Molecular Identification and Genotyping of Phytoplasmas Infecting Medicinal and Aromatic Plants in Northern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Surveys and Plant Sample Collection
2.2. Phytoplasma Identification
2.3. Molecular Typing of Identified Phytoplasmas
3. Results and Discussion
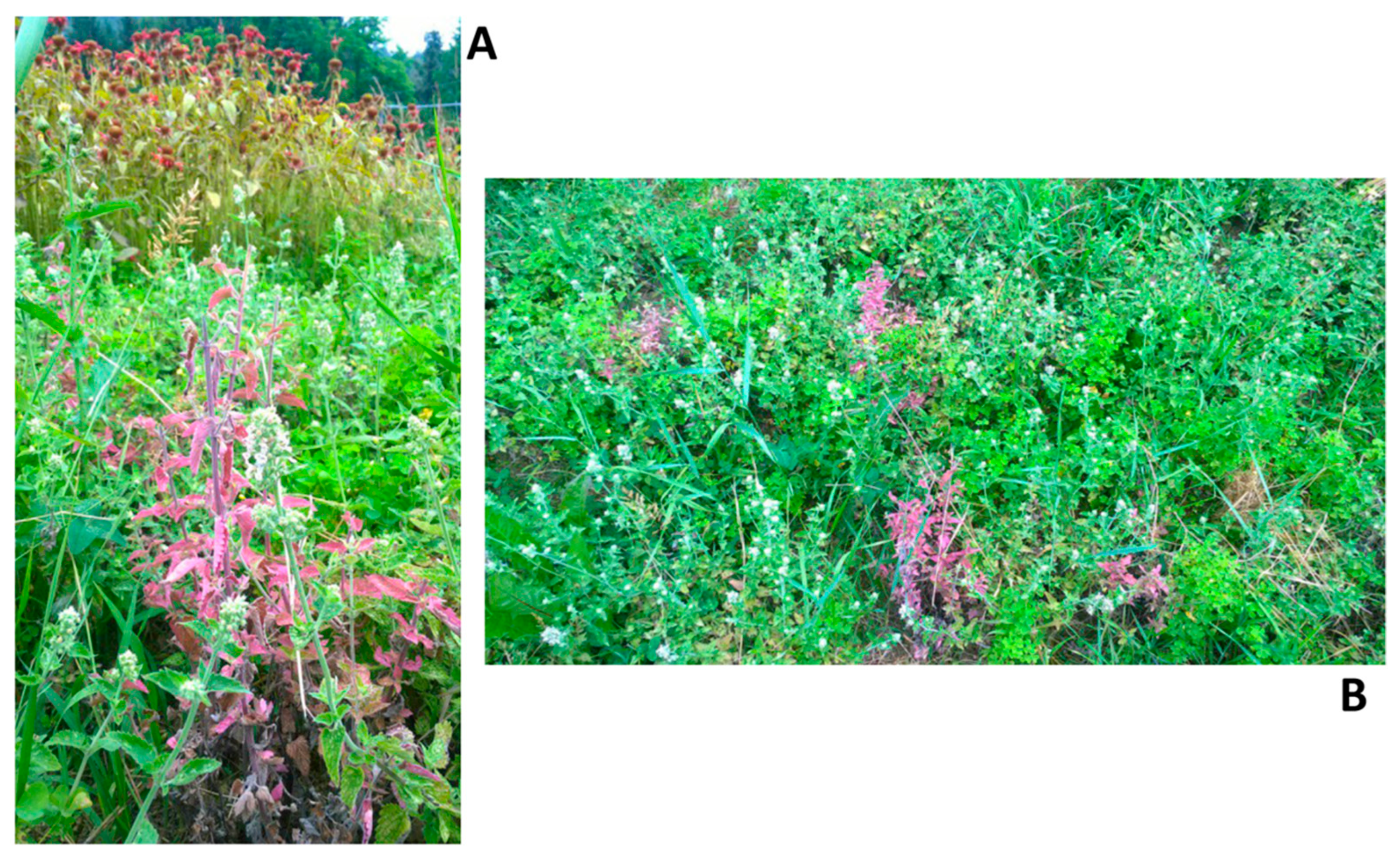
3.1. Phytoplasma-like Symptoms Observed During Field Surveys
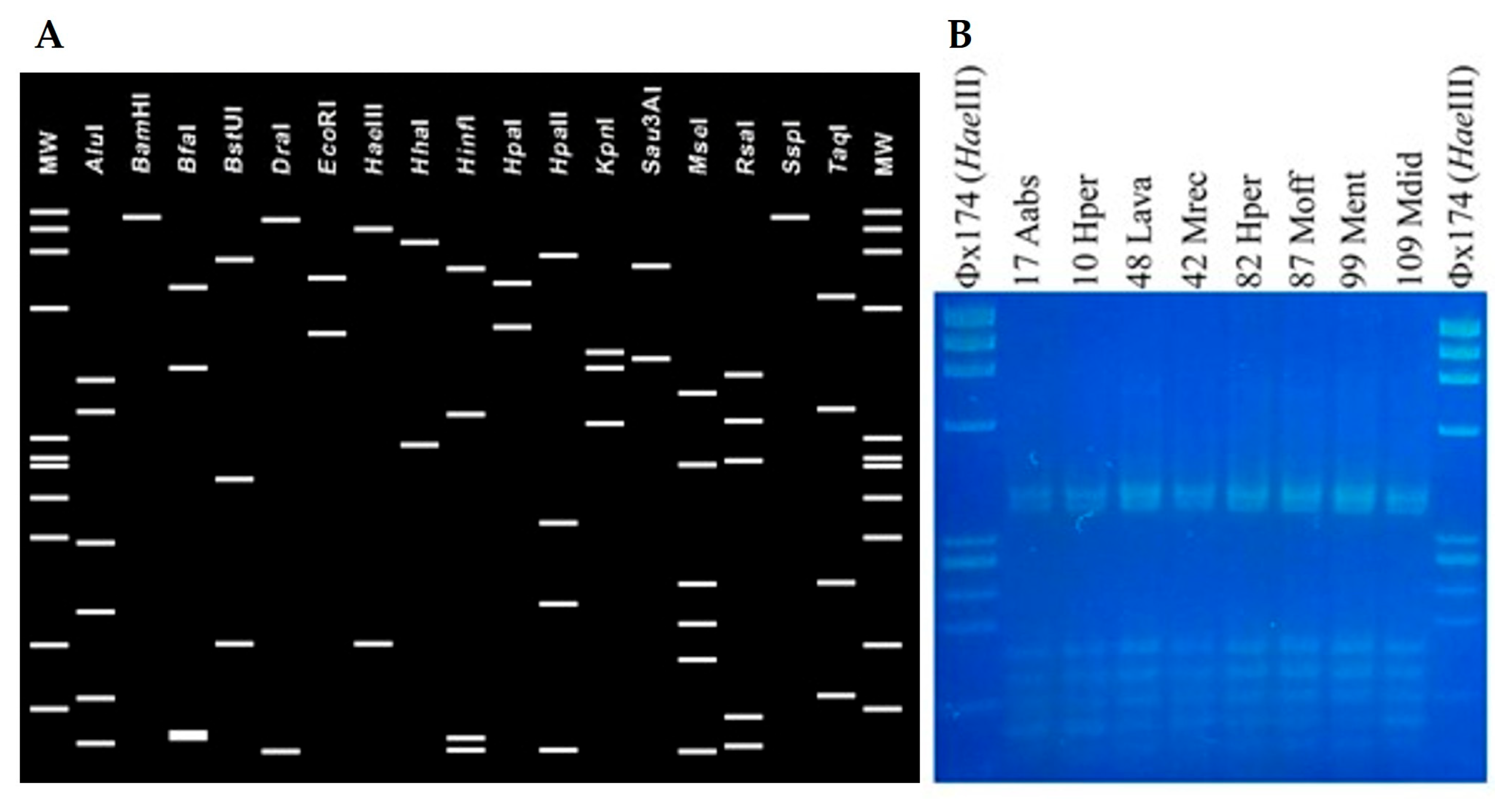
3.2. Identification of ‘Ca. Phytoplasma solani’ in Aromatic and Medicinal Plants
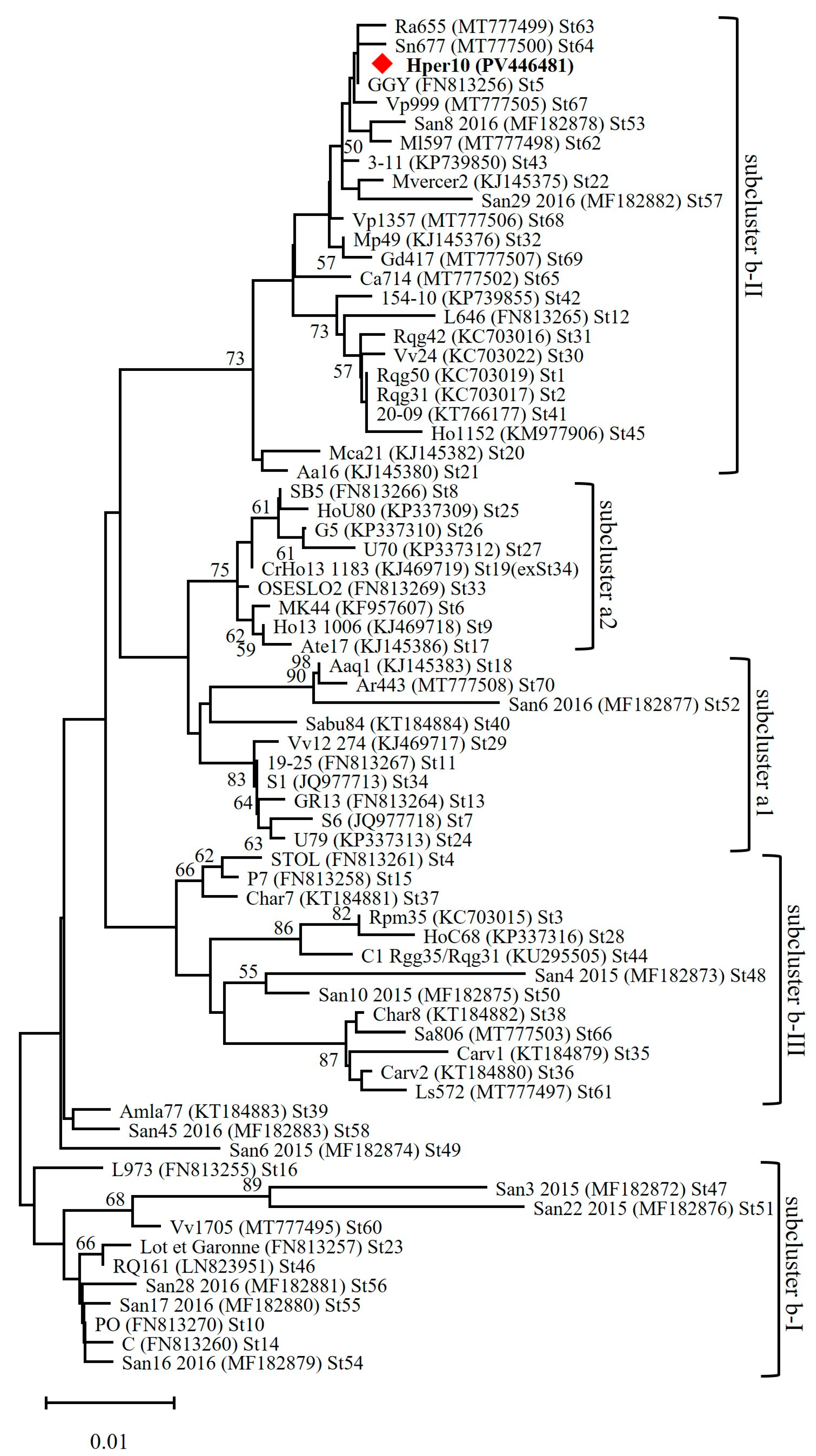
3.3. Molecular Typing Revealed a New ‘Ca. Phytoplasma solani’ Genotype Infecting MAPs
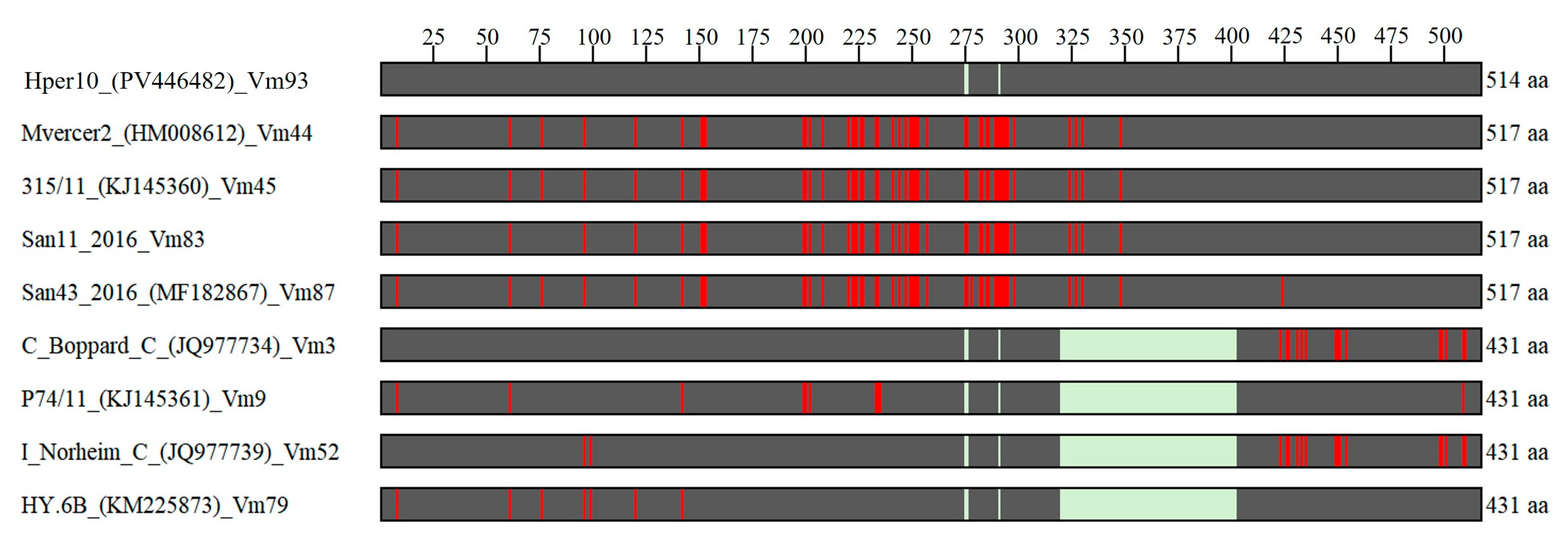
3.4. Vmp1 Gene Sequence Analyses Suggested Recombination Events Among ‘Ca. Phytoplasma solani’ Strains Identified in MAPs
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, E. Health and Wealth from Medicinal Aromatic Plants. FAO Diversification Booklet Number 17; Rural Infrastructure and Agro-Industries Division, Food and Agriculture Organization of the United Nations: Rome, Italy, 2012; Available online: https://www.fao.org/3/i2473e/i2473e.pdf (accessed on 23 May 2025).
- Pergola, M.; De Falco, E.; Belliggiano, A.; Ievoli, C. The Most Relevant Socio-Economic Aspects of Medicinal and Aromatic Plants through a Literature Review. Agriculture 2024, 14, 405. [Google Scholar] [CrossRef]
- Ozdal, T.; Tomas, M.; Toydemir, G.; Kamiloglu, S.; Capanoglu, E. Introduction to nutraceuticals, medicinal foods, and herbs. In Aromatic Herbs in Food—Bioactive Compounds, Processing, and Applications; Academic Press: Cambridge, MA, USA, 2021; pp. 1–34. [Google Scholar]
- De Falco, E.; Rigano, D.; Fico, V.; Vitti, A.; Barile, G.; Pergola, M. Spontaneous Officinal Plants in the Cilento, Vallo di Diano and Alburni National Park: Tradition, Protection, Enhancement, and Recovery. Plants 2023, 12, 465. [Google Scholar] [CrossRef] [PubMed]
- Meryani, L.M.; Janaki, P.; Suganthy, M.; Parameswari, E.; Krishnan, R.; Ramayer Latha, M. Potential of essential oils as natural herbicides: A comprehensive review of current developments. Plant Growth Regul. 2024, 104, 1307–1328. [Google Scholar] [CrossRef]
- Fincheira, P.; Jofré, I.; Espinoza, J.; Levío-Raimán, M.; Tortella, G.; Oliveira, H.C.; Diez, M.C.; Quiroz, A.; Rubilar, O. The efficient activity of plant essential oils for inhibiting Botrytis cinerea and Penicillium expansum: Mechanistic insights into antifungal activity. Microbiol. Res. 2023, 277, 127486. [Google Scholar] [CrossRef]
- Yadav, S.; Tiwari, K.S.; Gupta, C.; Tiwari, M.K.; Khan, A.; Sonkar, S.P. A brief review on natural dyes, pigments: Recent advances and future perspectives. Results Chem. 2023, 5, 100733. [Google Scholar] [CrossRef]
- Marcone, C.; Bellardi, M.G.; Bertaccini, A. Phytoplasma diseases of medicinal and aromatic plants. J. Plant Pathol. 2016, 98, 379–404. [Google Scholar] [CrossRef]
- Rao, G.P.; Marcone, C.; Bellardi, M.G.; Madhupriya. Phytoplasma Diseases of Medicinal Crops. In Phytoplasmas: Plant Pathogenic Bacteria—I, Characterisation and Epidemiology of Phytoplasma—Associated Diseases; Rao, G.P., Bertaccini, A., Fiore, N., Liefting, L.W., Eds.; Springer Nature: Singapore, 2018; pp. 235–266. [Google Scholar] [CrossRef]
- Namba, S. Molecular and biological properties of phytoplasmas. Proc. Jpn. Acad. Ser. B 2019, 95, 401–418. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Beanland, L. Insect vectors of phytoplasmas. Annu. Rev. Entomol. 2006, 51, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Contaldo, N.; Satta, E.; Zambon, Y.; Paltrinieri, S.; Bertaccini, A. Development and evaluation of different complex media for phytoplasma isolation and growth. J. Microbiol. Meth. 2016, 127, 105–110. [Google Scholar] [CrossRef]
- Bertaccini, A.; Arocha-Rosete, Y.; Contaldo, N.; Duduk, B.; Fiore, N.; Montano, H.G.; Kube, M.; Kuo, C.H.; Martini, M.; Oshima, K.; et al. Revision of the ‘Candidatus Phytoplasma’ species description guidelines. Int. J. Syst. Evol. Microbiol. 2022, 72, 005353. [Google Scholar] [CrossRef]
- Wei, W.; Zhao, Y. Phytoplasma Taxonomy: Nomenclature, Classification, and Identification. Biology 2022, 11, 1119. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues Jardim, B.; Tran-Nguyen, L.T.T.; Gambley, C.; Al-Sadi, A.M.; Al-Subhi, A.M.; Foissac, X.; Salar, P.; Cai, H.; Yang, J.Y.; Davis, R.; et al. The observation of taxonomic boundaries for the 16SrII and 16SrXXV phytoplasmas using genome—Based delimitation. Int. J. Syst. Evol. Microbiol. 2023, 73, 005977. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues Jardim, B.; Tran-Nguyen, L.T.T.; Gambley, C.; Webster, C.; Kehoe, M.; Bond, S.; Rodoni, B.; Constable, F.E. ‘Candidatus Phytoplasma vignae’, assigning a species description to a long-known phytoplasma occurring in northern Australia. Int. J. Syst. Evol. Microbiol. 2024, 74, 006502. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Lee, I.M.; Davis, R.E.; Suo, X.; Zhao, Y. Automated RFLP pattern comparison and similarity coefficient calculation for rapid delineation of new and distinct phytoplasma 16Sr subgroup lineages. Int. J. Syst. Evol. Microbiol. 2008, 58, 2368–2377. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an interactive online phytoplasma classification tool, iPhyClassifier, and its application in analysis of the peach X-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef]
- Panda, P.; Rao, G.P.; Sipahioğlu, H.M.; Hemmati, C.; Madhupriya; Kalita, M.K.; Oksal, H.D.; Usta, M.; Rastgou, M.; Alp, S.; et al. An update on phytoplasma diseases associated with ornamentals in Asia. In Phytoplasma Diseases in Asian Countries—Phytoplasma Diseases of Major Crops, Trees, and Weeds; Tiwari, A.K., Caglayan, K., Hoat, T.X., Al-Subhi, A., Nejat, N., Reddy, G., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 167–214. [Google Scholar] [CrossRef]
- Angelini, E.; Clair, D.; Borgo, M.; Bertaccini, A.; Boudon-Padieu, E. Flavescence dorée in France and Italy—Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 2001, 40, 79–86. [Google Scholar] [CrossRef]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA genes from culturable and nonculturable Mollicutes. J. Microbiol. Meth. 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Schneider, B.; Seemüller, E.; Smart, C.D.; Kirkpatrick, B.C. Phylogenetic classification of plant pathogenic mycoplasmalike organisms or phytoplasmas. In Molecular and Diagnostic Procedures in Mycoplasmology; Razin, S., Tully, J.G., Eds.; Academic Press: San Diego, CA, USA, 1995; pp. 369–380. [Google Scholar]
- Davis, R.E.; Lee, I.-M. Cluster-specific polymerase chain reaction amplification of 16S rDNA sequences for detection and identification of Mycoplasmalike organisms. Phytopathology 1993, 83, 1008–1011. [Google Scholar] [CrossRef]
- Lee, I.-M.; Hammond, R.W.; Davis, R.E.; Gundersen, D.E. Universal amplification and analysis of pathogen 16S rDNA for classification and identification of Mycoplasmalike organisms. Phytopathology 1993, 83, 834–842. [Google Scholar] [CrossRef]
- Hall, T.A. Bio Edit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Fabre, A.; Danet, J.L.; Foissac, X. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 2011, 472, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Cimerman, A.; Pacifico, D.; Salar, P.; Marzachì, C.; Foissac, X. Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Appl. Environ. Microbiol. 2009, 75, 2951–2957. [Google Scholar] [CrossRef]
- Fialová, R.; Válová, P.; Balakishiyeva, G.; Danet, J.L.; Sâfárová, D.; Foissac, X.; Navrátil, M. Genetic variability of Stolbur phytoplasma in annual crop and wild plant species in South Moravia. J. Plant Pathol. 2009, 91, 411–416. [Google Scholar]
- Quaglino, F.; Maghradze, D.; Casati, P.; Chkhaidze, N.; Lobjanidze, M.; Ravasio, A.; Passera, A.; Venturini, G.; Failla, O.; Bianco, P.A. Identification and characterization of new ‘Candidatus Phytoplasma solani’ strains associated with Bois noir disease in Vitis vinifera L. cultivars showing a range of symptoms severity in Georgia, the Caucasus Region. Plant Dis. 2016, 100, 904–915. [Google Scholar] [CrossRef]
- Pierro, R.; Passera, A.; Panattoni, A.; Casati, P.; Luvisi, A.; Rizzo, D.; Bianco, P.A.; Quaglino, F.; Materazzi, A. Molecular typing of ‘bois noir’ phytoplasma strains in the Chianti Classico area (Tuscany, central Italy) and their association with symptom severity in Vitis vinifera L. cv. Sangiovese. Phytopathology 2018, 108, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Pierro, R.; Passera, A.; Panattoni, A.; Rizzo, D.; Stefani, L.; Bartolini, L.; Casati, P.; Luvisi, A.; Quaglino, F.; Materazzi, A. Prevalence of a ‘Candidatus Phytoplasma solani’ strain, so far associated only with other hosts, in Bois noir-affected grapevines within Tuscan vineyards. Ann. Appl. Biol. 2018, 173, 202–212. [Google Scholar] [CrossRef]
- Quaglino, F.; Passera, A.; Faccincani, M.; Moussa, A.; Pozzebon, A.; Sanna, F.; Casati, P.; Bianco, P.A.; Mori, N. Molecular and spatial analyses reveal new insights on Bois noir epidemiology in Franciacorta vineyards. Ann. Appl. Biol. 2021, 179, 151–168. [Google Scholar] [CrossRef]
- Quaglino, F.; Zhao, Y.; Bianco, P.A.; Wei, W.; Casati, P.; Durante, G.; Davis, R.E. New 16Sr subgroups and distinct single nucleotide polymorphism lineages among grapevine Bois noir phytoplasma populations. Ann. Appl. Biol. 2009, 154, 279–289. [Google Scholar] [CrossRef]
- Mori, N.; Quaglino, F.; Tessari, F.; Pozzebon, A.; Bulgari, D.; Casati, P.; Bianco, P.A. Investigation on ‘bois noir’ epidemiology in north-eastern Italian vineyards through a multidisciplinary approach. Ann. Appl. Biol. 2015, 166, 75–89. [Google Scholar] [CrossRef]
- Pavlovic, S.; Jošic, D.; Starovic, M.; Stojanovic, S.; Aleksie, G.; Stojišin, V.; Radanovic, D. The first stolbur phytoplasma occurrence on two St. John’s worth species (Hypericum perforatum L. and Hypericum barhatum L.) in Serbia. J. Med. Plant Res. 2012, 6, 906–911. [Google Scholar] [CrossRef]
- Gaudin, J.; Semetey, O.; Foissac, X.; Eveillard, S. Phytoplasma titer in diseased lavender is not correlated to lavender tolerance to stolbur phytoplasma. Bull. Insectol. 2011, 64, S179–S180. [Google Scholar]
- Pavlovic, D.S.; Stojanovic, D.S.; Jošic, L.D.; Starovic, S.M. Phytoplasma disease of medicinal plants in Serbia. In Proceedings of the Eighth Conference on Medicinal and Aromatic Plants of Southeast European Countries (CMAPSEEC), Durrës, Albania, 19–22 May 2014; pp. 321–332. [Google Scholar]
- Mitrovic, P.; Trkulja, V.; Adamovic, D.; Balovic, I.; Milovac, Z.; Kovačic-Jošic, D.; Mihić Salapura, J. First report of stolbur phytoplasma on Mentha x piperita in Serbia. Plant Dis. 2016, 100, 853. [Google Scholar] [CrossRef]
- Contaldo, N.; Bellardi, M.G.; Cavicchi, L.; Epifano, F.; Genovese, S.; Curini, M.; Bertaccini, A. Phytochemical effects of phytoplasma infections on essential oil of Monarda fistulosa L. Bull. Insectol. 2011, 64, S177–S178. [Google Scholar]
- Contaldo, N.; Bertaccini, A.; Bozzano, G.; Cavicchi, L.; Bellardi, M.G. Detection and molecular characterization of phytoplasmas infecting Rosmarinus officinalis. J. Plant Pathol. 2012, 94, S4.60. [Google Scholar]
- Chuche, J.; Danet, J.L.; Rivoal, J.B.; Arricau-Bouvery, N.; Thiéry, D. Minor cultures as hosts for vectors of extensive crop diseases: Does Salvia sclarea act as a pathogen and vector reservoir for lavender decline? J. Pest Sci. 2018, 91, 145–155. [Google Scholar] [CrossRef]
- Langer, M.; Maixner, M. Molecular characterization of grapevine yellows associated phytoplasmas of the stolbur-group based on RFLP-analysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar] [CrossRef]
- Mitrovic, J.; Pavlovic, S.; Duduk, B. Survey and multigene characterization of stolbur phytoplasmas on various plant species in Serbia. Phytopathol. Mediterr. 2013, 52, 434–441. [Google Scholar] [CrossRef]
- Pavlovic, S.; Starovic, M.; Stojanovic, S.; Popovic, T.; Aleksie, G.; Dražic, S.; Jošic, D. Echinacea purpurea—A host of 16SrXII-A phytoplasma group in Serbia. Phytopath. Moll. 2011, 1, 35–39. [Google Scholar] [CrossRef]
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur- and bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef]
- Quaglino, F.; Sanna, F.; Moussa, A.; Faccincani, M.; Passera, A.; Casati, P.; Bianco, P.A.; Mori, N. Identification and ecology of alternative insect vectors of ‘Candidatus Phytoplasma solani’ to grapevine. Sci. Rep. 2019, 9, 19522. [Google Scholar] [CrossRef] [PubMed]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant ‘Candidatus Phytoplasma solani’ Stolbur tuf b phytoplasma strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef]
- Atanasova, B.; Jakovljević, M.; Spasov, D.; Jović, J.; Mitrović, M.; Toševski, I.; Cvrković, T. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur. J. Plant Pathol. 2015, 142, 759–770. [Google Scholar] [CrossRef]
- Plavec, J.; Križanac, I.; Budinšćak, Ž.; Škorić, D.; Šeruga Musić, M. A case study of FD and BN phytoplasma variability in Croatia: Multigene sequence analysis approach. Eur. J. Plant Pathol. 2015, 142, 591–601. [Google Scholar] [CrossRef]
- Flitman-Tene, R.; Mudahi-Orenstein, S.; Levisohn, S.; Yogev, D. Variable lipoprotein genes of Mycoplasma agalactiae are activated in vivo by promoter addition via site-specific DNA inversions. Infect. Immun. 2003, 71, 3821–3830. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Glew, M.; Papazisi, D.L.; Poumarat, F.; Bergonier, D.; Rosengarten, R.; Citti, C. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect. Immun. 2000, 68, 4539–4548. [Google Scholar] [CrossRef]
- Malembic-Maher, S.; Desqué, D.; Khalil, D.; Salar, P.; Bergey, B.; Danet, J.-L.; Duret, S.; Dubrana-Ourabah, M.-P.; Beven, L.; Ember, I.; et al. When a Palearctic bacterium meets a Nearctic insect vector: Genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLoS Path. 2020, 16, e1007967. [Google Scholar] [CrossRef]
- Beven, L.; Duret, S.; Batailler, B.; Dubrana, M.-P.; Saillard, C.; Renaudin, J.; Arricau-Buovery, N. The repetitive domain of ScARP3d triggers entry of Spiroplasma citri into cultured cells of the vector Circulifer haematoceps. PLoS ONE 2012, 7, e48606. [Google Scholar] [CrossRef]
- Bzymek, M.; Lovett, S.T. Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc. Nat. Acad. Sci. USA 2001, 98, 8319–8325. [Google Scholar] [CrossRef]
- Morag, A.S.; Saveson, C.J.; Lovett, S.T. Expansion of DNA repeats in Escherichia coli: Effects of recombination and replication functions. J. Mol. Biol. 1999, 289, 21–27. [Google Scholar] [CrossRef]
- Seruga Music, M.; Samarzija, I.; Hogenhout, S.A.; Haryono, M.; Cho, S.-T.; Kuo, C.H. The genome of ‘Candidatus Phytoplasma solani’ strain SA-1 is highly dynamic and prone to adopting foreign sequences. Syst. Appl. Microbiol. 2019, 42, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Nakamura-Tsuruta, N.; Smith, D.F.; Ongenaert, M.; Winter, H.C.; Rougé, P.; Goldstein, I.J.; Mo, H.; Kominami, J.; Culerrier, R.; et al. Phylogenetic and specificity studies of two-domain GNA-related lectins: Generation of multispecificity through domain duplication and divergent evolution. Biochem. J. 2007, 404, 51–61. [Google Scholar] [CrossRef] [PubMed]




| Farm—Location | Species | Family | No. of Plants | |
|---|---|---|---|---|
| Collected | PCR Positive | |||
| 1—Rovagnate (LC) | Achillea millefolium L. | Asteraceae | 1 | 0 |
| Artemisia absinthium L. | Asteraceae | 4 | 4 | |
| Calendula officinalis L. | Asteraceae | 5 | 0 | |
| Filipendula ulmaria (L.) Maxim. | Rosaceae | 5 | 4 | |
| Hypericum perforatum L. | Hypericaceae | 7 | 7 | |
| Inula salicina L. | Asteraceae | 5 | 0 | |
| Lavandula sp. L. | Lamiaceae | 4 | 3 | |
| Matricaria recutita L. | Asteraceae | 3 | 3 | |
| Melissa officinalis L. | Lamiaceae | 4 | 4 | |
| Mentha sp. L. | Lamiaceae | 4 | 3 | |
| Rosmarinus officinalis L. | Lamiaceae | 4 | 4 | |
| Salvia officinalis L. | Lamiaceae | 2 | 2 | |
| Urtica dioica L. | Urticaceae | 6 | 1 | |
| Valeriana officinalis L. | Caprifoliaceae | 6 | 5 | |
| Farm-1 total | 60 | 40 | ||
| 2—Solto Collina (BG) | Echinacea purpurea (L.) Moench | Asteraceae | 5 | 0 |
| Hypericum perforatum L. | Hypericaceae | 5 | 5 | |
| Lavandula sp. L. | Lamiaceae | 7 | 6 | |
| Levisticum officinale W.D.J.Koch | Apiaceae | 2 | 2 | |
| Melissa officinalis L. | Lamiaceae | 3 | 3 | |
| Mentha sp. L. | Lamiaceae | 5 | 5 | |
| Monarda didyma L. | Lamiaceae | 5 | 1 | |
| Nepeta cataria L. | Lamiaceae | 15 | 7 | |
| Urtica dioica L. | Urticaceae | 6 | 0 | |
| Farm-2 total | 53 | 29 | ||
| Overall total | 113 | 69 | ||
| vmp1 Gene Variant | No. SNPs (Sequence Identity) vs. vmp1 Sequence Variant Vm93 (1542 nt) | |
|---|---|---|
| with Gaps (Insertion/Deletion) (1551 nt) | Without Gaps (Insertion/Deletion) (1294 nt) | |
| Vm44 (1551 nt) | 77 (95.0%) | 68 (94.7%) |
| Vm45 (1551 nt) | 74 (95.2%) | 65 (95.0%) |
| Vm83 (1551 nt) | 73 (95.3%) | 64 (95.1%) |
| Vm87 (1551 nt) | 75 (95.1%) | 66 (94.9%) |
| Vm3 (1294 nt) | 266 (82.8%) | 18 (98.6%) |
| Vm9 (1294 nt) | 263 (83.0%) | 15 (98.8%) |
| Vm52 (1294 nt) | 271 (82.5%) | 23 (98.2%) |
| Vm79 (1294 nt) | 257 (83.4%) | 9 (99.3%) |
| vmp1 Gene Variant | Differences vs. vmp1/Vmp1 Sequence Variant Vm93 | |||
|---|---|---|---|---|
| Nt 1-957 | Aa 1-319 | Nt 958-1551 | Aa 320-517 | |
| No. SNPs * | No. aa-Change * | No. SNPs * | No. aa-Change * | |
| Vm44 | 71 (9) | 44 (3) | 6 (0) | 4 (0) |
| Vm45 | 68 (9) | 44 (3) | 6 (0) | 4 (0) |
| Vm83 | 67 (9) | 43 (3) | 6 (0) | 4 (0) |
| Vm87 | 68 (9) | 45 (3) | 7 (0) | 5 (0) |
| Vm3 | 0 (0) | 0 (0) | 266 (248) | 98 (83) |
| Vm9 | 14 (0) | 9 (0) | 249 (248) | 84 (83) |
| Vm52 | 5 (0) | 2 (0) | 266 (248) | 98 (83) |
| Vm79 | 9 (0) | 7 (0) | 248 (248) | 83 (83) |
| # | Vmp1 Domain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Vm93_R1_aa153-236 | ID | |||||||
| 2 | Vm3_R1_aa153-236 | 1 | ID | ||||||
| 3 | Vm44_R1_aa153-236 | 0.845 | 0.845 | ID | |||||
| 4 | Vm93_R2_aa237-319 | 0.69 | 0.69 | 0.738 | ID | ||||
| 5 | Vm3_R2_aa237-319 | 0.69 | 0.69 | 0.738 | 1 | ID | |||
| 6 | Vm44_R2_aa237-319 | 0.666 | 0.666 | 0.797 | 0.722 | 0.722 | ID | ||
| 7 | Vm93_R3_aa320-402 | 0.702 | 0.702 | 0.821 | 0.746 | 0.746 | 0.939 | ID | |
| 8 | Vm44_R3_aa320-402 | 0.666 | 0.666 | 0.785 | 0.722 | 0.722 | 0.987 | 0.951 | ID |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbieri, C.; Moussa, A.; Passera, A.; Casati, P.; Bianco, P.A.; Quaglino, F. Molecular Identification and Genotyping of Phytoplasmas Infecting Medicinal and Aromatic Plants in Northern Italy. Microorganisms 2025, 13, 1444. https://doi.org/10.3390/microorganisms13071444
Barbieri C, Moussa A, Passera A, Casati P, Bianco PA, Quaglino F. Molecular Identification and Genotyping of Phytoplasmas Infecting Medicinal and Aromatic Plants in Northern Italy. Microorganisms. 2025; 13(7):1444. https://doi.org/10.3390/microorganisms13071444
Chicago/Turabian StyleBarbieri, Camilla, Abdelhameed Moussa, Alessandro Passera, Paola Casati, Piero Attilio Bianco, and Fabio Quaglino. 2025. "Molecular Identification and Genotyping of Phytoplasmas Infecting Medicinal and Aromatic Plants in Northern Italy" Microorganisms 13, no. 7: 1444. https://doi.org/10.3390/microorganisms13071444
APA StyleBarbieri, C., Moussa, A., Passera, A., Casati, P., Bianco, P. A., & Quaglino, F. (2025). Molecular Identification and Genotyping of Phytoplasmas Infecting Medicinal and Aromatic Plants in Northern Italy. Microorganisms, 13(7), 1444. https://doi.org/10.3390/microorganisms13071444









