Synergistic Adaptations of Yak Rumen Microbiota, Metabolites and Host to Altitudinal
Abstract
1. Introduction
2. Materials and Methods
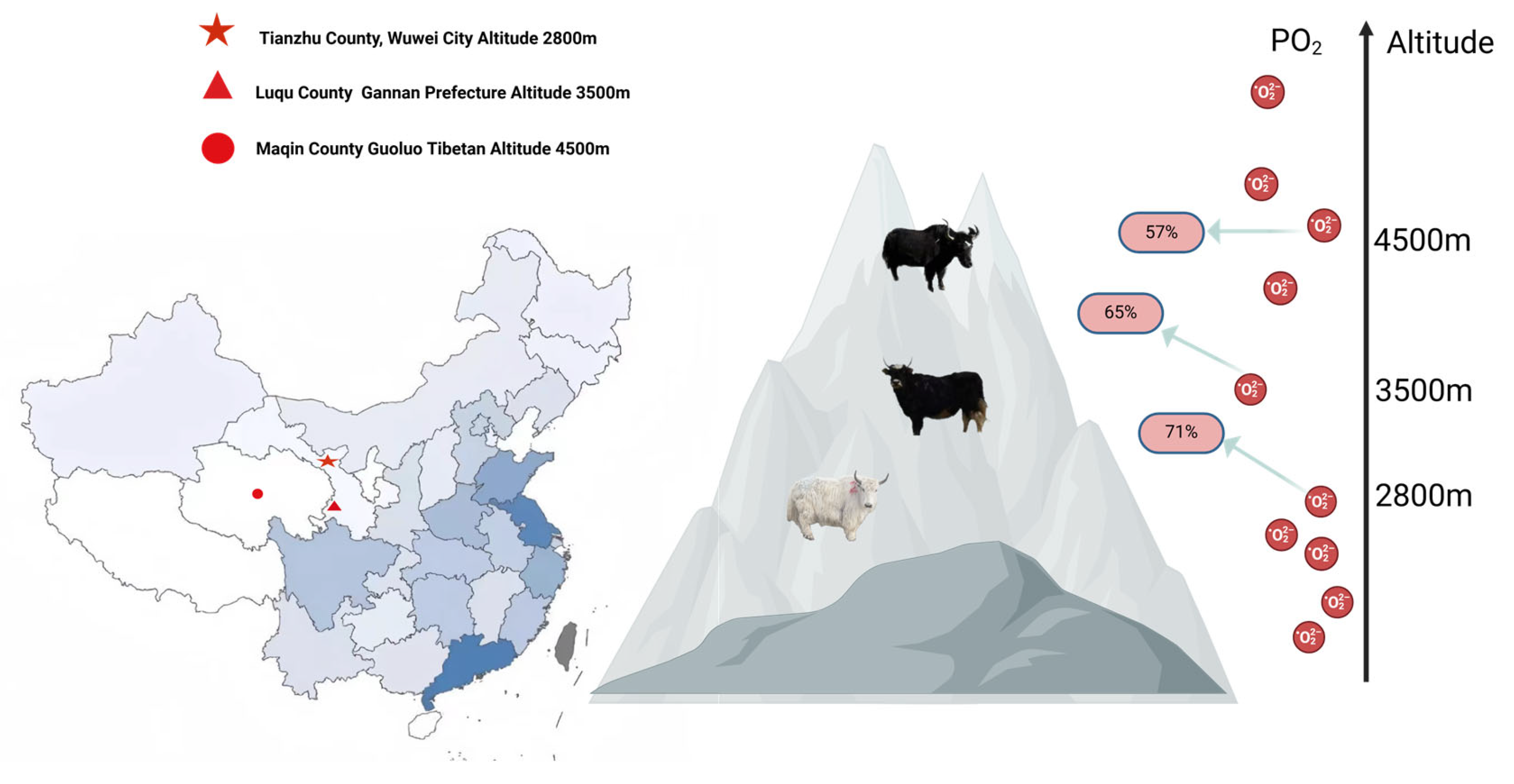
2.1. Experimental Sample Collection
2.2. Determination of Forage Nutrient Content
2.3. Determination of Rumen Fermentation Parameters
2.4. Metabolome Sequencing and Bioinformatics Analysis
2.5. Microbial Sequencing and Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Forage Nutrient Composition
3.2. Rumen Fermentation Parameters
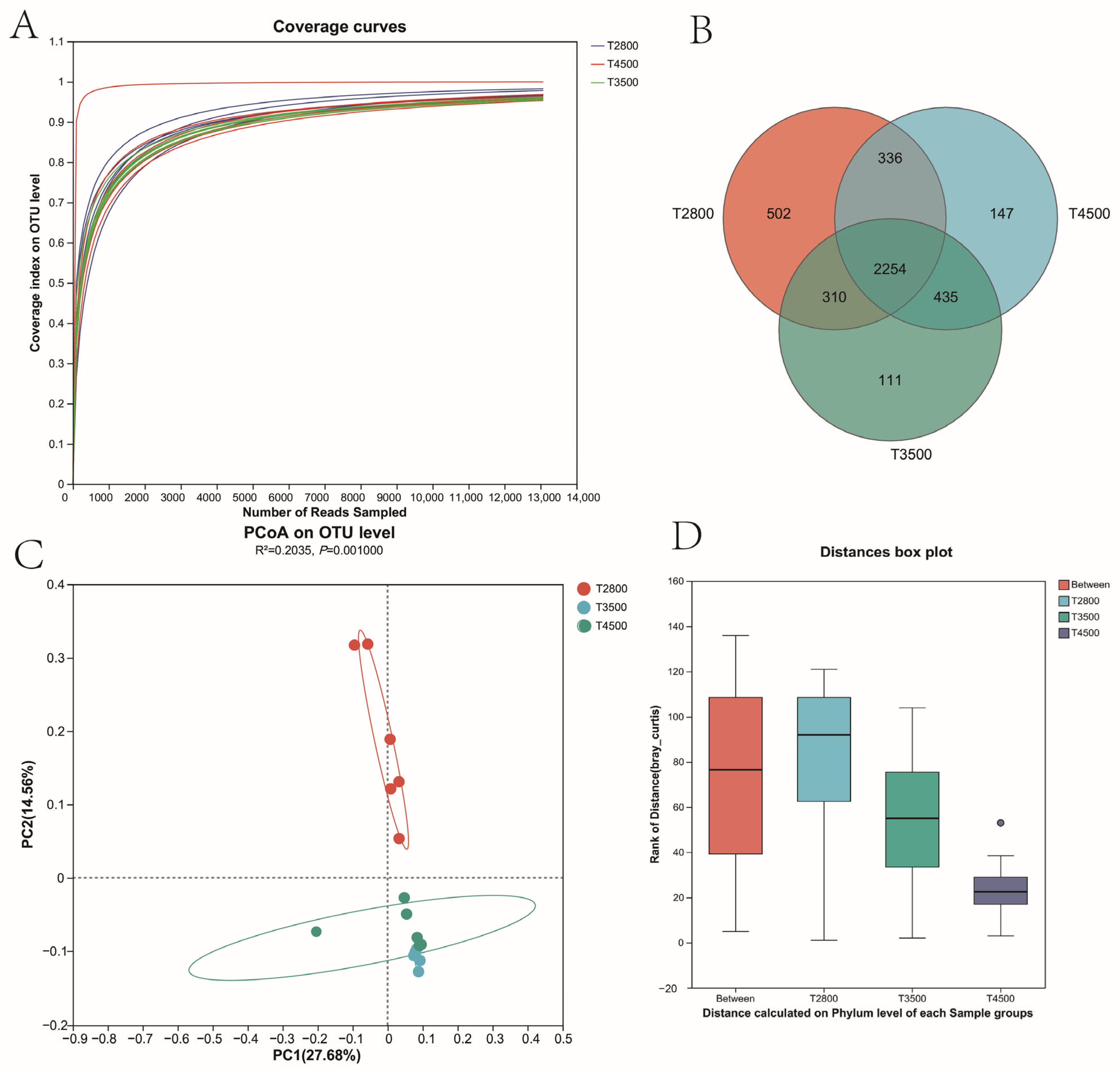
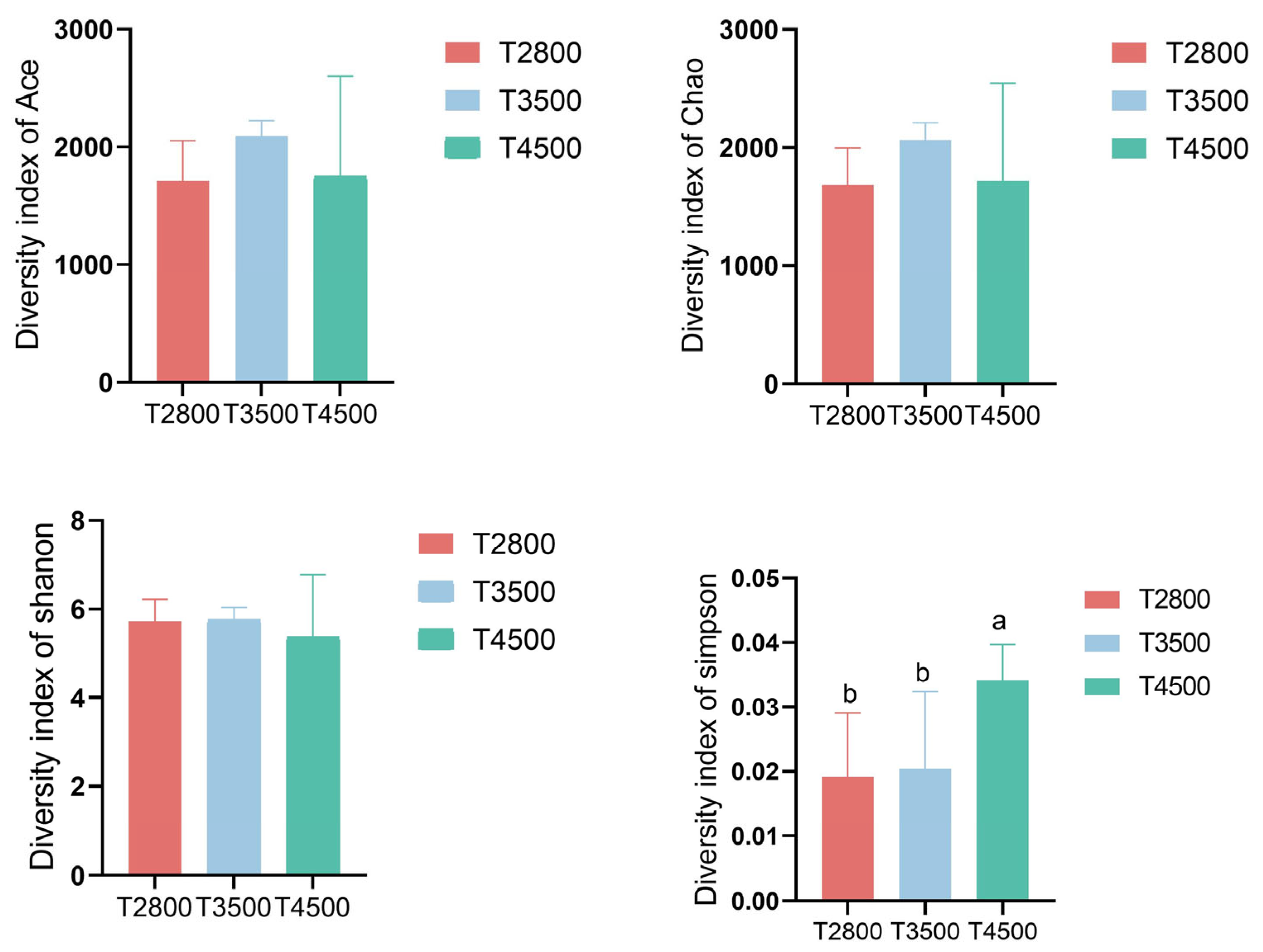
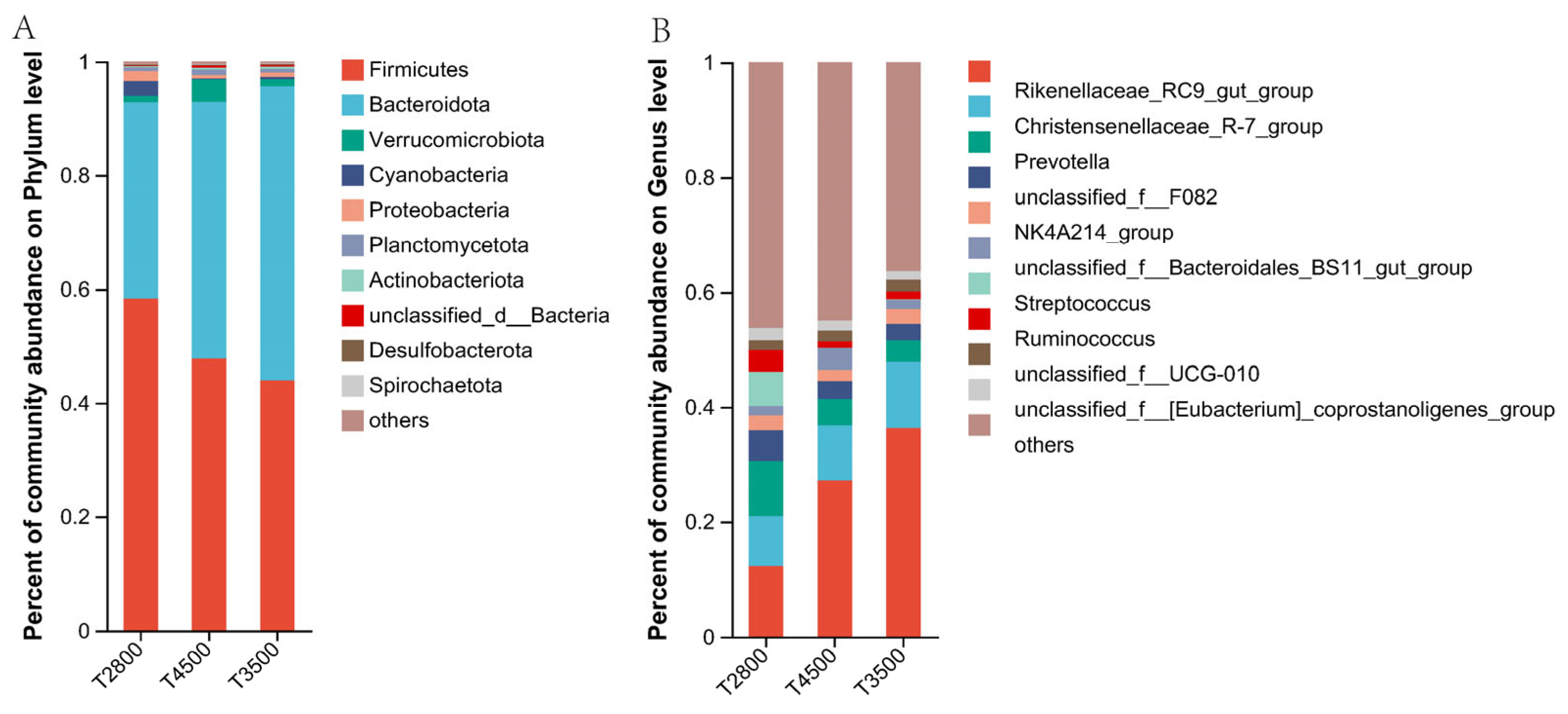
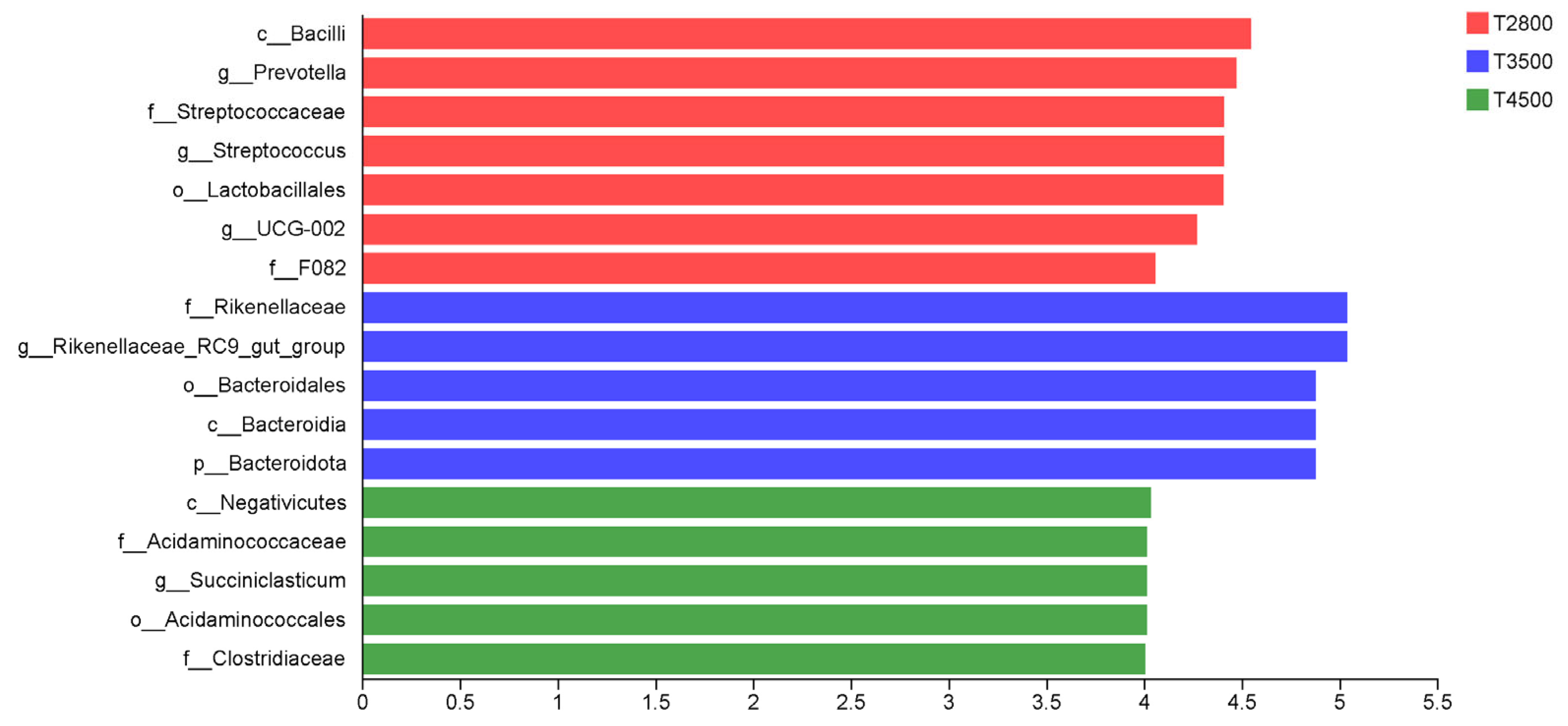
3.3. Analysis of Rumen Microbial Composition at Different Altitudes
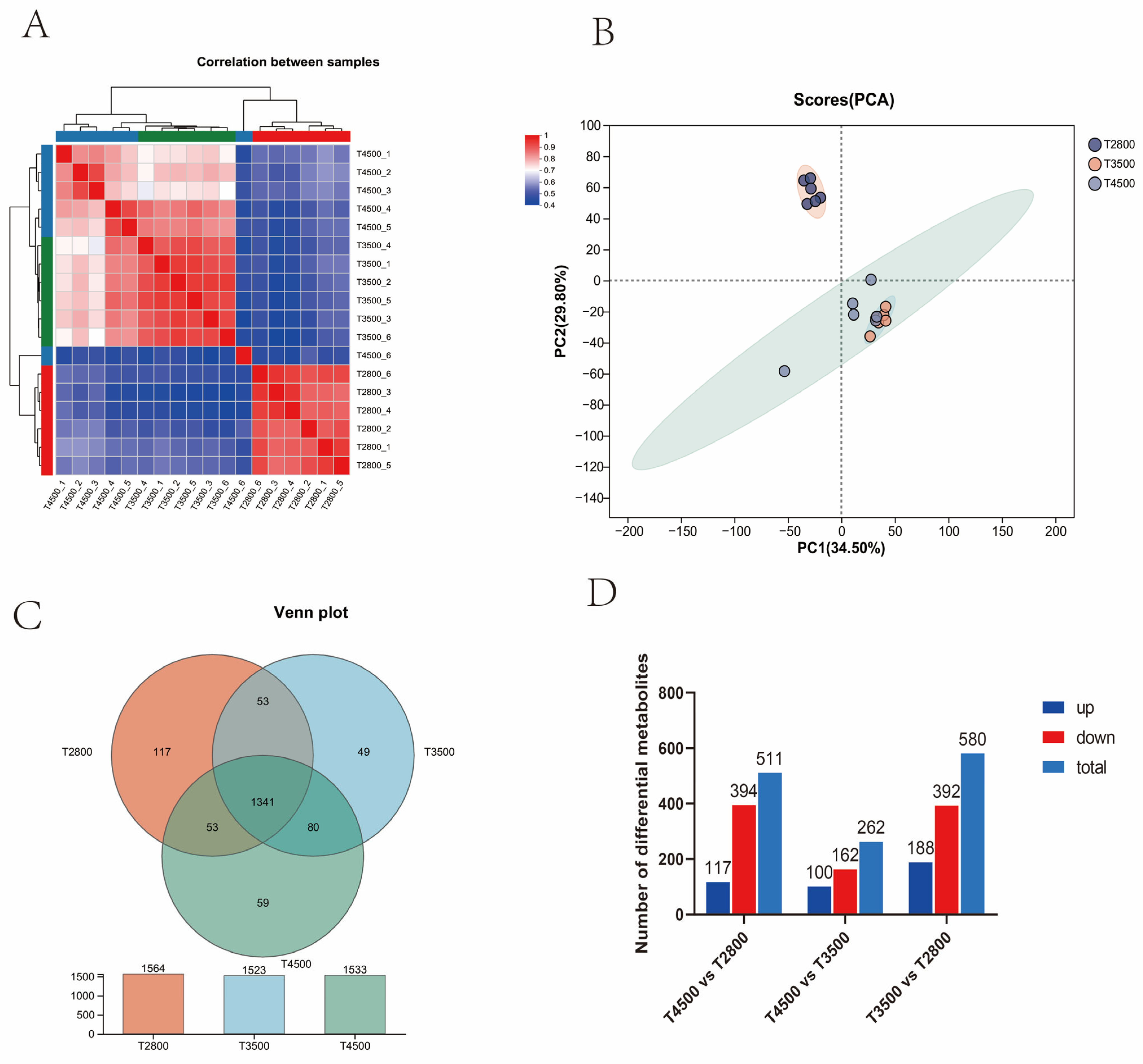
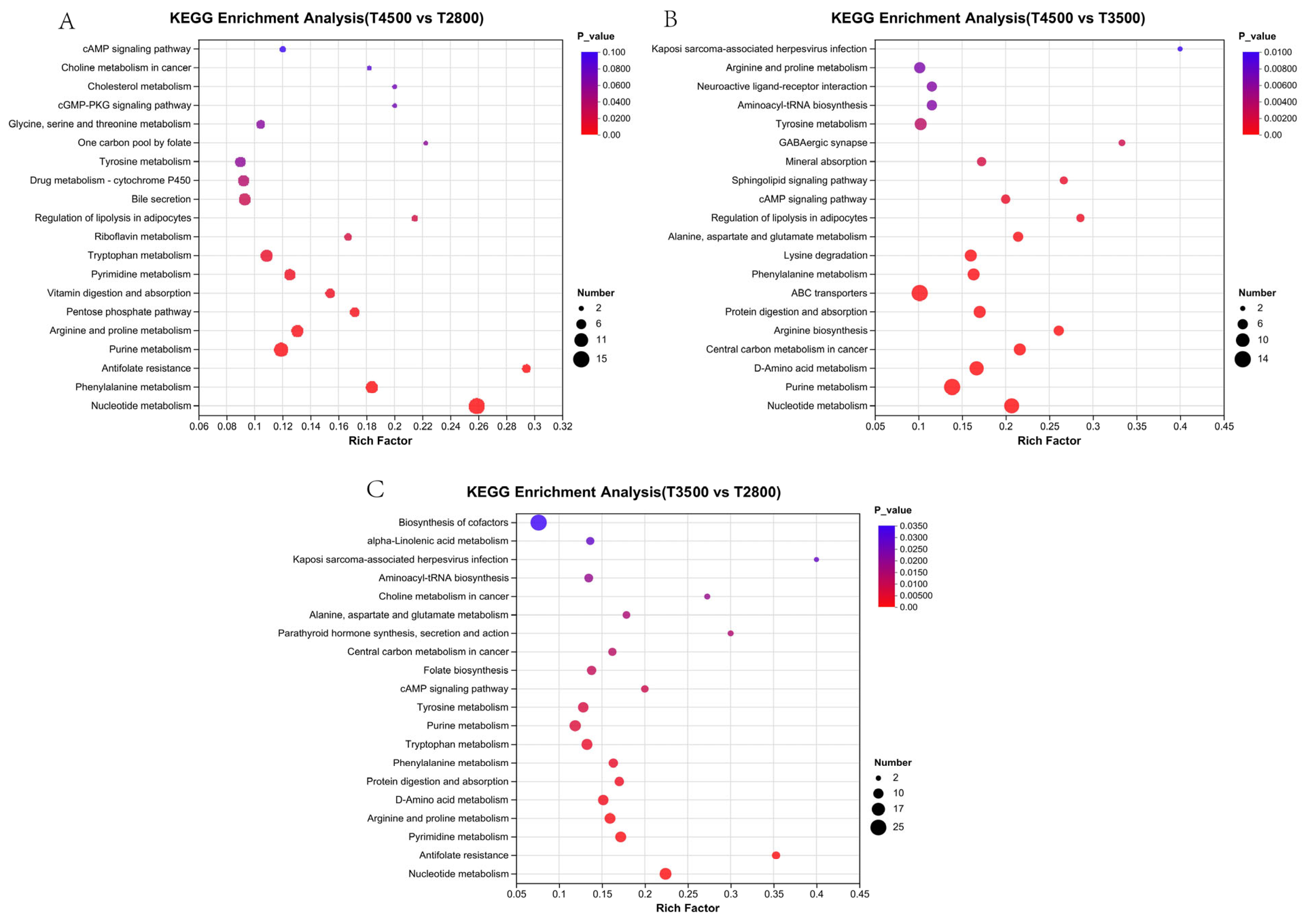
3.4. Characterization of Rumen Metabolites in Yaks at Different Altitudes
4. Discussion
4.1. Effect of Altitude on Rumen Microorganisms of Yak
4.2. Effect of Altitude on Microbial Diversity in the Rumen of Yak
4.3. Effect of Altitude on the Microbial Composition of the Rumen of Yaks
4.4. Effect of Altitude on Microbial Metabolites in the Rumen of Yak
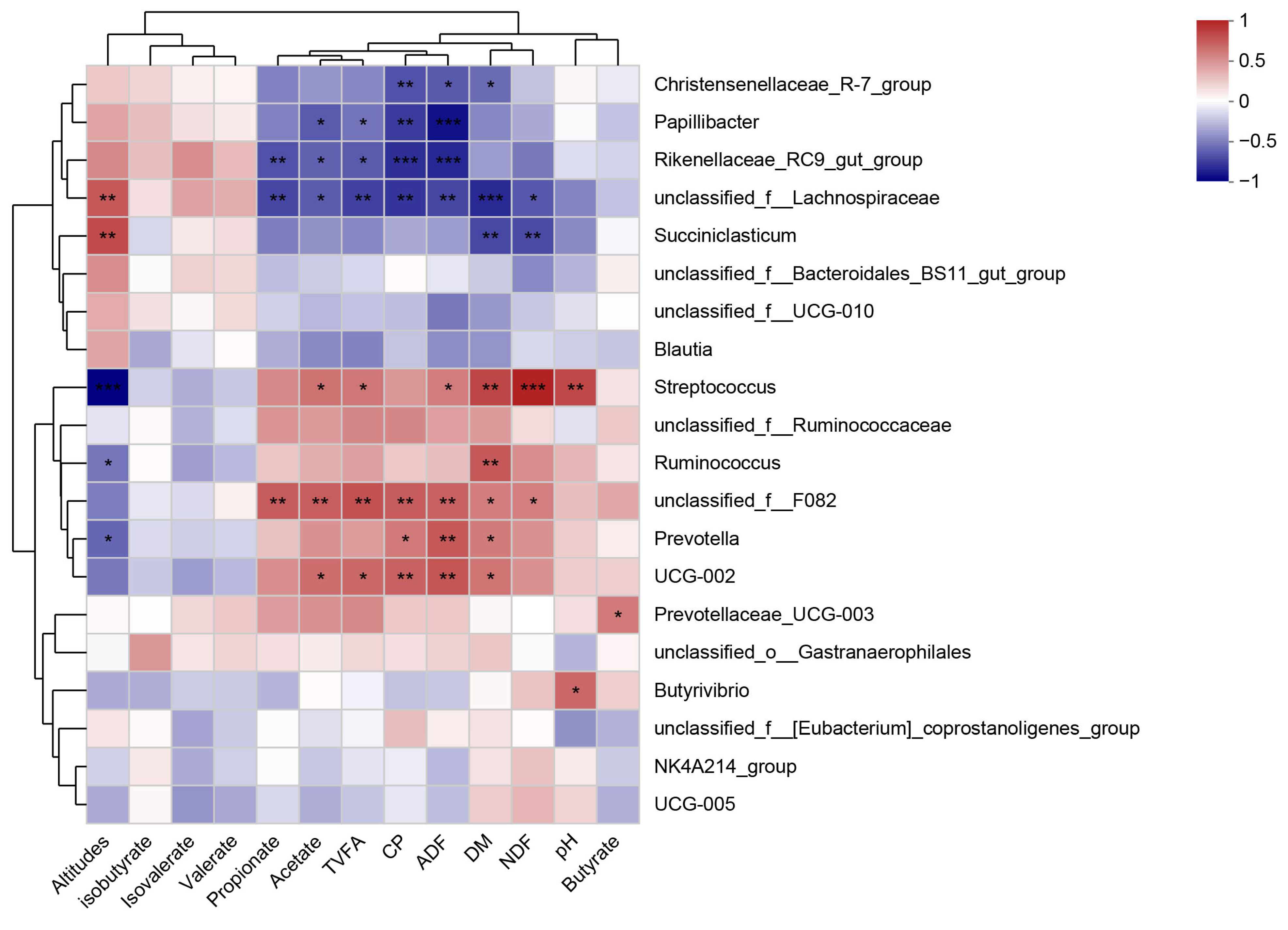
4.5. Correlation of Rumen Microorganisms, Metabolites, and Fermentation Products in Yak
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miao, F.H.; Guo, Z.G.; Xue, R.; Wang, X.Z.; Shen, Y.Y. Effects of Grazing and Precipitation on Herbage Biomass, Herbage Nutritive Value, and Yak Performance in an Alpine Meadow on the Qinghai-Tibetan Plateau. PLoS ONE 2015, 10, e0127275. [Google Scholar] [CrossRef] [PubMed]
- West, J.B. Recent Advances in High Altitude Medicine and Biology. High. Alt. Med. Biol. 2015, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Han, X.T.; Han, X.T.; Xie, A.Y.; Bi, X.C.; Liu, S.J.; Hu, L.H. Effects of high altitude and season on fasting heat production in the yak Bos grunniens or Poephagus grunniens. Br. J. Nutr. 2002, 88, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Pei, J.; Bao, P.; Chu, M.; Wu, X.; Ding, X.; Yan, P. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 2889–2890. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.J.; Ma, T.; Qian, W.B.; Wang, J.Y.; Ye, Z.Q.; Cao, C.C.; Hu, Q.J.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Chen, Y.B.; Fu, M.; Lan, D.L.; Li, J. Molecular Characterization and Expression Analysis of Insulin-like Growth Factor-1 and Insulin-like Growth Factor Binding Protein-1 Genes in Qinghai-Tibet Plateau Bos grunniens and Lowland Bos taurus. Asian-Australas. J. Anim. Sci. 2015, 28, 20–24. [Google Scholar] [CrossRef][Green Version]
- Han, N.; Pan, Z.; Liu, G.; Yang, R.; Yujing, B. Hypoxia: The “Invisible Pusher” of Gut Microbiota. Front. Microbiol. 2021, 12, 690600. [Google Scholar] [CrossRef]
- Wu, D.; Vinitchaikul, P.; Deng, M.; Zhang, G.; Sun, L.; Wang, H.; Gou, X.; Mao, H.; Yang, S. Exploration of the effects of altitude change on bacteria and fungi in the rumen of yak (Bos grunniens). Arch. Microbiol. 2021, 203, 835–846. [Google Scholar] [CrossRef]
- Rosenberg, E.; Zilber-Rosenberg, I. Do microbiotas warm their hosts? Gut Microbes 2016, 7, 283–285. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent Evolution of Rumen Microbiomes in High-Altitude Mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Zhou, Z.; Fang, L.; Meng, Q.; Li, S.; Chai, S.; Liu, S.; Schonewille, J.T. Assessment of Ruminal Bacterial and Archaeal Community Structure in Yak (Bos grunniens). Front. Microbiol. 2017, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wu, H.; Liu, S.; Chai, S.; Meng, Q.; Zhou, Z. Dynamic Alterations in Yak Rumen Bacteria Community and Metabolome Characteristics in Response to Feed Type. Front. Microbiol. 2019, 10, 1116. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J.; Rooke, J.A.; McKain, N.; Duthie, C.A.; Hyslop, J.J.; Ross, D.W.; Waterhouse, A.; Watson, M.; Roehe, R. The rumen microbial metagenome associated with high methane production in cattle. BMC Genom. 2015, 16, 839. [Google Scholar] [CrossRef]
- Li, F.; Hitch, T.C.A.; Chen, Y.; Creevey, C.J.; Guan, L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019, 7, 6. [Google Scholar] [CrossRef]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef]
- Midkiff, V.C. The History of Feed Analysis, as Chronicled in the Development of AOAC Official Methods, 1884–1984. J. Assoc. Off. Anal. Chem. 2020, 67, 851–860. [Google Scholar] [CrossRef]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Wang, X.; Shi, B.; Zuo, Z.; Qi, Y.; Zhao, S.; Zhang, X.; Lan, L.; Shi, Y.; Liu, X.; Li, S.; et al. Effects of Two Different Straw Pellets on Yak Growth Performance and Ruminal Microbiota during Cold Season. Animals 2023, 13, 335. [Google Scholar] [CrossRef]
- Sun, H.Z.; Wang, D.M.; Wang, B.; Wang, J.K.; Liu, H.Y.; Guan, L.L.; Liu, J.X. Metabolomics of four biofluids from dairy cows: Potential biomarkers for milk production and quality. J. Proteome Res. 2015, 14, 1287–1298. [Google Scholar] [CrossRef]
- Edwards, J.E.; McEwan, N.R.; Travis, A.J.; John Wallace, R. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 2004, 86, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Nauta, A.J.; Ben Amor, K.; Knippels, L.M.; Knol, J.; Garssen, J. Early life: Gut microbiota and immune development in infancy. Benef. Microbes 2010, 1, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Tantama, M.; Martinez-Francois, J.R.; Mongeon, R.; Yellen, G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 2013, 4, 2550. [Google Scholar] [CrossRef] [PubMed]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Penner, G.B.; Stumpff, F.; Gabel, G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J. Anim. Sci. 2011, 89, 1092–1107. [Google Scholar] [CrossRef]
- Sha, Y.; Ren, Y.; Zhao, S.; He, Y.; Guo, X.; Pu, X.; Li, W.; Liu, X.; Wang, J.; Li, S. Response of Ruminal Microbiota-Host Gene Interaction to High-Altitude Environments in Tibetan Sheep. Int. J. Mol. Sci. 2022, 23, 12430. [Google Scholar] [CrossRef]
- Zhou, M.; Peng, Y.J.; Chen, Y.; Klinger, C.M.; Oba, M.; Liu, J.X.; Guan, L.L. Assessment of microbiome changes after rumen transfaunation: Implications on improving feed efficiency in beef cattle. Microbiome 2018, 6, 62. [Google Scholar] [CrossRef]
- Nolan, J.V.; Leng, R.A.; Dobos, R.C.; Boston, R.C. The production of acetate, propionate and butyrate in the rumen of sheep: Fitting models to 14C- or 13C-labelled tracer data to determine synthesis rates and interconversions. Anim. Prod. Sci. 2014, 154, 2082–2088. [Google Scholar] [CrossRef]
- Xue, M.-Y.; Sun, H.-Z.; Wu, X.-H.; Liu, J.-X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 2049–2618. [Google Scholar] [CrossRef]
- Han, L.; Xue, W.; Cao, H.; Chen, X.; Qi, F.; Ma, T.; Tu, Y.; Diao, Q.; Zhang, C.; Cui, K. Comparison of Rumen Fermentation Parameters and Microbiota of Yaks From Different Altitude Regions in Tibet, China. Front. Microbiol. 2021, 12, 807512. [Google Scholar] [CrossRef]
- Zeng, B.; Zhang, S.Y.; Xu, H.L.; Kong, F.L.; Yu, X.Q.; Wang, P.; Yang, M.Y.; Li, D.Y.; Zhang, M.W.; Ni, Q.Y.; et al. Gut microbiota of Tibetans and Tibetan pigs varies between high and low altitude environments. Microbiol. Res. 2020, 235, 126447. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N.A.; Parks, D.H.; Vanwonterghem, I.; Morrison, M.; Tyson, G.W.; Hugenholtz, P. A Phylogenomic Analysis of the Bacterial Phylum Fibrobacteres. Front. Microbiol. 2016, 6, 1469. [Google Scholar]
- Khiaosa-ard, R.; Soliva, C.R.; Kreuzer, M.; Leiber, F. Effects of species-diverse high-alpine forage on ruminal fermentation when used as donor cow’s feed or directly incubated. Animal 2012, 6, 1764–1773. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Wu, Y.; Yao, Y.; Dong, M.; Xia, T.; Li, D.; Xie, M.; Wu, J.; Wen, A.; Wang, Q.; Zhu, G.; et al. Characterisation of the gut microbial community of rhesus macaques in high-altitude environments. BMC Microbiol. 2020, 20, 68. [Google Scholar] [CrossRef]
- Li, H.; Zhou, R.; Zhu, J.; Huang, X.; Qu, J. Environmental filtering increases with elevation for the assembly of gut microbiota in wild pikas. Microb. Biotechnol. 2019, 12, 976–992. [Google Scholar] [CrossRef]
- McCann, J.C.; Elolimy, A.A.; Loor, J.J. Rumen Microbiome, Probiotics, and Fermentation Additives. Vet. Clin. North. Am. Food Anim. Pract. 2017, 33, 539–553. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, S.; Chang, L.; Wang, H.J.; Ga, Q.; Ma, L.; Bai, Z.Z.; Shen, Y.Y.; Ge, R.L. Gut microbiota adaptation to high altitude in indigenous animals. Biochem. Biophys. Res. Commun. 2019, 516, 120–126. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, H.; Hao, L.; Cao, X.; Degen, A.; Zhou, J.; Zhang, C. Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine. Animals 2021, 11, 2425. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.Z.; Hu, J.; Shi, B.G.; Dingkao, R.Q.; Wang, J.Q.; Li, S.B.; Zhang, W.; Luo, Y.Z.; Liu, X. Characteristics and Functions of the Rumen Microbial Community of Cattle-Yak at Different Ages. Biomed. Res. Int. 2020, 2020, 3482692. [Google Scholar] [CrossRef] [PubMed]
- Cheviron, Z.A.; Bachman, G.C.; Connaty, A.D.; McClelland, G.B.; Storz, J.F. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc. Natl. Acad. Sci. USA 2012, 109, 8635–8640. [Google Scholar] [CrossRef] [PubMed]
- van Gylswyk, N.O. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 1995, 45, 297–300. [Google Scholar] [CrossRef]
- Raza, M.F.; Wang, Y.; Cai, Z.; Bai, S.; Yao, Z.; Awan, U.A.; Zhang, Z.; Zheng, W.; Zhang, H. Gut microbiota promotes host resistance to low-temperature stress by stimulating its arginine and proline metabolism pathway in adult Bactrocera dorsalis. PLoS Pathog. 2020, 16, e1008441. [Google Scholar] [CrossRef]
- Ge, R.-L.; Cai, Q.; Shen, Y.-Y.; San, A.; Ma, L.; Zhang, Y.; Yi, X.; Chen, Y.; Yang, L.; Huang, Y.; et al. Draft genome sequence of the Tibetan antelope. Nat. Commun. 2013, 4, 1858. [Google Scholar] [CrossRef]
- Evans, N.J.; Brown, J.M.; Murray, R.D.; Getty, B.; Birtles, R.J.; Hart, C.A.; Carter, S.D. Characterization of novel bovine gastrointestinal tract Treponema isolates and comparison with bovine digital dermatitis treponemes. Appl. Environ. Microbiol. 2011, 77, 138–147. [Google Scholar] [CrossRef]
- Ran, T.; Tang, S.X.; Yu, X.; Hou, Z.P.; Hou, F.J.; Beauchemin, K.A.; Yang, W.Z.; Wu, D.Q. Diets varying in ratio of sweet sorghum silage to corn silage for lactating dairy cows: Feed intake, milk production, blood biochemistry, ruminal fermentation, and ruminal microbial community. J. Dairy. Sci. 2021, 104, 12600–12615. [Google Scholar] [CrossRef]
- Malek, A.; Nasnas, R. An Unusual Presentation of Pseudothrombotic Microangiopathy in a Patient with Autoimmune Atrophic Gastritis. Case Rep. Hematol. 2016, 2016, 1087831. [Google Scholar] [CrossRef]
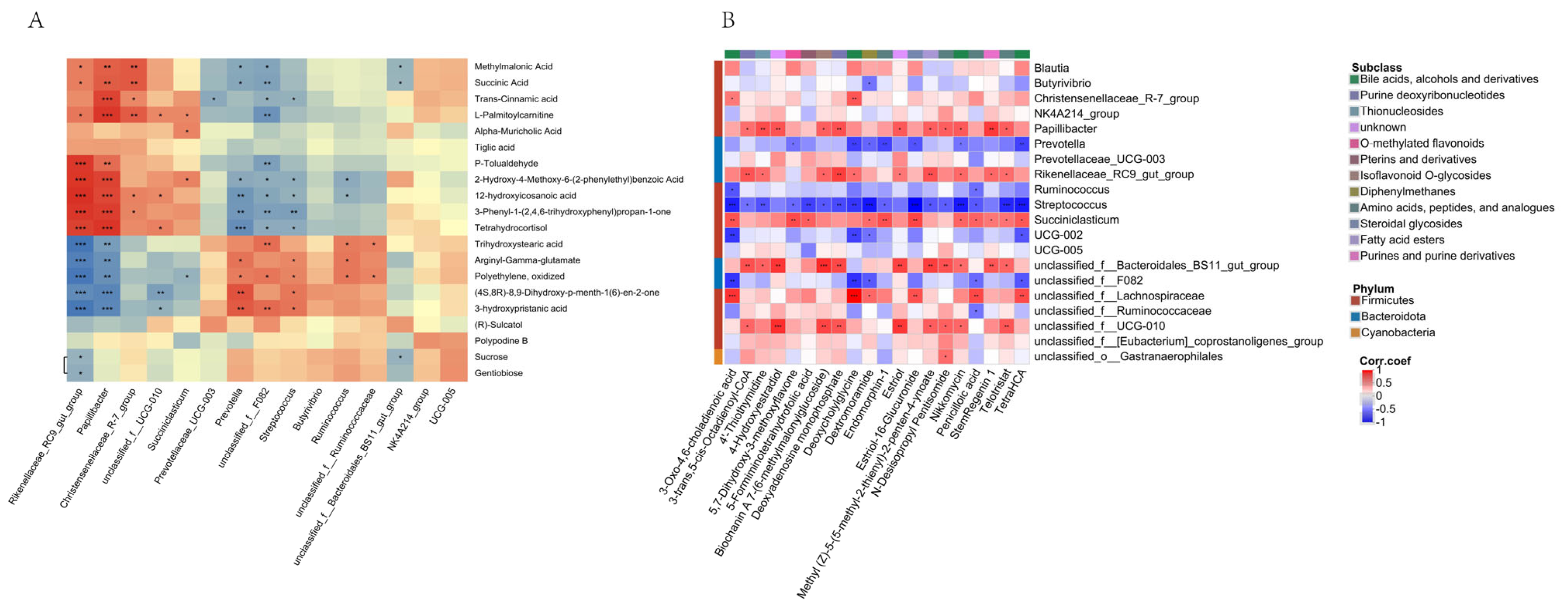
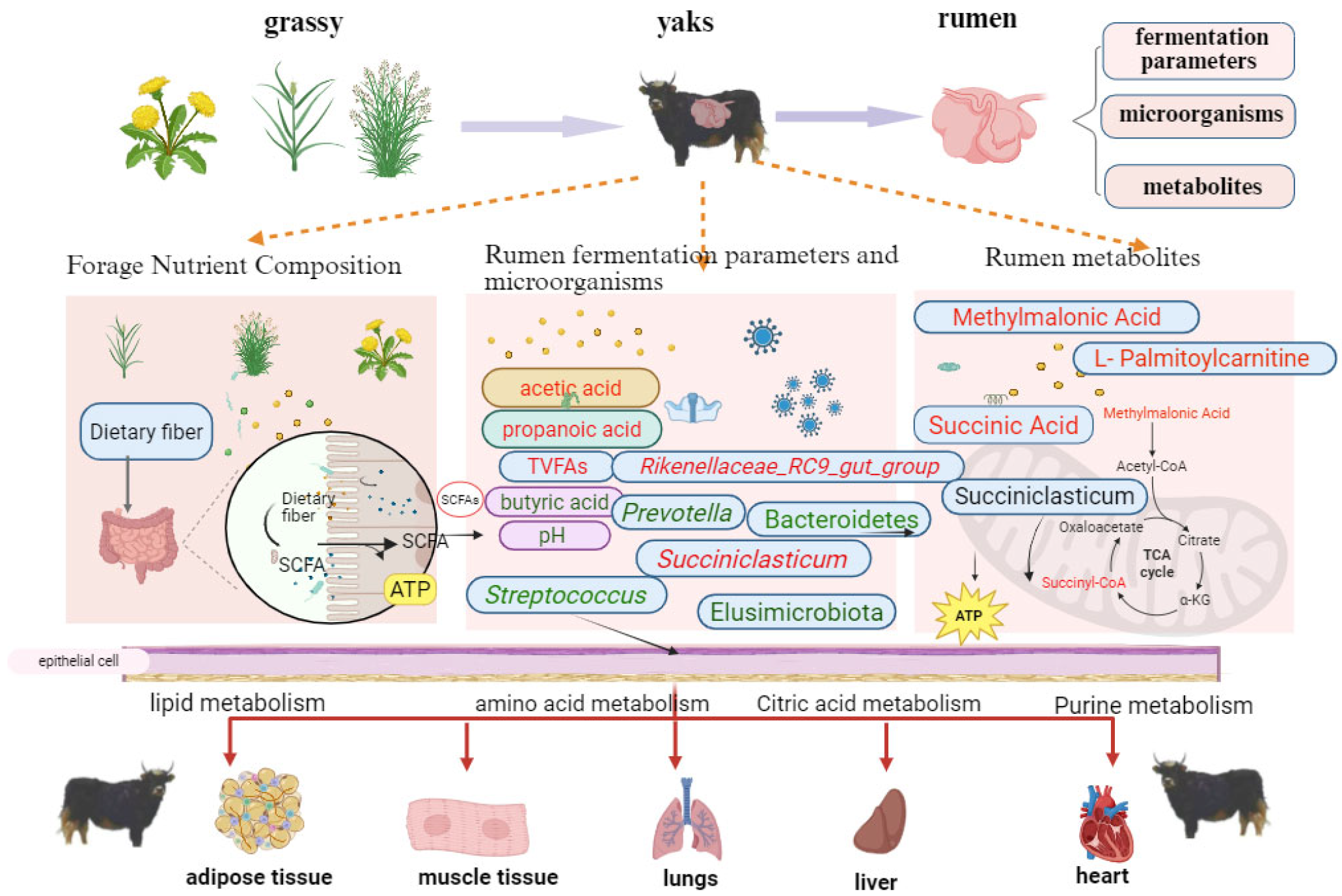
| Group | Dry Matter DM (%) | Crude Protein CP (%) | Ash (%) | Neutral Detergent Fiber NDF (%) | Acid Detergent Fiber ADF (%) | Ether Extract EE (%) |
|---|---|---|---|---|---|---|
| T2800 | 94.58 | 6.27 a | 5.88 b | 64.53 | 26.00 b | 8.03 a |
| T3500 | 93.73 | 4.67 c | 5.95 b | 67.60 | 20.33 c | 7.56 b |
| T4500 | 93.21 | 5.77 b | 7.64 a | 69.00 | 31.11 a | 8.06 a |
| SEM | 0.06 | 0.87 | 0.88 | 0.14 | 0.05 | 0.31 |
| p | 0.555 | 0.001 | 0.001 | 0.535 | 0.001 | 0.001 |
| Fermentation Parameters | T2800 | T3500 | T4500 | SEM | p-Value |
|---|---|---|---|---|---|
| pH | 7.69 a | 7.64 a | 7.22 b | 0.01 | 0.032 |
| NH3-N (mg/100 mL) | 0.30 | 0.42 | 0.52 | 0.02 | 0.053 |
| Acetic acid (%) | 18.169 b | 20.54 b | 27.53 a | 1.33 | 0.039 |
| Propionic acid (%) | 4.76 b | 4.10 b | 7.98 a | 0.48 | <0.001 |
| Isobutyrate (%) | 0.32 a | 0.16 b | 0.38 a | 0.18 | 0.037 |
| Butyric acid (%) | 3.13 | 2.96 | 3.04 | 0.08 | 0.965 |
| Isovalerate (%) | 0.31 b | 0.42 b | 1.19 a | 0.03 | <0.001 |
| Valerate (%) | 0.52 ± 0.01 ab | 0.27 ± 0.04 b | 0.75 ± 0.01 a | 0.01 | <0.001 |
| A/P | 4.61 ± 0.86 a | 5.05 ± 0.16 a | 3.46 ± 0.15 b | 0.29 | 0.042 |
| TVFA (mmol/L) | 28.27 ± 1.32 b | 28.47 ± 0.51 b | 39.65 ± 2.28 a | 0.90 | 0.045 |
| Phylum | T2800 | T3500 | T4500 | p | FDR |
|---|---|---|---|---|---|
| Bacteroidota | 34.51 ± 12.85 b | 51.27 ± 7.27 a | 45.09 ± 21.55 a | 0.048 | 0.100 |
| Fimicutes | 58.33 ± 8.12 a | 43.97 ± 6.91 b | 40.97 ± 2.70 b | 0.004 | 0.047 |
| p__unclassified_d__Bacteria | 0.09 ± 0.07 b | 0.26 ± 0.12 a | 0.35 ± 0.22 a | 0.042 | 0.047 |
| Campylobacterota | 0.07 ± 0.08 | 0.005 ± 0.003 | 0.01 ± 0.004 | 0.025 | 0.100 |
| WPS-2 | 0.00 ± 0.00 b | 0.03 ± 0.03 a | 0.06 ± 0.04 a | 0.008 | 0.047 |
| Genus | T2800 | T3500 | T4500 | p | FDR |
|---|---|---|---|---|---|
| Rikenellaceae_RC9_gut_group | 12.34 ± 6.40 b | 36.31 ± 6.78 a | 27.20 ± 14.17 a | 0.009 | 0.030 |
| Prevotella | 9.61 ± 5.46 a | 3.73 ± 1.35 b | 4.61 ± 3.83 b | 0.023 | 0.297 |
| Streptococcus | 6.02 ± 8.33 a | 0.13 ± 0.06 b | 0.04 ± 0.03 b | 0.001 | 0.201 |
| Succiniclasticum | 0.831 ± 0.16 b | 1.30 ± 0.66 b | 3.05 ± 3.34 a | 0.019 | 0.244 |
| Papillibacter | 0.59 ± 0.43 b | 1.67 ± 0.42 a | 1.12 ± 0.56 b | 0.011 | 0.087 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, J.; Ma, X.; Zhao, P.; Zhang, L.; Tao, S.; Wang, X.; Shi, B. Synergistic Adaptations of Yak Rumen Microbiota, Metabolites and Host to Altitudinal. Microorganisms 2025, 13, 1543. https://doi.org/10.3390/microorganisms13071543
Ren J, Ma X, Zhao P, Zhang L, Tao S, Wang X, Shi B. Synergistic Adaptations of Yak Rumen Microbiota, Metabolites and Host to Altitudinal. Microorganisms. 2025; 13(7):1543. https://doi.org/10.3390/microorganisms13071543
Chicago/Turabian StyleRen, Jianming, Xiong Ma, Pengfei Zhao, Lan Zhang, Shiyu Tao, Xiangyan Wang, and Bingang Shi. 2025. "Synergistic Adaptations of Yak Rumen Microbiota, Metabolites and Host to Altitudinal" Microorganisms 13, no. 7: 1543. https://doi.org/10.3390/microorganisms13071543
APA StyleRen, J., Ma, X., Zhao, P., Zhang, L., Tao, S., Wang, X., & Shi, B. (2025). Synergistic Adaptations of Yak Rumen Microbiota, Metabolites and Host to Altitudinal. Microorganisms, 13(7), 1543. https://doi.org/10.3390/microorganisms13071543






