Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures
Abstract
1. Introduction
| Salmonella Strain | Source | Biosafety Level (BSL) | Related Food | References |
|---|---|---|---|---|
| S. Typhimurium 53647 (Attenuated) | ATCC * (Avirulent strain) | 1 | ||
| S. Typhimurium 14028 | ATCC | 2 | Peanut butter, tomatoes, cantaloupes, cucumbers, basil, alfalfa, prepackaged salads, coconut, chicken salad, ground beef | [16] |
| S. Stanley H0558 | Outbreak in sprouts | 2 | Mushrooms, raw cashew cheese, peanuts | [16,17] |
| S. Senftenberg 8400 | ATTC | 2 | Infant formula, peanut butter, papayas, pistachios, smoked fish, tomatoes | [16,18,19,20] |
| S. Anatum F4317 | Outbreak in sprouts | 2 | Infant formula, basil, papaya, beef, pork, dried fish | [16,18,19,21,22] |
| S. Montevideo G4639 | Outbreak in tomato | 2 | Raw sprouts, pistachios, tahini sesame paste, Italian meats, salmon, cheese, turkey, chicken, eggs, sausage, beef, yogurt, pork | [16,19,23] |
| S. Enteritidis PT30 | Outbreak in raw almonds | 2 | Eggs, alfalfa, pine nuts, raw cookie dough, chicken products, peaches, bean sprouts, ground beef, cooked crab, mayonnaise, raw sausage, cake, pork, corn, potatoes, papaya, fish | [16,19,24] |
| S. Saint Paul 02-517-2 | Outbreak in cantaloupe | 2 | Ground beef, cucumbers, alfalfa, sprouts, raw produce, dried fish, turkey meat, paprika, cantaloupes, chicken, eggs, orange juice, confectionaries, jalapeño pepper | [16,19,25] |
2. Materials and Methods
2.1. Microbial Strains
2.2. Coupons
2.3. Biofilm Formation
2.4. Enumeration of Planktonic and Sessile Cells
2.5. Biofilm Index
2.6. Sequencing
2.7. Bioinformatics
2.8. Electron Microscopy
2.9. Statistical Analysis
3. Results and Discussion
3.1. Effect of Temperature
3.2. Effect of Food-Contact Surface
3.3. Effect of Bacterial Strain
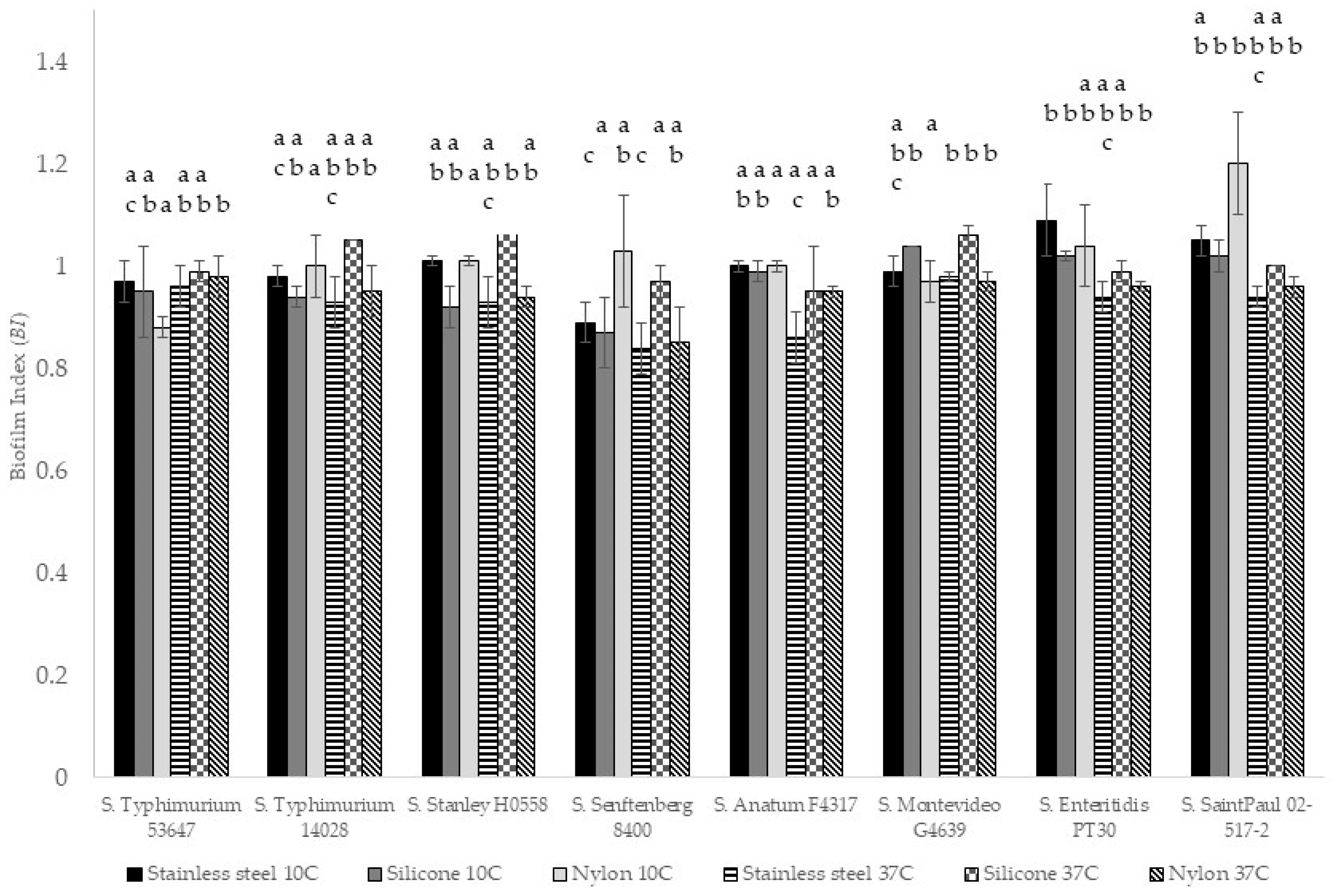
3.4. Biofilm Index (BI)
3.5. Genetic Variations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Statement
Abbreviations
| ATCC | American Type Culture Collection |
| BI | Biofilm index |
| BPW | Buffered peptone water |
| cfu | Colony-forming units |
| DNA | Deoxyribonucleic acid |
| g | Gravity |
| mL | Milliliter |
| µL | Microliter |
| ng | Nanogram |
| ONT | Oxford Nanopore Technologies |
| rpm | Revolutions per minute |
| SNP | Single nucleotide polymorphism |
| TSB | Tryptic soy broth |
| TSBYE | Tryptic soy broth with 0.6% yeast extract |
| XLD | Xylose lysine deoxycholate |
| YE | Yeast extract |
References
- Dhakal, J.; Sharma, C.; Nannapaneni, R.; McDaniel, C.; Kim, T.; Kiess, A. Effect of chlorine-induced sublethal oxidative stress on the biofilm-forming ability of Salmonella at different temperatures, nutrient conditions, and substrates. J. Food Prot. 2019, 82, 78–92. [Google Scholar] [CrossRef]
- Nguyen, H.D.N.; Yang, Y.S.; Yuk, H.G. Biofilm formation of Salmonella Typhimurium on stainless steel and acrylic surfaces as affected by temperature and pH level. LWT-Food Sci. Technol. 2014, 55, 383–388. [Google Scholar] [CrossRef]
- Roy, P.; Ha, A.; Mizan, M.; Hossain, M.; Ashrafudoulla, M.; Toushik, S.; Nahar, S.; Kim, Y.; Ha, S. Effects of environmental conditions (temperature, pH, and glucose) on biofilm formation of Salmonella enterica serotype Kentucky and virulence gene expression. Poult. Sci. 2021, 100, 101209. [Google Scholar] [CrossRef]
- Lianou, A.; Koustsoumanis, K. Strain variability of the biofilm-forming ability of Salmonella enterica under various experimental conditions. Int. J. Food Microbiol. 2012, 160, 171–178. [Google Scholar] [CrossRef]
- Niemira, B.A.; Boyd, G.; Sites, J. Cold plasma rapid decontamination of food contact surfaces contaminated with Salmonella biofilms. J. Food Sci. 2014, 79, M917–M922. [Google Scholar] [CrossRef]
- Niemira, B. Irradiation sensitivity of planktonic and biofilm-associated Escherichia coli O157:H7 isolates is influenced by culture conditions. Appl. Environ. Microbiol. 2007, 73, 3239–3244. [Google Scholar] [CrossRef]
- Solomon, E.; Niemira, B.; Sapers, G.; Annous, B. Biofilm formation, cellulose production, and curli biosynthesis by Salmonella originating from produce, animal and clinical sources. J. Food Prot. 2005, 68, 906–912. [Google Scholar] [CrossRef]
- Yang, Y.; Mikš-Krajnik, M.; Zheng, Q.; Lee, S.; Lee, S.; Yuk, H. Biofilm formation of Salmonella Enteritidis under food-related environmental stress conditions and its subsequent resistance to chlorine treatment. Food Microbiol. 2016, 54, 98–105. [Google Scholar] [CrossRef]
- Borges, K.; Furian, T.; Souza, S.; Menezes, R.; Tondo, E.; Salle, C.; Moraes, H.; Nascimento, V. Biofilm formation capacity of Salmonella serotypes at different temperature conditions. Pesqui. Vet. Bras. 2018, 38, 71–76. [Google Scholar] [CrossRef]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Römling, U. Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef]
- Gerstel, U.; Römling, U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 2003, 154, 659–667. [Google Scholar] [CrossRef]
- Pringent-Combaret, C.; Brombacher, E.; Vidal, O.; Ambert, A.; Lejeune, P.; Landini, P.; Dovel, C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 2001, 183, 7213–7223. [Google Scholar] [CrossRef]
- Romling, U.; Bian, Z.; Hammar, M.; Sierralta, W.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 1998, 180, 722–731. [Google Scholar] [CrossRef]
- Han, J.; Aljahdali, N.; Zhao, S.; Tang, H.; Harbottle, H.; Hoffmann, M.; Frye, J.; Foley, S. Infection biology of Salmonella enterica. EcoSal Plus 2024, 12, eesp-0001-2023. [Google Scholar] [CrossRef]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 2015, 520179. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention. 2025. Available online: www.cdc.gov (accessed on 11 March 2025).
- Kirk, M.; Little, C.; Lem, M.; Fufe, D.; Genobile, D.; Tan, A.; Threlfall, J.; Paccagnella, A.; Lightfoot, D.; Lyi, H. An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport—Sharing molecular information to solve international outbreaks. Epidemiol. Infect. 2004, 132, 571–577. [Google Scholar] [CrossRef]
- Leuschner, R.; Bew, J.; Boughtflower, M. A collaborative study to evaluate qualitatively powdered baby food validation samples artificially contaminated with Salmonella anatum. Int. J. Food Microbiol. 2004, 97, 43–51. [Google Scholar] [CrossRef]
- Heinitz, M.; Ruble, R.; Wagner, D.; Tatini, S. Incidence of Salmonella in fish and seafood. J. Food Prot. 2000, 63, 579–592. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control, European Food Safety Authority. Multi-Country Outbreak of Salmonella Senftenberg ST14 Infections, Possibly Linked to Cherry-like Tomatoes. 27 July 2023. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/ROA_S_Senftenberg-ST15_2023-FWD-00009.pdf (accessed on 3 March 2025).
- Reynoso, E.; Delgado-Suarez, E.; Hernandez-Perez, C.; Chavarin-Pineda, Y.; Godoy-Lozano, E.; Fierros-Zarate, G.; Aguilar-Vera, O.; Castillo-Ramirez, S.; Gomez-Pedroso, L.; Sanchez-Zamorano, L. Geography, antimicrobial resistance, and genomics of Salmonella enterica (Serotypes Newport and Anatum) from meat in Mexico (2021–2023). Microorganisms 2024, 12, 2485. [Google Scholar] [CrossRef]
- Pakalniskiene, J.; Falkenhorst, G.; Lisby, M.; Madsen, S.; Olsen, K.; Nielsen, E.; Mygh, A.; Boel, J.; Mølbak, K. A foodborne outbreak of enterotoxigenic E. coli and Salmonella Anatum infection after a high-school dinner in Denmark, November 2006. Epidemiol. Infect. 2009, 137, 396–401. [Google Scholar] [CrossRef]
- Dominguez, M.; Jourdan-Da Silva, N.; Vaillant, V.; Pihier, N.; Kermin, C.; Weill, F.; Delmas, G.; Kerouanton, A.; Brisabois, A.; de Valk, H. Outbreak of Salmonella enterica serovar Montevideo infections in France linked to consumption of cheese made from raw milk. Foodborne Pathog. Dis. 2009, 6, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Moratto Bergaimini, A.; Falcao, J. Genetic diversity, virulence genes, and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over 24-year period in Brazil. Food Microbiol. 2012, 32, 254–264. [Google Scholar] [CrossRef]
- Hayford, A.; Brown, E.; Zhao, S.; Mammel, M.; Gangiredla, J.; Abbott, J.; Friedman, S.; Ayers, S.; Lewis, J.; Lacher, D.; et al. Genetic and resistance phenotypic subtyping of Salmonella Saintpaul isolates from various food sources and humans: Phylogenetic concordance in combinatory analysis. Infect. Genet. Evol. 2015, 36, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Aguirre, D.; Uknalis, J.; Niemira, B.A. Attachment and removal of biofilm of Salmonella Typhimurium embedded in liquid whole egg on stainless steel, silicone, and nylon. Food Control 2025, 171, 111104. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Stepanović, S.; Ćirković, I.; Mijač, V.; Srabić-Vlahović, M. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Obe, T.; Richards, A.; Shariat, N. Differences in biofilm formation of Salmonella serovars on two surfaces under two temperature conditions. J. Appl. Microbiol. 2022, 132, 2410–2420. [Google Scholar] [CrossRef]
- Speranza, B.; Corbo, M.; Sinigaglia, M. Effects of nutritional and environmental conditions on Salmonella sp. biofilm formation. J. Food Sci. 2010, 76, M12–M16. [Google Scholar] [CrossRef]
- Joseph, B.; Otta, S.; Karunasagar, I.; Karunasagar, I. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food Microbiol. 2001, 64, 367–372. [Google Scholar] [CrossRef]
- Schlisselberg, D.; Yaron, S. The effects of stainless steel finish on Salmonella Typhimurium attachment, biofilm formation and sensitivity to chlorine. Food Microbiol. 2013, 35, 65–72. [Google Scholar] [CrossRef]
- Shatila, F.; Yaḉa, I.; Yalḉin, H.T. Biofilm formation by Salmonella enterica strains. Curr. Microbiol. 2021, 78, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Fabrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, K.; Daigle, F. Salmonella fimbriae: What is the clue to their hairdo? In Current Topics in Salmonella and Salmonellosis; InTech: London, UK, 2017. [Google Scholar]
- Yue, M.; Rankin, S.; Blanchet, R.; Nulton, J.; Edwards, R.; Schifferli, D. Diversification of the Salmonella fimbriae: A model of macro- and microevolution. PLoS ONE 2012, 7, e38596. [Google Scholar] [CrossRef]
- Barnhart, M.; Chapman, M. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef]
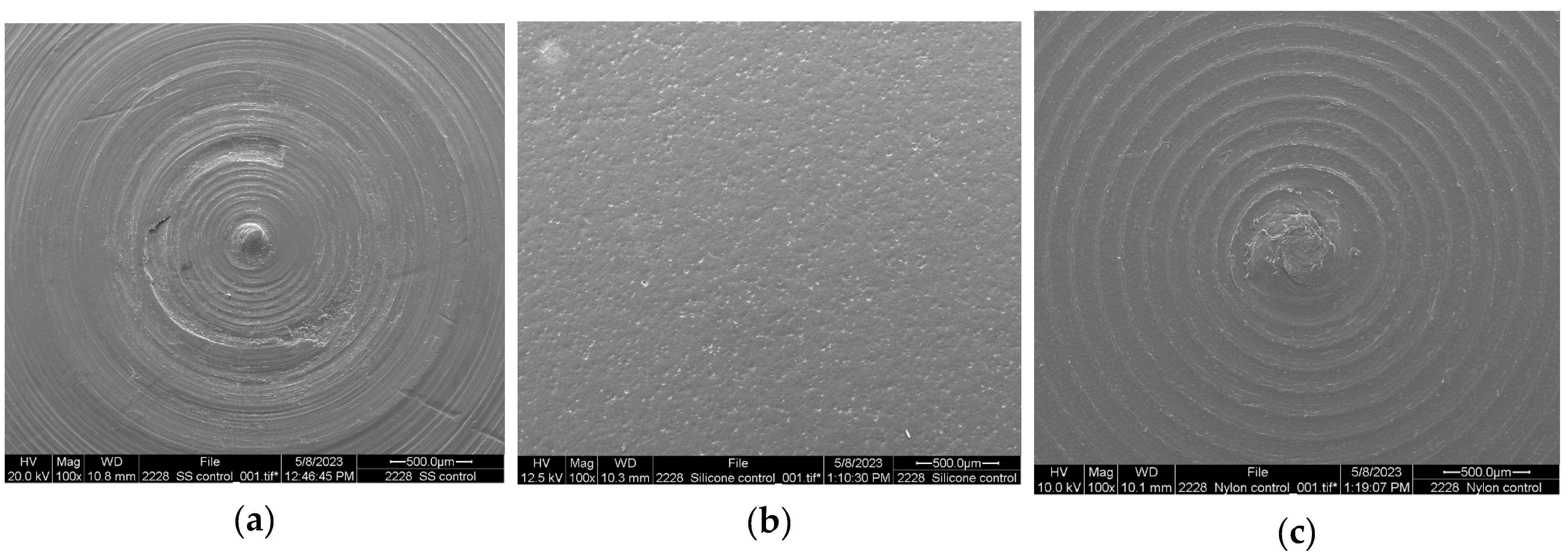
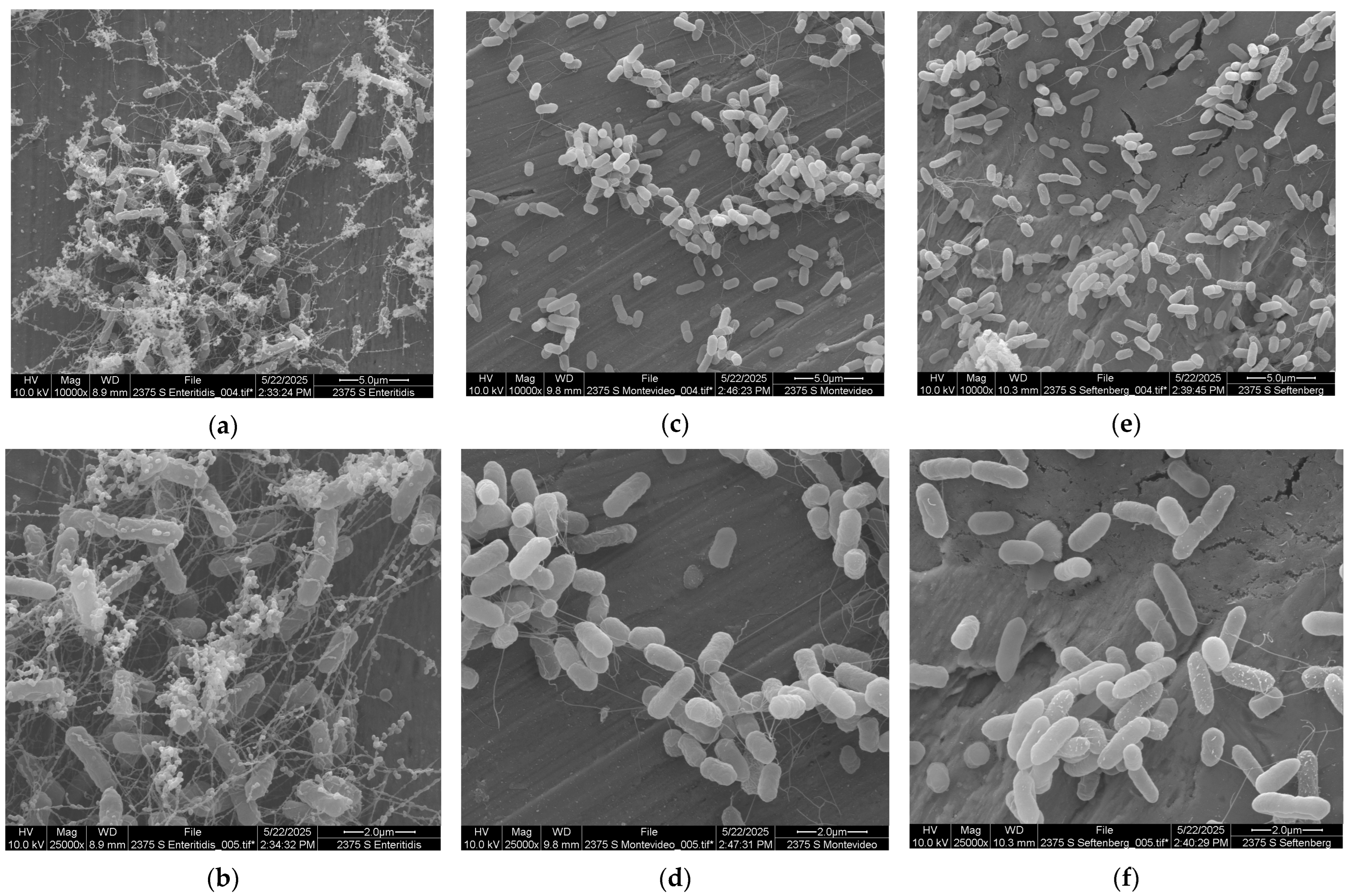
| 10 °C | Log N | Log N | Log N |
|---|---|---|---|
| Stainless Steel | Silicone | Nylon | |
| S. Typhimurium 53647 | 5.16 abA (±0.28) | 5.08 abA (±0.63) | 4.53 aA (±0.13) |
| S. Typhimurium 14028 | 5.06 abC (±0.14) | 5.00 abC (±0.22) | 4.80 aC (±0.47) |
| S. Stanley H0558 | 5.44 abB (±0.37) | 4.89 abB (±0.33) | 5.29 abB (±0.16) |
| S. Senftenberg 8400 | 4.77 aCD (±0.42) | 4.26 aC (±0.81) | 4.83 abACD (±0.42) |
| S. Anatum F4317 | 5.36 abAC (±0.18) | 5.23 abAC (±0.13) | 4.93 abA (±0.29) |
| S. Montevideo G4639 | 5.37 abA (±0.16) | 5.43 bA (±0.12) | 5.15 abA (±0.04) |
| S. Enteritidis PT30 | 5.53 bA (±0.11) | 5.27 abA (±0.30) | 5.27 abA (±0.30) |
| S. SaintPaul 02-517-2 | 5.49 bA (±0.01) | 5.47 bA (±0.15) | 5.56 bA (±0.13) |
| 37 °C | |||
| S. Typhimurium 53647 | 7.15 abB (±0.37) | 7.30 abB (±0.17) | 7.28 bB (±0.30) |
| S. Typhimurium 14028 | 6.65 abcA (±0.44) | 7.62 bB (±0.04) | 7.11 abAB (±0.23) |
| S. Stanley H0558 | 6.90 abcA (±0.19) | 7.72 b (±0.05) | 7.01 abA (±0.02) |
| S. Senftenberg 8400 | 6.20 acABD (±0.58) | 6.93 aB (±0.29) | 6.31 aAB (±0.68) |
| S. Anatum F4317 | 6.03 cC (±0.43) | 6.96 aB (±0.52) | 6.91 abB (±0.13) |
| S. Montevideo G4639 | 7.37 bB (±0.28) | 7.78 bB (±0.07) | 7.29 bB (±0.01) |
| S. Enteritidis PT30 | 7.19 bB (±0.08) | 7.33 abB (±0.00) | 7.27 bB (±0.01) |
| S. SaintPaul 02-517-2 | 7.06 abB (±0.03) | 7.43 abB (±0.18) | 7.23 bB (±0.25) |
| Serovar | Number | faeC | faeD | faeE | lpfA | lpfB | lpfC | lpfE |
|---|---|---|---|---|---|---|---|---|
| S. Typhimurium 53647 (Attenuated) | 117 | − | − | − | + | + | + | + |
| S. Typhimurium 14028 | 118 | − | − | − | + | + | + | + |
| S. Stanley H0558 | 105 | + | + | + | + | + | + | + |
| S. Senftenberg 8400 | 101 | − | − | − | + | + | + | + |
| S. Anatum F4317 | 111 | − | − | − | + | + | + | + |
| S. Montevideo G4639 | 99 | + | + | + | − | − | − | − |
| S. Enteritidis PT30 | 112 | − | − | − | + | + | + | + |
| S. Saint Paul 02-517-2 | 109 | + | + | + | + | + | + | + |
| Strain | Gene | Single Nucleotide Polymorphism Type |
|---|---|---|
| S. Typhimurium 53647 (Attenuated) | ||
| S. Typhimurium 14028 | ||
| S. Stanley H0558 | ||
| S. Senftenberg 8400 | csgF lpfA | 1 noncoding 5 noncoding, 1 synonymous |
| S. Anatum F4317 | ||
| S. Montevideo G4639 | csgB csgC | 2 missense 2 missense, 1 synonymous |
| S. Enteritidis PT30 | ||
| S. Saint Paul 02-517-2 | csgB | 1 missense |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Counihan, K.L.; Tilman, S.; Uknalis, J.; Mukhopadhyay, S.; Niemira, B.A.; Bermudez-Aguirre, D. Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures. Microorganisms 2025, 13, 1446. https://doi.org/10.3390/microorganisms13071446
Counihan KL, Tilman S, Uknalis J, Mukhopadhyay S, Niemira BA, Bermudez-Aguirre D. Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures. Microorganisms. 2025; 13(7):1446. https://doi.org/10.3390/microorganisms13071446
Chicago/Turabian StyleCounihan, Katrina L., Shannon Tilman, Joseph Uknalis, Sudarsan Mukhopadhyay, Brendan A. Niemira, and Daniela Bermudez-Aguirre. 2025. "Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures" Microorganisms 13, no. 7: 1446. https://doi.org/10.3390/microorganisms13071446
APA StyleCounihan, K. L., Tilman, S., Uknalis, J., Mukhopadhyay, S., Niemira, B. A., & Bermudez-Aguirre, D. (2025). Attachment and Biofilm Formation of Eight Different Salmonella Serotypes on Three Food-Contact Surfaces at Different Temperatures. Microorganisms, 13(7), 1446. https://doi.org/10.3390/microorganisms13071446






