Microbial Interconnections in One Health: A Critical Nexus Between Companion Animals and Human Microbiomes
Abstract
1. Introduction
1.1. One Health: A Concept with Deep Roots in History
1.2. So, What Is New About the One Health Concept?
1.3. The Adventures of a Definition
1.4. Humans and Companion Animals: A Long Story Made Short
1.5. One Health: Humans and Canines–Felines
2. Microbiotas, Microbiomes and a Paradigm Shift
3. Pets, Owners and Their Microbiomes
3.1. Skin
3.2. Urinary
3.3. Placental Microbiomes
| Anatomical Site | Humans | Dogs | Cats |
|---|---|---|---|
| Male Urogenital System | Dominant genera: Prevotella, Finegoldia, Peptoniphilus, Staphylococcus, Corynebacterium, Anaerococcus [52,53,54] | No studies available. | No studies available. |
| Urine | Nine phyla: Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Fusobacteria, Proteobacteria, Synergistetes, Tenericutes [55] | Actinobacteria, Bacteroidota, Proteobacteria, Firmicutes [56]. Dominant: Proteobacteria (Pseudomonas, Sphingobium, Acinetobacter johnsonii) [57]. Species: Corynebacterium auriscanis, Streptococcus parasanguinis [70]. Some found urine sterile [58] | No studies reported for cats in urine microbiome. |
| Placental Microbiomes | Dominant phyla: Firmicutes, Proteobacteria, Bacteroidetes, Actinobacterium [71]. Debate exists about sterile womb [63,67] | Bacillus spp., Pseudomonas spp., Staphylococcus spp., Micrococcus spp., Acinetobacter spp. in placenta, amniotic fluid, meconium [68,69] | Staphylococcus epidermidis, Pseudomonas aeruginosa on placenta–uterus surface, P. aeruginosa in amniotic fluid, Psychrobacter sanguinis in meconium [68] |
| Vaginal and Uterine Microbiomes | Vagina: Dominated by Lactobacilli; influenced by ethnicity, diet, stress [72,73]. Uterus: Lactobacillus, Pseudomonas, Acinetobacter, Vagococcus [74,75,76,77,78] | Vagina: Highly diverse (300+ OTUs). Dominant phyla: Bacteroidetes, Proteobacteria, Tenericutes, Firmicutes [79]. Common: Echerichia coli, beta-hemolytic Streptococci, Staphylococci, Pasteurella [80]. Uterus: Pseudomonas, Staphylococcus, Corynebacterium [58] | Dominant genera: Escherichia-Shigella, Streptococcus, Pasteurella, Bacteroides, Staphylococcus [68]. Common species: hemolytic E. coli, S. canis, Streptococcus felis, Enterococcus spp. [68]. Stage of estrous cycle, age, and body condition score did not affect diversity; domestic vs. feral environment did [68] |
3.4. Vaginal and Uterine Microbiomes
3.5. Respiratory Microbiome
3.6. Ocular Microbiome
3.7. Oral Microbiome
3.8. The Gastrointestinal Tract Microbiome
3.8.1. Esophagus
3.8.2. Stomach
3.8.3. The Gut
- -
- Gut Microbiome Toxicity in the Context of One Health
4. Can We Support the One Health Approach in Terms of Microbiomes?
- -
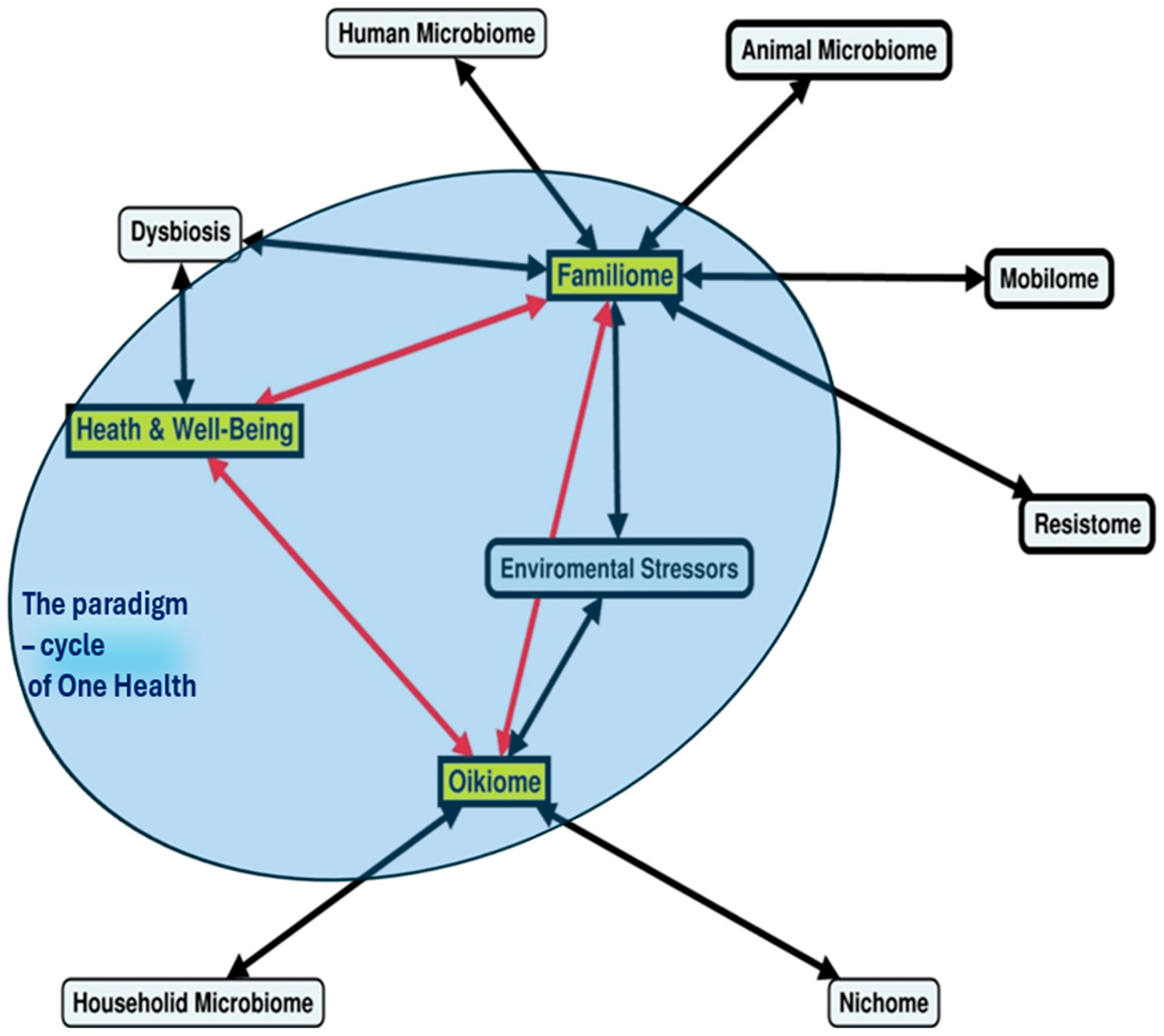
- The familiome and the oikiome
5. The Transition from the Microbiome to the Resistome
6. Summary and Outlooks—One Health Approach for Pets and Humans
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jouanna, J. Water, Health and Disease in the Hippocratic Treatise Airs, Waters, Places. In Greek Medicine from Hippocrates to Galen; Brill: Leiden, The Netherlands, 2012. [Google Scholar] [CrossRef]
- Capua, I.; Cattoli, G. One Health (r)Evolution: Learning from the Past to Build a New Future. Viruses 2018, 10, 725. [Google Scholar] [CrossRef] [PubMed]
- Gyles, C. One Medicine, One Health, One World. Can Vet J. 2016, 57, 345–346. [Google Scholar] [PubMed] [PubMed Central]
- Schultz, M. Rudolf Virchow. Emerg. Infect. Dis. 2008, 14, 1480–1481. [Google Scholar] [CrossRef][Green Version]
- Brown, H.L.; Pursley, I.G.; Horton, D.L.; La Ragione, R.M. One health: A structured review and commentary on trends and themes. One Health Outlook 2024, 6, 17. [Google Scholar] [CrossRef]
- Karesh, W.B.; Cook, A.R. One world—One health. Clin. Med. 2009, 9, 259–260. [Google Scholar] [CrossRef]
- Pitt, S.J.; Gunn, A. The One Health Concept. Br. J. Biomed. Sci. 2024, 81, 12366. [Google Scholar] [CrossRef]
- Olympio, K.P.K.; Salles, F.J.; Ferreira, A.P.S.d.S.; Pereira, E.C.; de Oliveira, A.S.; Leroux, I.N.; Vieira, F.B.A. The human exposome unraveling the impact of environment on health: Promise or reality? Rev. De Saude Publica 2019, 53, 6. [Google Scholar] [CrossRef]
- Available online: https://www.cdc.gov/one-health/about/index.html (accessed on 10 April 2025).
- Available online: https://www.avma.org/KB/Resources/Reports/Documents/onehealth_final.pdf (accessed on 10 April 2025).
- Mackenzie, J.S.; Jeggo, M. The One Health Approach—Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://ohi.vetmed.ucdavis.edu/ (accessed on 1 April 2025).
- Available online: https://www.fao.org/one-health/overview/one-health-overview/en (accessed on 10 April 2025).
- Perri, A.R.; Feuerborn, T.R.; Frantz, L.A.F.; Larson, G.; Malhi, R.S.; Meltzer, D.J.; Witt, K.E. Dog domestication and the dual dispersal of people and dogs into the Americas. Proc. Natl. Acad. Sci. USA 2021, 118, e2010083118. [Google Scholar] [CrossRef]
- Xuan, J. The origin and evolution of cats. Anim. Mol. Breed. 2023, 13, 1–6. [Google Scholar] [CrossRef]
- Driscoll, C.A.; Macdonald, D.W.; O’BRien, S.J. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl. Acad. Sci. USA 2009, 106, 9971–9978. [Google Scholar] [CrossRef]
- Available online: https://europeanpetfood.org/wp-content/uploads/2023/07/FEDIAF_Annual-Report_2023.pdf (accessed on 1 April 2025).
- Available online: https://www.statista.com/statistics/515475/dog-ownership-european-union-eu-by-country/ (accessed on 10 April 2025).
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Ana, M.; Meghan, D.; Amanda, T.; Brendan, H.; Pam, T.; Baofeng, H.; Irving, N.; Ebbing, L.; Daniel, M.; Elizabeth, G. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 2015, 3, 2. [Google Scholar] [CrossRef]
- Oh, C.; Lee, K.; Cheong, Y.; Lee, S.-W.; Park, S.-Y.; Song, C.-S.; Choi, I.-S.; Lee, J.-B.; White, B.A. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PLoS ONE 2015, 10, e0131468. [Google Scholar] [CrossRef] [PubMed]
- BSc, B.M.J.D. One Health: The small animal dimension. Veter. Rec. 2010, 167, 847–849. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Zoonotic Diseases. 2023. Available online: https://www.cdc.gov/one-health/about/about-zoonotic-diseases.html (accessed on 1 April 2025).
- Tannock, G.W. The normal microflora: An introduction. In Medical Importance of the Normal Microflora; Tannock, G.W., Ed.; Springer: Boston, MA, USA, 1999. [Google Scholar] [CrossRef]
- Deng, P.; Swanson, K.S. Gut microbiota of humans, dogs and cats: Current knowledge and future opportunities and challenges. Br. J. Nutr. 2014, 113, S6–S17. [Google Scholar] [CrossRef]
- Grześkowiak, Ł.; Endo, A.; Beasley, S.; Salminen, S. Microbiota and probiotics in canine and feline welfare. Anaerobe 2015, 34, 14–23. [Google Scholar] [CrossRef]
- Dogra, S.K.; Martin, F.-P.; Donnicola, D.; Julita, M.; Berger, B.; Sprenger, N. Human Milk Oligosaccharide-Stimulated Bifidobacterium Species Contribute to Prevent Later Respiratory Tract Infections. Microorganisms 2021, 9, 1939. [Google Scholar] [CrossRef]
- Hernandez, J.; Rhimi, S.; Kriaa, A.; Mariaule, V.; Boudaya, H.; Drut, A.; Jablaoui, A.; Mkaouar, H.; Saidi, A.; Biourge, V.; et al. Domestic Environment and Gut Microbiota: Lessons from Pet Dogs. Microorganisms 2022, 10, 949. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.; Suchodolski, J. Understanding the canine intestinal microbiota and its modification by pro-, pre- and synbiotics—What is the evidence? Vet Med Sci. 2016, 2, 71–94. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Hoffmann, A.R.; Proctor, L.M.; Surette, M.G.; Suchodolski, J.S. The Microbiome: The Trillions of Microorganisms That Maintain Health and Cause Disease in Humans and Companion Animals. Veter. Pathol. 2015, 53, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Abdolghanizadeh, S.; Salmeh, E.; Mirzakhani, F.; Soroush, E.; Siadat, S.D.; Tarashi, S. Microbiota insights into pet ownership and human health. Res. Veter. Sci. 2024, 171, 105220. [Google Scholar] [CrossRef]
- McBurney, M.I.; Davis, C.; Fraser, C.M.; Schneeman, B.O.; Huttenhower, C.; Verbeke, K.; Walter, J.; Latulippe, M.E. Establishing What Constitutes a Healthy Human Gut Microbiome: State of the Science, Regulatory Considerations, and Future Directions. J. Nutr. 2019, 149, 1882–1895. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Khan, I.; Yasir, M.; Azhar, E.I.; Kumosani, T.; Barbour, E.K.; Bibi, F.; Kamal, M.A. Implication of Gut Microbiota in Human Health. CNS Neurol. Disord.—Drug Targets 2014, 13, 1325–1333. [Google Scholar] [CrossRef]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Mitev, K.; Taleski, V. Association between the Gut Microbiota and Obesity. Open Access Maced J. Med. Sci. 2019, 7, 2050–2056. Available online: https://oamjms.eu/index.php/mjms/article/view/oamjms.2019.586 (accessed on 10 April 2025). [CrossRef]
- Yamasaki, Y.; Nomura, R.; Nakano, K.; Naka, S.; Matsumoto-Nakano, M.; Asai, F.; Ooshima, T. Distribution of periodontopathic bacterial species in dogs and their owners. Arch. Oral Biol. 2012, 57, 1183–1188. [Google Scholar] [CrossRef]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef]
- Reynoso-García, J.; Miranda-Santiago, A.E.; Meléndez-Vázquez, N.M.; Acosta-Pagán, K.; Sánchez-Rosado, M.; Díaz-Rivera, J.; Rosado-Quiñones, A.M.; Acevedo-Márquez, L.; Cruz-Roldán, L.; Tosado-Rodríguez, E.L.; et al. A complete guide to human microbiomes: Body niches, transmission, development, dysbiosis, and restoration. Front. Syst. Biol. 2022, 2, 951403. [Google Scholar] [CrossRef]
- Boost, M.V.; O’DOnoghue, M.M.; James, A. Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol. Infect. 2007, 136, 953–964. [Google Scholar] [CrossRef]
- Bierowiec, K.; Płoneczka-Janeczko, K.; Rypuła, K.; de Lencastre, H. Is the Colonisation of Staphylococcus aureus in Pets Associated with Their Close Contact with Owners? PLoS ONE 2016, 11, e0156052. [Google Scholar] [CrossRef]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human Skin Microbiome: Impact of Intrinsic and Extrinsic Factors on Skin Microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Renaud, G.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Wolfsberg, T.G.; Turner, M.L.; Segre, J.A. A diversity profile of the human skin microbiota. Genome Res. 2008, 18, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Cundell, A.M. Microbial Ecology of the Human Skin. Microb. Ecol. 2018, 76, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Murillo, N.; Raoult, D. Skin Microbiota: Overview and Role in the Skin Diseases Acne Vulgaris and Rosacea. Futur. Microbiol. 2013, 8, 209–222. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Buerger, S. The skin and oral microbiome: An examination of overlap and potential interactions between microbiome communities. Ski. Microbiome Handb. Basic Res. Prod. Dev. 2020, 45–57. [Google Scholar] [CrossRef]
- Naz, G.; Rasheed, M.; Sarwar, A.; Mehmood, S.; Farooq, W.; Imran, U.; Uroos, A.; Urwa, J. A one-health approach to combat common pet-associated fungal zoonosis. In Zoonosis; Altaf, S., Khan, A., Abbas, R.Z., Eds.; Unique Scientific Publishers: Faisalabad, Pakistan, 2023. [Google Scholar] [CrossRef]
- Pereira, A.M.; Clemente, A. Dogs’ Microbiome From Tip to Toe. Top. Companion Anim. Med. 2021, 45, 100584. [Google Scholar] [CrossRef]
- Older, C.E.; Diesel, A.B.; Lawhon, S.D.; Queiroz, C.R.R.; Henker, L.C.; Hoffmann, A.R.; Dawson, T.L. The feline cutaneous and oral microbiota are influenced by breed and environment. PLoS ONE 2019, 14, e0220463. [Google Scholar] [CrossRef]
- Nelson, D.E.; Dong, Q.; Van Der Pol, B.; Toh, E.; Fan, B.; Katz, B.P.; Mi, D.; Rong, R.; Weinstock, G.M.; Sodergren, E.; et al. Bacterial Communities of the Coronal Sulcus and Distal Urethra of Adolescent Males. PLoS ONE 2012, 7, e36298. [Google Scholar] [CrossRef]
- Liu, C.M.; Hungate, B.A.; Tobian, A.A.R.; Serwadda, D.; Ravel, J.; Lester, R.; Kigozi, G.; Aziz, M.; Galiwango, R.M.; Nalugoda, F.; et al. Male Circumcision Significantly Reduces Prevalence and Load of Genital Anaerobic Bacteria. mBio 2013, 4, e00076-13. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Fernandes, Â.R.; Rodrigues, A.G.; Lisboa, C. Microbiome in Male Genital Mucosa (Prepuce, Glans, and Coronal Sulcus): A Systematic Review. Microorganisms 2022, 10, 2312. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.A.; Brown, R.; Williams, J.; White, P.; Jacobson, S.K.; Marchesi, J.R.; Drake, M.J. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front. Cell Infect. Microbiol. 2013, 3, 41. [Google Scholar] [CrossRef]
- Coffey, E.L.; Gomez, A.M.; Ericsson, A.C.; Burton, E.N.; Granick, J.L.; Lulich, J.P.; Furrow, E. The impact of urine collection method on canine urinary microbiota detection: A cross-sectional study. BMC Microbiol. 2023, 23, 101. [Google Scholar] [CrossRef] [PubMed]
- Burton, E.N.; Cohn, L.A.; Reinero, C.N.; Rindt, H.; Moore, S.G.; Ericsson, A.C.; Dong, Q. Characterization of the urinary microbiome in healthy dogs. PLoS ONE 2017, 12, e0177783. [Google Scholar] [CrossRef] [PubMed]
- Gronsfeld, V.; Brutinel, F.; Egyptien, S.; Porsmoguer, C.; Hamaide, A.; Taminiau, B.; Daube, G.; Van de Weerdt, M.-L.; Deleuze, S.; Noel, S. Evaluation of the vaginal and urinary microbiota of healthy cycling bitches. BMC Veter. Res. 2024, 20, 315. [Google Scholar] [CrossRef]
- Doyle, R.M.; Harris, K.; Kamiza, S.; Harjunmaa, U.; Ashorn, U.; Nkhoma, M.; Dewey, K.G.; Maleta, K.; Ashorn, P.; Klein, N.; et al. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS ONE 2017, 12, e0180167. [Google Scholar] [CrossRef]
- La, X.; Wang, Y.; Xiong, X.; Shen, L.; Chen, W.; Zhang, L.; Yang, F.; Cai, X.; Zheng, H.; Jiang, H. The Composition of Placental Microbiota and Its Association With Adverse Pregnancy Outcomes. Front. Microbiol. 2022, 13, 911852. [Google Scholar] [CrossRef]
- Cohen, J.M.; Hernández-Díaz, S.M.; Bateman, B.T.M.; Park, Y.M.; Desai, R.J.; Gray, K.J.; Patorno, E.M.; Mogun, H.; Huybrechts, K.F. Placental Complications Associated With Psychostimulant Use in Pregnancy. Obstet. Gynecol. 2017, 130, 1192–1201. [Google Scholar] [CrossRef]
- Gomez de Agüero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Perez-Muñoz, M.E.; Arrieta, M.-C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef]
- Lauder, A.P.; Roche, A.M.; Sherrill-Mix, S.; Bailey, A.; Laughlin, A.L.; Bittinger, K.; Leite, R.; Elovitz, M.A.; Parry, S.; Bushman, F.D. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Parnell, L.A.; Briggs, C.M.; Cao, B.; Delannoy-Bruno, O.; Schrieffer, A.E.; Mysorekar, I.U. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci. Rep. 2017, 7, 11200. [Google Scholar] [CrossRef]
- Sterpu, I.; Fransson, E.; Hugerth, L.W.; Du, J.; Pereira, M.; Cheng, L.; Radu, S.A.; Calderón-Pérez, L.; Zha, Y.; Angelidou, P.; et al. No evidence for a placental microbiome in human pregnancies at term. Am. J. Obstet. Gynecol. 2021, 224, 296.e1–296.e23. [Google Scholar] [CrossRef] [PubMed]
- Panzer, J.J.; Romero, R.; Greenberg, J.M.; Winters, A.D.; Galaz, J.; Gomez-Lopez, N.; Theis, K.R. Is there a placental microbiota? A critical review and re-analysis of published placental microbiota datasets. BMC Microbiol. 2023, 23, 76. [Google Scholar] [CrossRef]
- Banchi, P.; Colitti, B.; Del Carro, A.; Corrò, M.; Bertero, A.; Ala, U.; Del Carro, A.; Van Soom, A.; Bertolotti, L.; Rota, A. Challenging the Hypothesis of in Utero Microbiota Acquisition in Healthy Canine and Feline Pregnancies at Term: Preliminary Data. Veter. Sci. 2023, 10, 331. [Google Scholar] [CrossRef]
- Rota, A.; Del Carro, A.; Bertero, A.; Del Carro, A.; Starvaggi Cucuzza, A.; Banchi, P.; Corrò, M. Does bacteria colonization of canine newborns start in the uterus? Animals 2021, 11, 1415. [Google Scholar] [CrossRef]
- Melgarejo, T.; Sharp, N.; Krumbeck, J.A.; Wu, G.; Kim, Y.J.; Linde, A. The Urinary Resistome of Clinically Healthy Companion Dogs: Potential One Health Implications. Antibiotics 2022, 11, 780. [Google Scholar] [CrossRef]
- Zakis, D.R.; Paulissen, E.; Kornete, L.; Kaan, A.; Nicu, E.A.; Zaura, E. The evidence for placental microbiome and its composition in healthy pregnancies: A systematic review. J. Reprod. Immunol. 2022, 149, 103455. [Google Scholar] [CrossRef]
- Smith, S.B.; Ravel, J. The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 2016, 595, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Holdcroft, A.M.; Ireland, D.J.; Payne, M.S. The Vaginal Microbiome in Health and Disease—What Role Do Common Intimate Hygiene Practices Play? Microorganisms 2023, 11, 298. [Google Scholar] [CrossRef]
- Chen, C.; Song, X.; Wei, W.; Zhong, H.; Dai, J.; Lan, Z.; Li, F.; Yu, X.; Feng, Q.; Wang, Z.; et al. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat. Commun. 2017, 8, 875. [Google Scholar] [CrossRef]
- Mitchell, C.M.; Haick, A.; Nkwopara, E.; Garcia, R.; Rendi, M.; Agnew, K.; Fredricks, D.N.; Eschenbach, D. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am. J. Obstet. Gynecol. 2015, 212, 611.e1–611.e9. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.M.; Chase, D.M.; Herbst-Kralovetz, M.M. Uterine Microbiota: Residents, Tourists, or Invaders? Front. Immunol. 2018, 9, 208. [Google Scholar] [CrossRef]
- Medina-Bastidas, D.; Camacho-Arroyo, I.; García-Gómez, E. Current findings in endometrial microbiome: Impact on uterine diseases. Reproduction 2022, 163, R81–R96. [Google Scholar] [CrossRef]
- Moosa, Y.; Kwon, D.; de Oliveira, T.; Wong, E.B. Determinants of Vaginal Microbiota Composition. Front. Cell Infect. Microbiol. 2020, 10, 467. [Google Scholar] [CrossRef]
- Lyman, C.C.; Holyoak, G.R.; Meinkoth, K.; Wieneke, X.; Chillemi, K.A.; DeSilva, U.; Wade, C. Canine endometrial and vaginal microbiomes reveal distinct and complex ecosystems. PLoS ONE 2019, 14, e0210157. [Google Scholar] [CrossRef] [PubMed]
- Leps, A.S.; Klein, B.; Schneider, M.; Meyer, C.; Šoba, A.; Simon, C.; Dyachenko, V.; Siesenop, U.; Verspohl, J.; Goericke-Pesch, S. The Canine Vaginal Flora: A Large-Cohort Retrospective Study. Veter. Sci. 2024, 11, 55. [Google Scholar] [CrossRef]
- Miller, E.A.; Beasley, D.E.; Dunn, R.R.; Archie, E.A. Lactobacilli Dominance and Vaginal pH: Why Is the Human Vaginal Microbiome Unique? Front. Microbiol. 2016, 7, 1936. [Google Scholar] [CrossRef]
- Stokholm, J.; Schjørring, S.; Pedersen, L.; Bischoff, A.L.; Følsgaard, N.; Carson, C.G.; Chawes, B.; Bønnelykke, K.; Mølgaard, A.; Krogfelt, K.A.; et al. Living with Cat and Dog Increases Vaginal Colonization with E. coli in Pregnant Women. PLoS ONE 2012, 7, e46226. [Google Scholar] [CrossRef] [PubMed]
- Faner, R.; Sibila, O.; Agustí, A.; Bernasconi, E.; Chalmers, J.D.; Huffnagle, G.B.; Manichanh, C.; Molyneaux, P.L.; Paredes, R.; Brocal, V.P.; et al. The microbiome in respiratory medicine: Current challenges and future perspectives. Eur. Respir. J. 2017, 49, 1602086. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.L.; de Koff, E.M.; Bogaert, D. Characterising the respiratory microbiome. Eur. Respir. J. 2019, 53, 1801711. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L.; Clemente, J.C. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef]
- Vientós-Plotts, A.I.; Ericsson, A.C.; McAdams, Z.L.; Rindt, H.; Reinero, C.R. Respiratory dysbiosis in cats with spontaneous allergic asthma. Front. Veter. Sci. 2022, 9, 930385. [Google Scholar] [CrossRef] [PubMed]
- Vangrinsven, E.; Fastrès, A.; Taminiau, B.; Frédéric, B.; Daube, G.; Clercx, C. Variations in facial conformation are associated with differences in nasal microbiota in healthy dogs. BMC Veter. Res. 2021, 17, 361. [Google Scholar] [CrossRef]
- Dorn, E.S.; Tress, B.; Suchodolski, J.S.; Nisar, T.; Ravindran, P.; Weber, K.; Hartmann, K.; Schulz, B.S.; Smidt, H. Bacterial microbiome in the nose of healthy cats and in cats with nasal disease. PLoS ONE 2017, 12, e0180299. [Google Scholar] [CrossRef]
- Vientós-Plotts, A.I.; Ericsson, A.C.; Reinero, C.R. The respiratory microbiota and its impact on health and disease in dogs and cats: A One Health perspective. J. Veter. Intern. Med. 2023, 37, 1641–1655. [Google Scholar] [CrossRef]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P.; Xu, J. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef]
- Santacroce, L.; Charitos, I.A.; Ballini, A.; Inchingolo, F.; Luperto, P.; De Nitto, E.; Topi, S. The Human Respiratory System and its Microbiome at a Glimpse. Biology 2020, 9, 318. [Google Scholar] [CrossRef]
- Invernizzi, R.; Lloyd, C.M.; Molyneaux, P.L. Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology 2020, 160, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Rajasekaran, J.J.; Krishnamurthy, H.K.; Bosco, J.; Jayaraman, V.; Krishna, K.; Wang, T.; Bei, K. Oral Microbiome: A Review of Its Impact on Oral and Systemic Health. Microorganisms 2024, 12, 1797. [Google Scholar] [CrossRef]
- Tress, B.; Dorn, E.S.; Suchodolski, J.S.; Nisar, T.; Ravindran, P.; Weber, K.; Hartmann, K.; Schulz, B.S.; Smidt, H. Bacterial microbiome of the nose of healthy dogs and dogs with nasal disease. PLoS ONE 2017, 12, e0176736. [Google Scholar] [CrossRef]
- Isaiah, A.; Hoffmann, A.R.; Kelley, R.; Mundell, P.; Steiner, J.M.; Suchodolski, J.S.; He, Z. Characterization of the nasal and oral microbiota of detection dogs. PLoS ONE 2017, 12, e0184899. [Google Scholar] [CrossRef] [PubMed]
- Vientós-Plotts, A.I.; Ericsson, A.C.; Rindt, H.; Reinero, C.R. Oral Probiotics Alter Healthy Feline Respiratory Microbiota. Front. Microbiol. 2017, 8, 1287. [Google Scholar] [CrossRef]
- Dickson, R.P.; Singer, B.H.; Newstead, M.W.; Falkowski, N.R.; Erb-Downward, J.R.; Standiford, T.J.; Huffnagle, G.B. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 2016, 1, 16113. [Google Scholar] [CrossRef]
- Drigot, Z.G.; Clark, E.S. Insights into the role of the respiratory tract microbiome in defense against bacterial pneumonia. Curr. Opin. Microbiol. 2024, 77, 102428. [Google Scholar] [CrossRef] [PubMed]
- Puiu, R.; Motoc, N.S.; Lucaciu, S.; Ruta, M.V.; Rajnoveanu, R.-M.; Todea, D.A.; Man, M.A. The Role of Lung Microbiome in Fibrotic Interstitial Lung Disease—A Systematic Review. Biomolecules 2024, 14, 247. [Google Scholar] [CrossRef]
- Lu, L.J.; Liu, J. Human Microbiota and Ophthalmic Disease. Yale J Biol Med. 2016, 89, 325–330. [Google Scholar]
- Ozkan, J.; Nielsen, S.; Diez-Vives, C.; Coroneo, M.; Thomas, T.; Willcox, M. Temporal Stability and Composition of the Ocular Surface Microbiome. Sci. Rep. 2017, 7, 9880. [Google Scholar] [CrossRef]
- Petrillo, F.; Pignataro, D.; Lavano, M.A.; Santella, B.; Folliero, V.; Zannella, C.; Astarita, C.; Gagliano, C.; Franci, G.; Avitabile, T.; et al. Current Evidence on the Ocular Surface Microbiota and Related Diseases. Microorganisms 2020, 8, 1033. [Google Scholar] [CrossRef] [PubMed]
- Leis, M.L.; Costa, M.O. Initial description of the core ocular surface microbiome in dogs: Bacterial community diversity and composition in a defined canine population. Veter. Ophthalmol. 2018, 22, 337–344. [Google Scholar] [CrossRef]
- Rogers, C.M.; Scott, E.M.; Sarawichitr, B.; Arnold, C.; Suchodolski, J.S.; Clegg, S. Evaluation of the bacterial ocular surface microbiome in ophthalmologically normal dogs prior to and following treatment with topical neomycin-polymyxin-bacitracin. PLoS ONE 2020, 15, e0234313. [Google Scholar] [CrossRef] [PubMed]
- Torikachvili, M.; da Silva, M.S.; Petersen, M.B.; Mayer, F.Q.; Budaszewski, R.d.F.; Weber, M.N.; Pigatto, J.A.T.; Canal, C.W.; Siqueira, F.M. Determination of microbial diversity in the ocular conjunctiva of healthy dogs by total DNA sequencing. Cienc. Anim. Bras. 2024, 25, 77549E. [Google Scholar] [CrossRef]
- Darden, J.E.; Scott, E.M.; Arnold, C.; Scallan, E.M.; Simon, B.T.; Suchodolski, J.S.; Oakley, B.B. Evaluation of the bacterial ocular surface microbiome in clinically normal cats before and after treatment with topical erythromycin. PLoS ONE 2019, 14, e0223859. [Google Scholar] [CrossRef]
- Lucyshyn, D.R.; Vernau, W.; Maggs, D.J.; Murphy, C.J.; Leonard, B.C. Correlations between clinical signs and corneal cytology in feline eosinophilic keratoconjunctivitis. Veter. Ophthalmol. 2021, 24, 620–626. [Google Scholar] [CrossRef]
- Chiang, M.-C.; Chern, E. Ocular surface microbiota: Ophthalmic infectious disease and probiotics. Front. Microbiol. 2022, 13, 952473. [Google Scholar] [CrossRef]
- Dong, Q.; Brulc, J.M.; Iovieno, A.; Bates, B.; Garoutte, A.; Miller, D.; Revanna, K.V.; Gao, X.; Antonopoulos, D.A.; Slepak, V.Z.; et al. Diversity of Bacteria at Healthy Human Conjunctiva. Investig. Opthalmology Vis. Sci. 2011, 52, 5408–5413. [Google Scholar] [CrossRef]
- Zhou, Y.; Holland, M.J.; Makalo, P.; Joof, H.; Roberts, C.H.; Mabey, D.C.; Bailey, R.L.; Burton, M.J.; Weinstock, G.M.; Burr, E.S. The conjunctival microbiome in health and trachomatous disease: A case control study. Genome Med. 2014, 6, 99. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, B.; Li, W. Defining the normal core microbiome of conjunctival microbial communities. Clin. Microbiol. Infect. 2016, 22, 643.e7–643.e12. [Google Scholar] [CrossRef]
- Doan, T.; Akileswaran, L.; Andersen, D.; Johnson, B.; Ko, N.; Shrestha, A.; Shestopalov, V.; Lee, C.S.; Lee, A.Y.; Van Gelder, R.N. Paucibacterial Microbiome and Resident DNA Virome of the Healthy Conjunctiva. Investig. Opthalmology Vis. Sci. 2016, 57, 5116–5126. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Miao, L.; Deng, Y.; Bible, P.W.; Hu, X.; Zou, Y.; Liu, Y.; Guo, S.; Liang, J.; Chen, T.; et al. The Influence of Age and Sex on Ocular Surface Microbiota in Healthy Adults. Investig. Opthalmology Vis. Sci. 2017, 58, 6030–6037. [Google Scholar] [CrossRef]
- Li, Z.; Gong, Y.; Chen, S.; Li, S.; Zhang, Y.; Zhong, H.; Wang, Z.; Chen, Y.; Deng, Q.; Jiang, Y.; et al. Comparative portrayal of ocular surface microbe with and without dry eye. J. Microbiol. 2019, 57, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Sutani, T.; Nakai, H.; Shirahige, K.; Kinoshita, S. The Microbiome of the Meibum and Ocular Surface in Healthy Subjects. Investig. Opthalmology Vis. Sci. 2020, 61, 18. [Google Scholar] [CrossRef] [PubMed]
- Büttner, J.N.; Schneider, M.; Csokai, J.; Müller, E.; Eule, J.C. Microbiota of the conjunctival sac of 120 healthy cats. Veter. Ophthalmol. 2018, 22, 328–336. [Google Scholar] [CrossRef]
- Welch, J.L.M.; Ramírez-Puebla, S.T.; Borisy, G.G. Oral Microbiome Geography: Micron-Scale Habitat and Niche. Cell Host Microbe 2020, 28, 160–168. [Google Scholar] [CrossRef]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef]
- Lisjak, A.; Lopes, B.C.; Pilla, R.; Nemec, A.; Suchodolski, J.S.; Tozon, N. A Comparison of the Oral Microbiota in Healthy Dogs and Dogs with Oral Tumors. Animals 2023, 13, 3594. [Google Scholar] [CrossRef]
- Flancman, R.; Singh, A.; Weese, J.S.; Trackman, P.C. Evaluation of the impact of dental prophylaxis on the oral microbiota of dogs. PLoS ONE 2018, 13, e0199676. [Google Scholar] [CrossRef]
- Ruparell, A.; Warren, M.; Staunton, R.; Deusch, O.; Dobenecker, B.; Wallis, C.; O’FLynn, C.; McGenity, P.; Holcombe, L.J. Effect of feeding a daily oral care chew on the composition of plaque microbiota in dogs. Res. Veter. Sci. 2020, 132, 133–141. [Google Scholar] [CrossRef]
- Oba, P.M.; Carroll, M.Q.; Alexander, C.; Somrak, A.J.; Keating, S.C.J.; Sage, A.M.; Swanson, K.S. Dental chews positively shift the oral microbiota of adult dogs. J. Anim. Sci. 2021, 99, skab100. [Google Scholar] [CrossRef] [PubMed]
- Oba, P.M.; Sieja, K.M.; Keating, S.C.J.; Hristova, T.; Somrak, A.J.; Swanson, K.S. Oral microbiota populations of adult dogs consuming wet or dry foods. J. Anim. Sci. 2022, 100, skac200. [Google Scholar] [CrossRef] [PubMed]
- Kislik, G.; Zhou, L.; Rubbi, L.; Pellegrini, M. Age-correlated changes in the canine oral microbiome. Front. Microbiol. 2024, 15, 1426691. [Google Scholar] [CrossRef]
- Sharma, N.; Bhatia, S.; Sodhi, A.S.; Batra, N. Oral microbiome and health. AIMS Microbiol. 2018, 4, 42–66. [Google Scholar] [CrossRef]
- Sturgeon, A.; Pinder, S.L.; Costa, M.C.; Weese, J.S. Characterization of the oral microbiota of healthy cats using next-generation sequencing. Vet. J. 2014, 201, 223–229. [Google Scholar] [CrossRef]
- Adler, C.J.; Malik, R.; Browne, G.V.; Norris, J.M. Diet may influence the oral microbiome composition in cats. Microbiome 2016, 4, 23. [Google Scholar] [CrossRef]
- Whyte, A.; Gracia, A.; Bonastre, C.; Tejedor, M.T.; Whyte, J.; Monteagudo, L.V.; Simón, C. Oral Disease and Microbiota in Free-Roaming Cats. Top. Companion Anim. Med. 2017, 32, 91–95. [Google Scholar] [CrossRef]
- Krumbeck, J.A.; Reiter, A.M.; Pohl, J.C.; Tang, S.; Kim, Y.J.; Linde, A.; Prem, A.; Melgarejo, T. Characterization of Oral Microbiota in Cats: Novel Insights on the Potential Role of Fungi in Feline Chronic Gingivostomatitis. Pathogens 2021, 10, 904. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.G.; Rojas, C.A.; Scarsella, E.; Entrolezo, Z.; Jospin, G.; Hoffman, S.L.; Force, J.; MacLellan, R.H.; Peak, M.; Shope, B.H.; et al. The Oral Microbiome across Oral Sites in Cats with Chronic Gingivostomatitis, Periodontal Disease, and Tooth Resorption Compared with Healthy Cats. Animals 2023, 13, 3544. [Google Scholar] [CrossRef]
- Vuik, F.E.; Nieuwenburg, S.A.; Bardou, M.; Lansdorp-Vogelaar, I.; Dinis-Ribeiro, M.; Bento, M.J.; Zadnik, V.; Pellisé, M.; Esteban, L.; Kaminski, M.F.; et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut 2019, 68, 1820–1826. [Google Scholar] [CrossRef]
- Li, D.; He, R.; Hou, G.; Ming, W.; Fan, T.; Chen, L.; Zhang, L.; Jiang, W.; Wang, W.; Lu, Z.; et al. Characterization of the Esophageal Microbiota and Prediction of the Metabolic Pathways Involved in Esophageal Cancer. Front. Cell. Infect. Microbiol. 2020, 10, 268. [Google Scholar] [CrossRef]
- Li, Z.; Dou, L.; Zhang, Y.; He, S.; Zhao, D.; Hao, C.; Song, G.; Zhang, W.; Liu, Y.; Wang, G. Characterization of the Oral and Esophageal Microbiota in Esophageal Precancerous Lesions and Squamous Cell Carcinoma. Front. Cell Infect. Microbiol. 2021, 11, 714162. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.; Figueiredo, C.; Ferreira, R.M. The Role of the Microbiota in Esophageal Cancer. Cancers 2023, 15, 2576. [Google Scholar] [CrossRef] [PubMed]
- Hitch, T.C.; Bisdorf, K.; Afrizal, A.; Riedel, T.; Overmann, J.; Strowig, T.; Clavel, T. A taxonomic note on the genus Prevotella: Description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst. Appl. Microbiol. 2022, 45, 126354. [Google Scholar] [CrossRef]
- Delgado, S.; Cabrera-Rubio, R.; Mira, A.; Suárez, A.; Mayo, B. Microbiological Survey of the Human Gastric Ecosystem Using Culturing and Pyrosequencing Methods. Microb. Ecol. 2013, 65, 763–772. [Google Scholar] [CrossRef]
- Liu, X.; Shao, L.; Liu, X.; Ji, F.; Mei, Y.; Cheng, Y.; Liu, F.; Yan, C.; Li, L.; Ling, Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019, 40, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Nardone, G.; Compare, D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 2015, 3, 255–260. [Google Scholar] [CrossRef]
- Sung, J.; Kim, N.; Kim, J.; Jo, H.J.; Park, J.H.; Nam, R.H.; Seok, Y.-J.; Kim, Y.-R.; Lee, D.H.; Jung, H.C. Comparison of Gastric Microbiota Between Gastric Juice and Mucosa by Next Generation Sequencing Method. J. Cancer Prev. 2016, 21, 60–65. [Google Scholar] [CrossRef]
- Zilberstein, B.; Quintanilha, A.G.; Santos, M.A.A.; Pajecki, D.; De Moura, E.G.; Alves, P.R.A.; Filho, F.M.; De Souza, J.A.U.; Gama-Rodrigues, J. Digestive tract microbiota in healthy volunteers. Clin. Sci. 2007, 62, 47–54. [Google Scholar] [CrossRef]
- Bilello, J.; Okereke, I. Impact of Environmental and Pharmacologic Changes on the Upper Gastrointestinal Microbiome. Biomedicines 2021, 9, 617. [Google Scholar] [CrossRef]
- Pappas-Gogos, G.; Tepelenis, K.; Fousekis, F.; Katsanos, K.; Pitiakoudis, M.; Vlachos, K. The Implication of Gastric Microbiome in the Treatment of Gastric Cancer. Cancers 2022, 14, 2039. [Google Scholar] [CrossRef] [PubMed]
- Nardone, G.; Compare, D.; Rocco, A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol. Hepatol. 2017, 2, 298–312. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, X.; Liu, X.; Ling, Z.; Ji, F. Role of the Gastric Microbiome in Gastric Cancer: From Carcinogenesis to Treatment. Front. Microbiol. 2021, 12, 641322. [Google Scholar] [CrossRef] [PubMed]
- Deschamps, C.; Humbert, D.; Zentek, J.; Denis, S.; Priymenko, N.; Apper, E.; Blanquet-Diot, S. From Chihuahua to Saint-Bernard: How did digestion and microbiota evolve with dog sizes. Int. J. Biol. Sci. 2022, 18, 5086–5102. [Google Scholar] [CrossRef]
- Lee, J.H.; Kuhar, S.; Seo, J.-H.; Pasricha, P.J.; Mittal, R. Computational modeling of drug dissolution in the human stomach: Effects of posture and gastroparesis on drug bioavailability. Phys. Fluids 2022, 34, 081904. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, G.; Argentini, C.; Milani, C.; Turroni, F.; Cristina Ossiprandi, M.; van Sinderen, D.; Ventura, M. Catching a glimpse of the bacterial gut community of companion animals: A canine and feline perspective. Microb Biotechnol. 2020, 13, 1708–1732. [Google Scholar] [CrossRef]
- Rindels, E.J.; Loman, B.R. Gut microbiome—The key to our pets’ health and happiness? Anim. Front. 2024, 14, 46–53. [Google Scholar] [CrossRef]
- Wernimont, S.M.; Radosevich, J.; Jackson, M.I.; Ephraim, E.; Badri, D.V.; MacLeay, J.M.; Jewell, D.E.; Suchodolski, J.S. The Effects of Nutrition on the Gastrointestinal Microbiome of Cats and Dogs: Impact on Health and Disease. Front. Microbiol. 2020, 11, 1266. [Google Scholar] [CrossRef]
- Nayak, R.R.; Orellana, D.A. The impact of the human gut microbiome on the treatment of autoimmune disease. Immunol. Rev. 2024, 325, 107–130. [Google Scholar] [CrossRef]
- Bradley, E.; Haran, J. The human gut microbiome and aging. Gut Microbes 2024, 16, 2359677. [Google Scholar] [CrossRef]
- Jiang, C.; Cui, Z.; Fan, P.; Du, G.; Liu, G. Effects of dog ownership on the gut microbiota of elderly owners. PLoS ONE 2022, 17, e0278105. [Google Scholar] [CrossRef]
- Du, G.; Huang, H.; Zhu, Q.; Ying, L.; Zoetendal, E.G. Effects of cat ownership on the gut microbiota of owners. PLoS ONE 2021, 16, e0253133. [Google Scholar] [CrossRef] [PubMed]
- Kates, A.E.; Jarrett, O.; Skarlupka, J.H.; Sethi, A.; Duster, M.; Watson, L.; Suen, G.; Poulsen, K.; Safdar, N. Household Pet Ownership and the Microbial Diversity of the Human Gut Microbiota. Front. Cell Infect. Microbiol. 2020, 10, 73. [Google Scholar] [CrossRef]
- Baatz, A.; Bidgood, A.; Taylor, G.; Young, R. The trouble with a cuddle: Families’ experiences of supervising interactions between children in middle childhood and the family dog. Hum. Anim. Interact. 2023, 233, 83–89. [Google Scholar] [CrossRef]
- Arenas-Montes, J.; Perez-Martinez, P.; Vals-Delgado, C.; Romero-Cabrera, J.L.; Cardelo, M.P.; Leon-Acuña, A.; Quintana-Navarro, G.M.; Alcala-Diaz, J.F.; Lopez-Miranda, J.; Camargo, A.; et al. Owning a Pet Is Associated with Changes in the Composition of Gut Microbiota and Could Influence the Risk of Metabolic Disorders in Humans. Animals 2021, 11, 2347. [Google Scholar] [CrossRef]
- Koontz, J.M.; Dancy, B.C.R.; Horton, C.L.; Stallings, J.D.; DiVito, V.T.; Lewis, J.A. The Role of the Human Microbiome in Chemical Toxicity. Int. J. Toxicol. 2019, 38, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: A major player in the toxicity of environmental pollutants? Npj Biofilms Microbiomes 2016, 2, 16003. [Google Scholar] [CrossRef]
- Scully, T.; Ettela, A.; LeRoith, D.; Gallagher, E.J. Obesity, Type 2 Diabetes, and Cancer Risk. Front. Oncol. 2021, 10, 615375. [Google Scholar] [CrossRef]
- Collins, S.L.; Patterson, A.D. The gut microbiome: An orchestrator of xenobiotic metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef]
- Koppel, N.; Rekdal, V.M.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, 6344. [Google Scholar] [CrossRef]
- Tu, P.; Chi, L.; Bodnar, W.; Zhang, Z.; Gao, B.; Bian, X.; Stewart, J.; Fry, R.; Lu, K. Gut Microbiome Toxicity: Connecting the Environment and Gut Microbiome-Associated Diseases. Toxics 2020, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Ritchie, L.E.; Steiner, J.M.; Suchodolski, J.S. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol. Ecol. 2008, 66, 590–598. [Google Scholar] [CrossRef]
- Suchodolski, J.S. Analysis of the gut microbiome in dogs and cats. Veter. Clin. Pathol. 2021, 50, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.S.; Camacho, J.; Steiner, J.M. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol. Ecol. 2008, 66, 567–578. [Google Scholar] [CrossRef]
- Middelbos, I.S.; Boler, B.M.V.; Qu, A.; White, B.A.; Swanson, K.S.; Fahey, G.C., Jr. Phylogenetic Characterization of Fecal Microbial Communities of Dogs Fed Diets with or without Supplemental Dietary Fiber Using 454 Pyrosequencing. PLoS ONE 2010, 5, e9768. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2010, 5, 639–649. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Lanerie, D.J.; Dowd, S.E.; Paddock, C.G.; Grützner, N.; Steiner, J.M.; Ivanek, R.; Suchodolski, J.S. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiol. Ecol. 2011, 78, 542–554. [Google Scholar] [CrossRef]
- Bermingham, E.N.; Young, W.; Kittelmann, S.; Kerr, K.R.; Swanson, K.S.; Roy, N.C.; Thomas, D.G. Dietary format alters fecal bacterial populations in the domestic cat (Felis catus). Microbiologyopen 2013, 2, 173–181. [Google Scholar] [CrossRef]
- Shanahan, F.; Ghosh, T.S.; O’tOole, P.W. The Healthy Microbiome—What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]
- Heil, B.A.; Paccamonti, D.L.; Sones, J.L. Role for the mammalian female reproductive tract microbiome in pregnancy outcomes. Physiol Genom. 2019, 51, 390–399. [Google Scholar] [CrossRef]
- Acinas, S.G.; Sarma-Rupavtarm, R.; Klepac-Ceraj, V.; Polz, M.F. PCR-Induced Sequence Artifacts and Bias: Insights from Comparison of Two 16S rRNA Clone Libraries Constructed from the Same Sample. Appl. Environ. Microbiol. 2005, 71, 8966–8969. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.-S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef]
- Trinh, P.; Zaneveld, J.R.; Safranek, S.; Rabinowitz, P.M. One Health Relationships Between Human, Animal, and Environmental Microbiomes: A Mini-Review. Front. Public Health 2018, 6, 235. [Google Scholar] [CrossRef] [PubMed]
- Scoresby, K.J.; Strand, E.B.; Ng, Z.; Brown, K.C.; Stilz, C.R.; Strobel, K.; Barroso, C.S.; Souza, M. Pet Ownership and Quality of Life: A Systematic Review of the Literature. Vet Sci. 2021, 8, 332. [Google Scholar] [CrossRef]
- Bhandary, R.; Venugopalan, G.; Ramesh, A.; Tartaglia, G.M.; Singhal, I.; Khijmatgar, S. Microbial Symphony: Navigating the Intricacies of the Human Oral Microbiome and Its Impact on Health. Microorganisms 2024, 12, 571. [Google Scholar] [CrossRef]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-W.; Cha, C.-J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef]
- Zhang, H.; Rahman, S.; Li, W.; Fu, G.; Kaur, P. Characterization of a novel domain ‘GATE’ in the ABC protein DrrA and its role in drug efflux by the DrrAB complex. Biochem. Biophys. Res. Commun. 2015, 459, 148–153. [Google Scholar] [CrossRef]
- Gillieatt, B.F.; Coleman, N.V. Unravelling the mechanisms of antibiotic and heavy metal resistance co-selection in environmental bacteria. FEMS Microbiol. Rev. 2024, 48, fuae017. [Google Scholar] [CrossRef]
- Ho, J.; Yeoh, Y.K.; Barua, N.; Chen, Z.; Lui, G.; Wong, S.H.; Yang, X.; Chan, M.C.; Chan, P.K.; Hawkey, P.M.; et al. Systematic review of human gut resistome studies revealed variable definitions and approaches. Gut Microbes 2020, 12, 1700755. [Google Scholar] [CrossRef]
- Mouiche, M.M.M.; Mpouam, S.E.; Moffo, F.; Nkassa, C.M.N.; Mbah, C.K.; Mapiefou, N.P.; Awah-Ndukum, J. Prescription Pattern of Antimicrobial Use in Small Animal Veterinary Practice in Cameroon. Top. Companion Anim. Med. 2021, 44, 100540. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.; Denis, K.S.; Collins, S.; Dowgray, N.; Ellis, S.L.; Heath, S.; Rodan, I.; Ryan, L. 2022 ISFM/AAFP Cat Friendly Veterinary Environment Guidelines. J. Feline Med. Surg. 2022, 24, 1133–1163. [Google Scholar] [CrossRef]
- Bhat, A.H. Bacterial zoonoses transmitted by household pets and as reservoirs of antimicrobial resistant bacteria. Microb. Pathog. 2021, 155, 104891. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hu, X.; Cai, S.; Hu, N.; Yuan, Y.; Wu, Y.; Wang, Y.; Mi, J.; Liao, X. Pet cats may shape the antibiotic resistome of their owner’s gut and living environment. Microbiome 2023, 11, 235. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dou, Q.; Smalla, K.; Wang, Y.; Johnson, T.A.; Brandt, K.K.; Mei, Z.; Liao, M.; Hashsham, S.A.; Schäffer, A.; et al. Gut microbiota research nexus: One Health relationship between human, animal, and environmental resistomes. mLife 2023, 2, 350–364. [Google Scholar] [CrossRef]
- Inda-Díaz, J.S.; Lund, D.; Parras-Moltó, M.; Johnning, A.; Bengtsson-Palme, J.; Kristiansson, E. Latent antibiotic resistance genes are abundant, diverse, and mobile in human, animal, and environmental microbiomes. Microbiome 2023, 11, 44. [Google Scholar] [CrossRef]
- Chen, C.; Li, Y.; Wu, Z.; Ruan, Y.; Long, T.; Wang, X.; Li, W.; Ren, H.; Liao, X.; Liu, Y.; et al. Cat and dog feces as reservoirs of diverse novel antibiotic resistance genes. Environ. Res. 2024, 261, 119690. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, X.; Li, W.; Li, L.; Liao, X.; Xing, S. Abundance, diversity and diffusion of antibiotic resistance genes in cat feces and dog feces. Environ. Pollut. (Barking Essex 1987) 2021, 292, 118364. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Stankiewicz, K.; Czernecka, N.; Ratajewicz, A.; Bulanda, K.; Heliasz, M.; Sosińska, D.; Dworak, K.; Ciesielska, D.; Siemińska, I.; et al. Wounds of Companion Animals as a Habitat of Antibiotic-Resistant Bacteria That Are Potentially Harmful to Humans—Phenotypic, Proteomic and Molecular Detection. Int. J. Mol. Sci. 2024, 25, 3121. [Google Scholar] [CrossRef]
- Jin, M.; Osman, M.; Green, B.A.; Yang, Y.; Ahuja, A.; Lu, Z.; Cazer, C.L. Evidence for the transmission of antimicrobial resistant bacteria between humans and companion animals: A scoping review. One Health 2023, 17, 100593. [Google Scholar] [CrossRef]
- Ma, X.; Brinker, E.; Lea, C.R.; Delmain, D.; Chamorro, E.D.; Martin, D.R.; Graff, E.C.; Wang, X. Evaluation of fecal sample collection methods for feline gut microbiome profiling: Fecal loop vs. litter box. Front. Microbiol. 2024, 15, 1337917. [Google Scholar] [CrossRef] [PubMed]
- Šakarnytė, L.; Šiugždinienė, R.; Žymantienė, J.; Ruzauskas, M. Comparison of Oral Microbial Composition and Determinants Encoding Antimicrobial Resistance in Dogs and Their Owners. Antibiotics 2023, 12, 1554. [Google Scholar] [CrossRef]
- Tsang, W.; Linde, A.; Krumbeck, J.A.; Wu, G.; Kim, Y.J.; Lushington, G.H.; Melgarejo, T. Occurrence of Antimicrobial Resistance Genes in the Oral Cavity of Cats with Chronic Gingivostomatitis. Animals 2021, 11, 3589. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moein, K.A.; El-Hariri, M.D.; Wasfy, M.O.; Samir, A. Occurrence of ampicillin-resistant Enterococcus faecium carrying esp gene in pet animals: An upcoming threat for pet lovers. J. Glob. Antimicrob. Resist. 2017, 9, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Abad- Oro, A.; Martín-Burriel, I.; Moreno, B.; Morales, M.; Bolea, R. Multidrug resistance in pathogenic Escherichia coli isolates from urinary tract infections in dogs, Spain. Front. Vet. Sci. 2024, 11, 1325072. [Google Scholar] [CrossRef]
- Røken, M.; Forfang, K.; Wasteson, Y.; Haaland, A.H.; Eiken, H.G.; Hagen, S.B.; Bjelland, A.M. Antimicrobial resistance—Do we share more than companionship with our dogs? J. Appl. Microbiol. 2022, 133, 1027–1039. [Google Scholar] [CrossRef]
- Zhao, R.; Hao, J.; Yang, J.; Tong, C.; Xie, L.; Xiao, D.; Zeng, Z.; Xiong, W. The co-occurrence of antibiotic resistance genes between dogs and their owners in families. Imeta 2022, 1, e21. [Google Scholar] [CrossRef]
- Naziri, Z.; Poormaleknia, M.; Oliyaei, A.G. Risk of sharing resistant bacteria and/or resistance elements between dogs and their owners. BMC Veter. Res. 2022, 18, 203. [Google Scholar] [CrossRef]
- Walas, N.; Müller, N.F.; Parker, E.; Henderson, A.; Capone, D.; Brown, J.; Barker, T.; Graham, J.P. Application of phylodynamics to identify spread of antimicrobial-resistant Escherichia coli between humans and canines in an urban environment. Sci. Total. Environ. 2024, 916, 170139. [Google Scholar] [CrossRef]
- Buranasinsup, S.; Wiratsudakul, A.; Chantong, B.; Maklon, K.; Suwanpakdee, S.; Jiemtaweeboon, S.; Sakcamduang, W. Prevalence and characterization of antimicrobial-resistant Escherichia coli isolated from veterinary staff, pets, and pet owners in Thailand. J. Infect. Public Health 2023, 16, 194–202. [Google Scholar] [CrossRef]
- Abdullahi, I.N.; Lozano, C.; González-Azcona, C.; Zarazaga, M.; Torres, C. Genetic Diversification and Resistome of Coagulase-Negative Staphylococci from Nostrils of Healthy Dogs and Dog-Owners in La Rioja, Spain. Pathogens 2024, 13, 229. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.M.; Yan, Y.S.; Wang, H.; Zhong, Y.; Inam; Gao, Y.H.; Li, G.M.; Mu, G.D.; Dong, H.F.; Li, Y.; et al. Transmission of human-pet antibiotic resistance via aerosols in pet hospitals of Changchun. One Health 2024, 18, 100765. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Next-generation sequencing to monitor the spread of antimicrobial resistance. Genome Med. 2017, 9, 68. [Google Scholar] [CrossRef]
- Belas, A.; Marques, C.; Pomba, C. The gut microbiome and antimicrobial resistance in companion animals. In Advances in Animal Health, Medicine and Production: A Research Portrait of the Centre for Interdisciplinary Research in Animal Health (CIISA), University of Lisbon, Portugal; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 233–245. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Domrazek, K.; Jurka, P. Application of Next-Generation Sequencing (NGS) Techniques for Selected Companion Animals. Animals 2024, 14, 1578. [Google Scholar] [CrossRef]

| Dominant Phyla | Common Families/Genera | Factors Influencing Diversity | Observations | References | |
|---|---|---|---|---|---|
| Humans | Actinobacteria, Firmicutes, Proteobacteria, Bacteroidetes | Coagulase negative Staphylococci, Cutibacterium spp., Corynebacterium spp., Micrococcus spp., Streptococcus spp., Acinetobacter spp. | Skin location, moisture | Colonized by 103–104 CFU/cm2 of bacteria; 16 phyla identified; fungal infections rising among pet owners due to close contact | [19,43,44,45,46,47,48,49] |
| Dogs | Proteobacteria, Oxalobacteriaceae, Fusobacteria, Actinobacteria, Bacteroidetes, Firmicutes, Tenericutes, Cyanobacteria | Porphyromonadaceae, Moraxellaceae, Pasteurellaceae, Pseudomonadaceae | Skin region, gender, body site, physiology, individual factors | Diversity similar to human microbiota in families; mouth-to-skin transmission of Betaproteobacteria observed | [26,28,50] |
| Cats | Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes | - | Less biodiversity compared to dogs | Limited comparison to canine microbiota | [51] |
| Section | Humans | Dogs | Cats |
|---|---|---|---|
| Upper Respiratory Microbiome | Oropharynx is major source of bacterial spread; Actinobacteria (Corynebacterium, Propionibacterium), Firmicutes (Streptococcus in children, Staphylococcus in adults), Bacteroidetes (low density), Moraxellaceae (children), Gamma-proteobacteria (adults) [90,91,92,93,94] | Upper respiratory system and inhaled air influence lower respiratory microbiome; nasal cavity: Proteobacteria, Bacteroidetes, Firmicutes, Tenericutes [94,95]. Proteobacteria dominant in oropharynx and lower respiratory tract; Pasteurellaceae, Moraxellaceae, Pseudomonaceae [86]. | Nasal cavity and oropharynx: Proteobacteria, Bacteroidetes, Firmicutes; nasal: Moraxellaceae, Bradyrhizobiaceae; oropharynx: Pasteurellaceae, Moraxellaceae, Porphyromonadaceae, Pseudomonadaceae; lungs: Proteobacteria dominant [88,96] |
| Lung Microbiome | Prevotella, Sphingomonas, Pseudomonas, Acinetobacter, Fusobacterium, Megasphaera, Veillonella, Staphylococcus, Streptococcus, Porphyromonas, Haemophilus; fungi: Aspergillus, Cladosporium, Penicillium; Enterobacteriaceae; Tropheryma whipplei [92,97,98,99] | Pulmonary microbiota: dominated by Pseudomonaceae and Moraxellaceae; Cutibacterium, Streptococcus, Acinetobacter, Pseudomonas abundant in lower airways [86] | Proteobacteria (Pseudomonadaceae, Sphingobacteriaceae, Bradyrhizobiaceae) dominant [86,96] |
| Anatomical Site | Humans | Dogs | Cats |
|---|---|---|---|
| Conjunctiva and cornea | Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Deinococcus-Thermes are the dominant phyla [102,108]. Pseudomonas, Propionibacterium, Bradyrhizobium, Corynebacterium, Acinetobacter, Brevundimonas, Staphylococcus, Aquabacterium, Sphingomonas, Streptococcus, Streptophyta, Ralstonia, Anaerococcus, Finegoldia, Simonsiella, Veillonella, Milisia, Massilia, Rothia, Neisseria and Methylobacterium [101,105,106,107,108,109,110,111,112,113,114,115]. | Proteobacteria (alfa-, beta-,gamma-), Actinobacteria, Firmicutes, Bacteroidetes, Fusobacteria. Dominant families: Pseudomonadaceae, Micrococcaceae, Pasteurellaceae, Microbacteriaceae, Enterobacteriaceae, Neisseriaceae, Moraxellaceae, Bifidobacteriaceae, Lachnospiraceae and Corynebacteriaceae [103,104,105] | Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Fusobacteria and Chlamydiae [106,107]. Corynebacteriaceae, Helicobacteraceae, Moraxellaceae and Comamonadaceae are the most common families [106]. Mycoplasma, Streptococcus, Pseudomonas Staphylococcus felis and Moraxella osloensis are often isolated from the ocular surface of cats [107,116] |
| Species | Phyla | Dominant Families/Genera | Study | Notes |
|---|---|---|---|---|
| Human | Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria | Actinomyces, Atopobium, Corynebacterium, Rothia, Campylobacter, Cardiobacterium, Haemophilus, Neisseria, Fusobacterium, Streptococcus, Veilonella, Prevotella | [117,125] | Describes stomatotypes based on genus abundance. |
| Canine | Firmicutes, Proteobacteria, Bacteroidetes, Spirochaetes, Synergistetes, Actinobacteria, Fusobacteria, Tenericutes | - | [119] | Distinct microbiota in different oral sites; diet affects composition. |
| Canine | 26 phyla, mainly: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Spirochaetes | - | [120] | |
| Canine | - | Fretibacterium fastidiosum, Filifactor alocis, Treponema medium, Tannerella forsythia, Porphyromonas canaris, Porphyromonas gingivalis | [123] | Detrimental bacteria more abundant in dogs fed wet food. |
| Feline | 18 phyla | Proteobacteria, Bacteroidetes, Spirochaetes, Fusobacteria, Firmicutes, Actinobacteria | [126] | Most abundant genera include Pasteurellaceae, Moraxella, Thermomonas, Comamonadaceae and Neisseria. |
| Feline | Bacteroidetes, Firmicutes, Proteobacteria | - | [127] | Most abundant genera include Porphyromonas, Treponema and Fusibacter. |
| Feline | - | Streptococcus, Staphylococcus, Neisseria, Pasteurella | [128] | |
| Feline | - | Porphyromonas gulae, Porphyromonas circumdentaria, Moraxella spp., Bacteroidales spp. | [129] | Detected 249 bacterial genera and 186 genera of fungi; most prevalent fungus was Sacharomyces cerevisiae. |
| Feline | - | Flavobacterium, Moraxella, Conchiformibius, Neisseria, Bergeyella, Streptococcus, Catonella, Actinobacillus | [130] | Influenced by breed, sex, and environment. |
| Section/ Samples | Species | Dominant Phyla | Dominant Genera | Factors Affecting Microbiome | References |
|---|---|---|---|---|---|
| Esophagus | Humans | Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria | Streptococcus, Ralstonia, Fusobacterium, Neisseria, Haemophilus, Prevotella, Porphyromonas, Actinobacillus, Veillonella, Tissierella, Staphylococcus | Alcohol consumption, diet, medicines, smoking, BMI | [131,132,133,134] |
| Stomach | Humans | Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria | Helicobacter, Prevotella, Pseudomonas, Streptococcus, Veillonella, Rothia, Haemophilus | Age, ethnicity, gender, diet, lifestyle, medicines, H. pylori | [19,137,141,142,143,144] |
| Stomach | Dogs | Proteobacteria | Helicobacter spp., Lactobacillus spp. | Less bacterial load compared to intestines | [145,146] |
| Gut | Humans | Bacteroidetes, Firmicutes | Escherichia coli, Klebsiella, Enterococcus, Bacteroides, Ruminococcus, Dorea | Species, breed, age, diseases, diet, gender, genetics | [25,148] |
| Gut | Cats | Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria | Clostridiales, Lactobacilli, Bacteroidales | Species, breed, age, diseases, diet, gender, genetics | [25,164] |
| Gut | Dogs | Firmicutes | Clostridiales, Lactobacilli, Enterobacteriales | Species, breed, age, diseases, diet, gender, genetics | [26,165,166] |
| Fecal Samples | Dogs | Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria | Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, Enterococcus | Methodology differences affect species detection | [167,168] |
| Fecal Samples | Cats | Firmicutes, Actinobacteria | Proteobacteria, Bacteroidetes, Fusobacteria | Overall population distribution in healthy cats | [169,170] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skoufos, S.; Stavropoulou, E.; Tsigalou, C.; Voidarou, C. Microbial Interconnections in One Health: A Critical Nexus Between Companion Animals and Human Microbiomes. Microorganisms 2025, 13, 1564. https://doi.org/10.3390/microorganisms13071564
Skoufos S, Stavropoulou E, Tsigalou C, Voidarou C. Microbial Interconnections in One Health: A Critical Nexus Between Companion Animals and Human Microbiomes. Microorganisms. 2025; 13(7):1564. https://doi.org/10.3390/microorganisms13071564
Chicago/Turabian StyleSkoufos, Stylianos, Elisavet Stavropoulou, Christina Tsigalou, and Chrysoula (Chrysa) Voidarou. 2025. "Microbial Interconnections in One Health: A Critical Nexus Between Companion Animals and Human Microbiomes" Microorganisms 13, no. 7: 1564. https://doi.org/10.3390/microorganisms13071564
APA StyleSkoufos, S., Stavropoulou, E., Tsigalou, C., & Voidarou, C. (2025). Microbial Interconnections in One Health: A Critical Nexus Between Companion Animals and Human Microbiomes. Microorganisms, 13(7), 1564. https://doi.org/10.3390/microorganisms13071564






