant | 2447 | |
| 21_VP_1545 | blaCARB-21 | AMX, AMP, PIP | Beta-lactam | Resistant | 114 | |
| 22_VP_1570 | blaCARB-48 | AMX, AMP, PIP | Beta-lactam | Resistant | Unknown | 3085, 281 |
| 22_VP_1577 | blaCARB-45 | AMX, AMP, PIP | Beta-lactam | Resistant | 917 | |
| 22_VP_1578 | blaCARB-46 | AMX, AMP, PIP | Beta-lactam | Resistant | 1256 | |
| 22_VP_1579 | blaCARB-33 | AMX, AMP, PIP | Beta-lactam | Resistant | Unknown | 992, 396, 3223 |
| 22_VP_1580 | blaCARB-33 | AMX, AMP, PIP | Beta-lactam | Resistant | 1823 | |
| 22_VP_1581 | blaCARB-33 | AMX, AMP, PIP | Beta-lactam | Resistant | Unknown | 2590, 2621, 2125, 1956, 358 |
| VF Class | Virulence Factor | Related Genes |
|---|---|---|
| Adherence | Mannose-sensitive hemagglutinin (MSHA type IV pilus) | mshA, mshE, mshF, mph, mshH, mshI, mshJ, mshK, mshL, mshM, mshN |
| Type IV pilus | pilB, pilC, pilD | |
| Antiphagocytosis | Capsular polysaccharide | cpsA, cpsB, cpsD, cpsE, cpsF, cpsG, cpsH, cpsI, cpsJ, wbfV/wcvB |
| Chemotaxis and motility | Flagella | cheA, cheB, cheR, cheV, cheW, cheY, cheZ, filM, flaA, flaB, flaI, flgD, flgE, flgF, flgG, flgH, flgI, flgJ, flgK, flgL, flhA, flhB, flhF, flhG, fliA, fliD, fliE, fliF, fliG, fliH, fliI, fliK, fliL, fliN, fliO, fliP, fliR, fliS, flrA, flrB, flrC, motA, motB, motX, motY |
| Iron uptake | Enterobactin receptors | irgA, vctA |
| Heme receptors | hutA, hutR | |
| Periplasmic binding protein-dependent ABC transport systems | vctC, vctG, vctP | |
| Quorum sensing | Cholerae autoinducer-1 | cqsA |
| Secretion system | EPS type II secretion system | epsC, epsE, epsF, epsG, epsH, epsJ, epsK, epsL, epsM, epsN, gspD |
| T3SS1 secreted effectors | vopQ, vopR, vopS | |
| T3SS1 | sycN, tyeA, vcrD, vcrG, vcrV, virF, vopB, vopD, vopN, vscA, vscC, vscD, vscG, vscH, vscI, vscJ, vscK, vscL, vscN, vscO, vscP, vscQ, vscR, vscT, vscU, vxsC | |
| Toxin | Thermolabile hemolysin | tlh |
| VF Class | Virulence Factor | Related Genes | 2021 | 2022 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1533 | VP1536 | VP1544 | VP1545 | VP1570 | VP1577 | VP1578 | VP1579 | VP1580 | VP1581 | |||
| Adherence | Mannose-sensitive hemagglutinin (MSHA type IV pilus) | mshC | ||||||||||
| mshD | ||||||||||||
| Type IV pilus | pilA | |||||||||||
| Tight adherence locus (Haemophilus) | tadA | |||||||||||
| Antiphagocytosis | Capsular polysaccharide | cpsC | ||||||||||
| rmlA | ||||||||||||
| rmlB | ||||||||||||
| rmlC | ||||||||||||
| wbfT | ||||||||||||
| wbfU | ||||||||||||
| wbfV/wcvB | ||||||||||||
| wbfY | ||||||||||||
| wbjD/wecB | ||||||||||||
| wecA | ||||||||||||
| wecC | ||||||||||||
| wza | ||||||||||||
| wzb | ||||||||||||
| wzc | ||||||||||||
| Chemotaxis and motility | Flagella | flaD | ||||||||||
| flaG | ||||||||||||
| flgA | ||||||||||||
| flgB | ||||||||||||
| flgC | ||||||||||||
| flgM | ||||||||||||
| flgN | ||||||||||||
| fliJ | ||||||||||||
| fliQ | ||||||||||||
| Iron uptake | Periplasmic binding protein-dependent ABC transport systems | vctD | ||||||||||
| Quorum sensing | Autoinducer-2 | luxS | ||||||||||
| Secretion system | EPS type II secretion system | epsI | ||||||||||
| epsM | ||||||||||||
| T3SS1 | vcrH | |||||||||||
| vcrR | ||||||||||||
| virG | ||||||||||||
| vscB | ||||||||||||
| vscF | ||||||||||||
| vscS | ||||||||||||
| vscX | ||||||||||||
| vscY | ||||||||||||
| VAS effector proteins | hcp-2 | |||||||||||
| vgrG-2 | ||||||||||||
| vgrG-3 | ||||||||||||
| VAS type VI secretion system | vasA | |||||||||||
| vasB | ||||||||||||
| vasD | ||||||||||||
| vasE | ||||||||||||
| vasG | ||||||||||||
| vasH | ||||||||||||
| vasJ | ||||||||||||
| vasK | ||||||||||||
| TTSS (SPI-1 encode) | invF | |||||||||||
| Toxin | Phytotoxin phaseolotoxin | cysC1 | ||||||||||
| Colonization and Immune evasion | Capsule biosynthesis and transport | kpsF | ||||||||||
| Endotoxin | Lipooligosaccharide | lgtF | ||||||||||
| Immune evasion | Lipopolysaccharide | acpXL | ||||||||||
| Others | O-antigen | fcl | ||||||||||
| manB | ||||||||||||
| cpsB | ||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Kim, H.; Kang, H.; Park, Y.; Joo, I.; Kim, H. Antibiotic Resistance and Characteristics of Vibrio parahaemolyticus Isolated from Seafood Distributed in South Korea from 2021 to 2022. Microorganisms 2025, 13, 1566. https://doi.org/10.3390/microorganisms13071566
Lee J, Kim H, Kang H, Park Y, Joo I, Kim H. Antibiotic Resistance and Characteristics of Vibrio parahaemolyticus Isolated from Seafood Distributed in South Korea from 2021 to 2022. Microorganisms. 2025; 13(7):1566. https://doi.org/10.3390/microorganisms13071566
Chicago/Turabian StyleLee, Jonghoon, Hansol Kim, Haiseong Kang, Yongchjun Park, Insun Joo, and Hyochin Kim. 2025. "Antibiotic Resistance and Characteristics of Vibrio parahaemolyticus Isolated from Seafood Distributed in South Korea from 2021 to 2022" Microorganisms 13, no. 7: 1566. https://doi.org/10.3390/microorganisms13071566
APA StyleLee, J., Kim, H., Kang, H., Park, Y., Joo, I., & Kim, H. (2025). Antibiotic Resistance and Characteristics of Vibrio parahaemolyticus Isolated from Seafood Distributed in South Korea from 2021 to 2022. Microorganisms, 13(7), 1566. https://doi.org/10.3390/microorganisms13071566






