Complementary Rhizosphere Microbial Strategies Drive Functional Specialization in Coastal Halophyte Succession: Differential Adaptation of Suaeda glauca and Phragmites communis to Saline–Alkali Stress
Abstract
1. Introduction
2. Materials and Methods
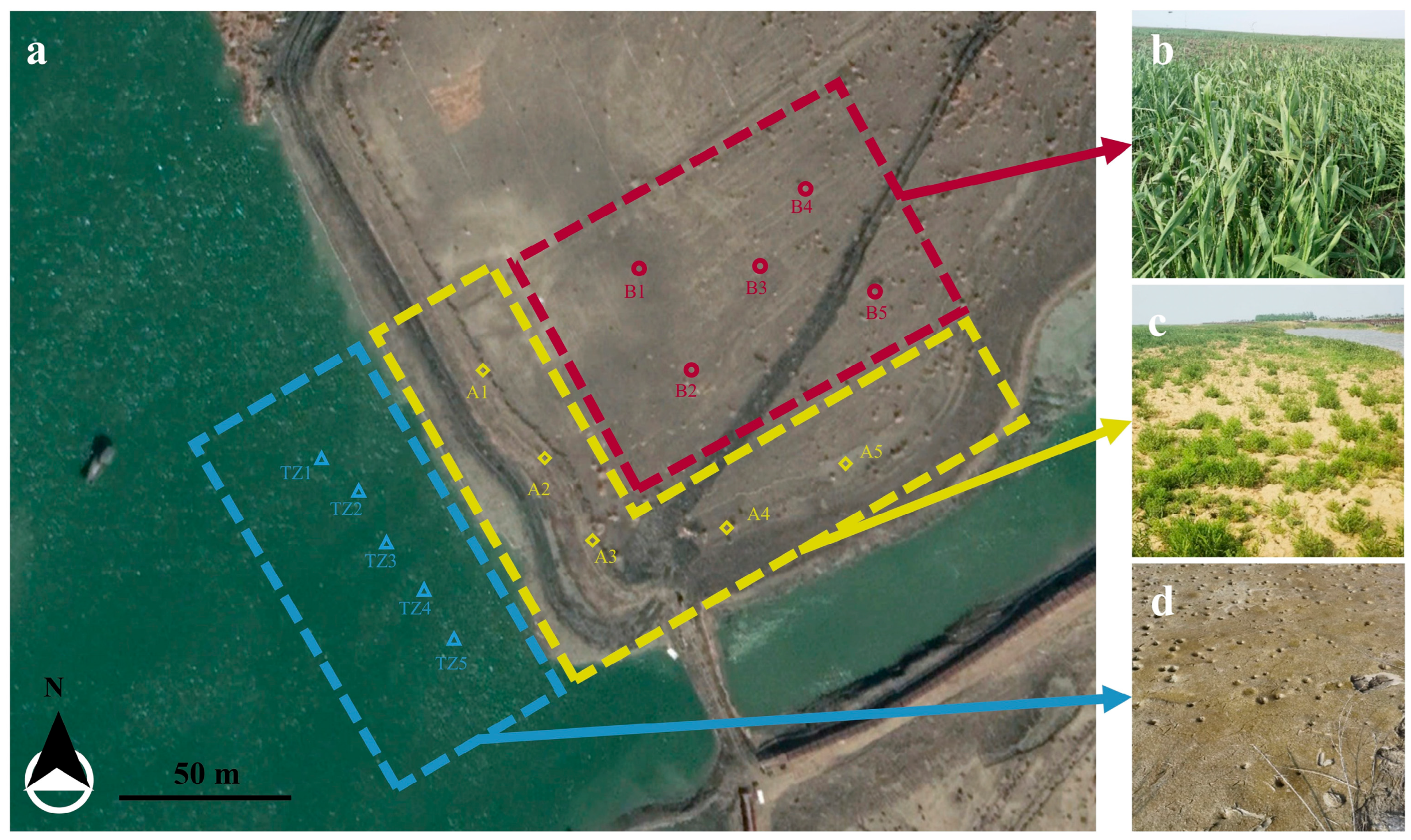
2.1. Study Region
2.2. Sample Collection
2.3. Soil Physicochemical Analysis
2.4. Plant Compartment Separation
2.5. Extracellular DNA Removal
2.6. DNA Extraction
2.7. Library Construction, Sequencing, and Statistical Analysis
3. Results
3.1. Analysis of Soil Physicochemical Properties
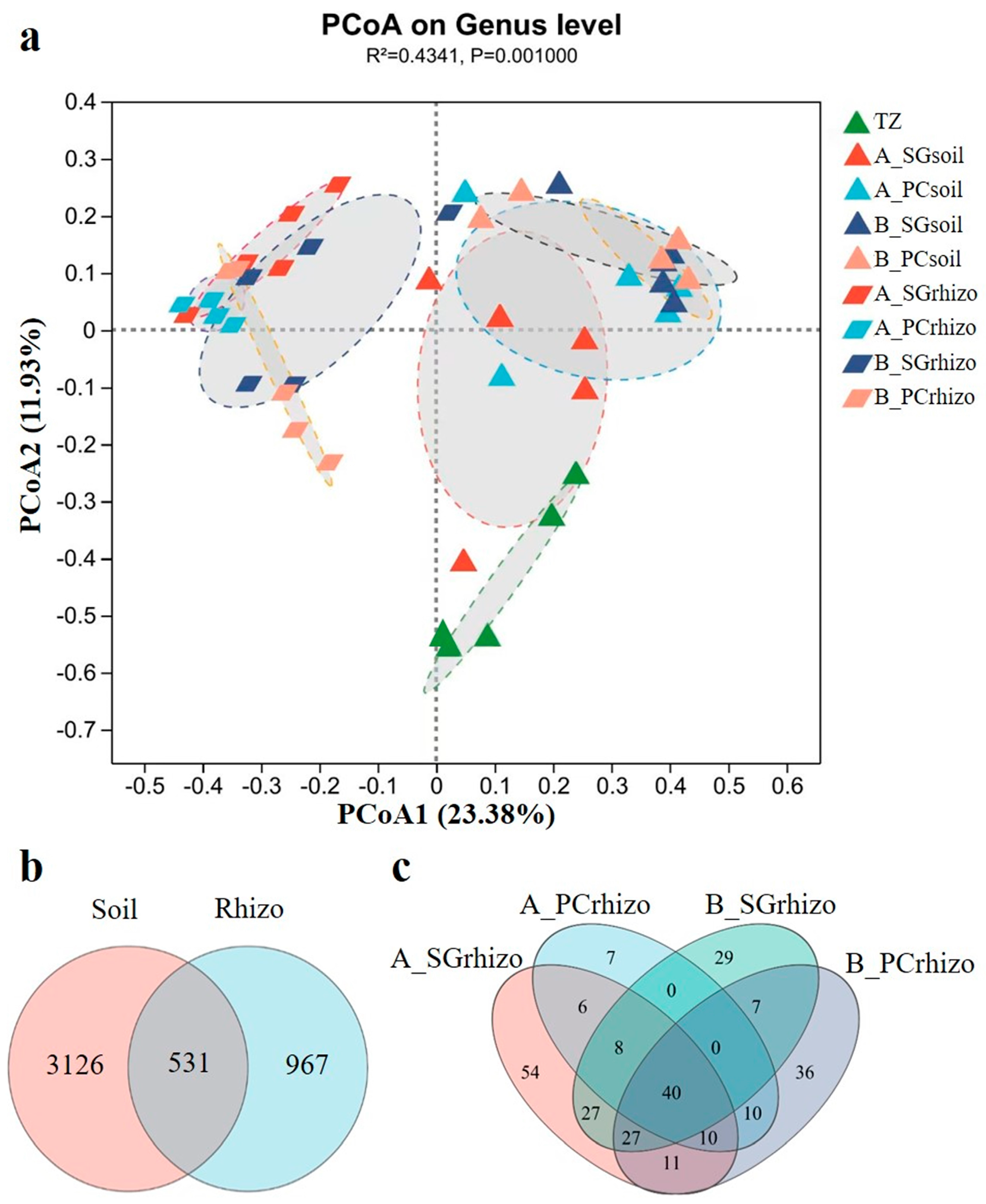
3.2. Analysis of Bacterial Community Diversity
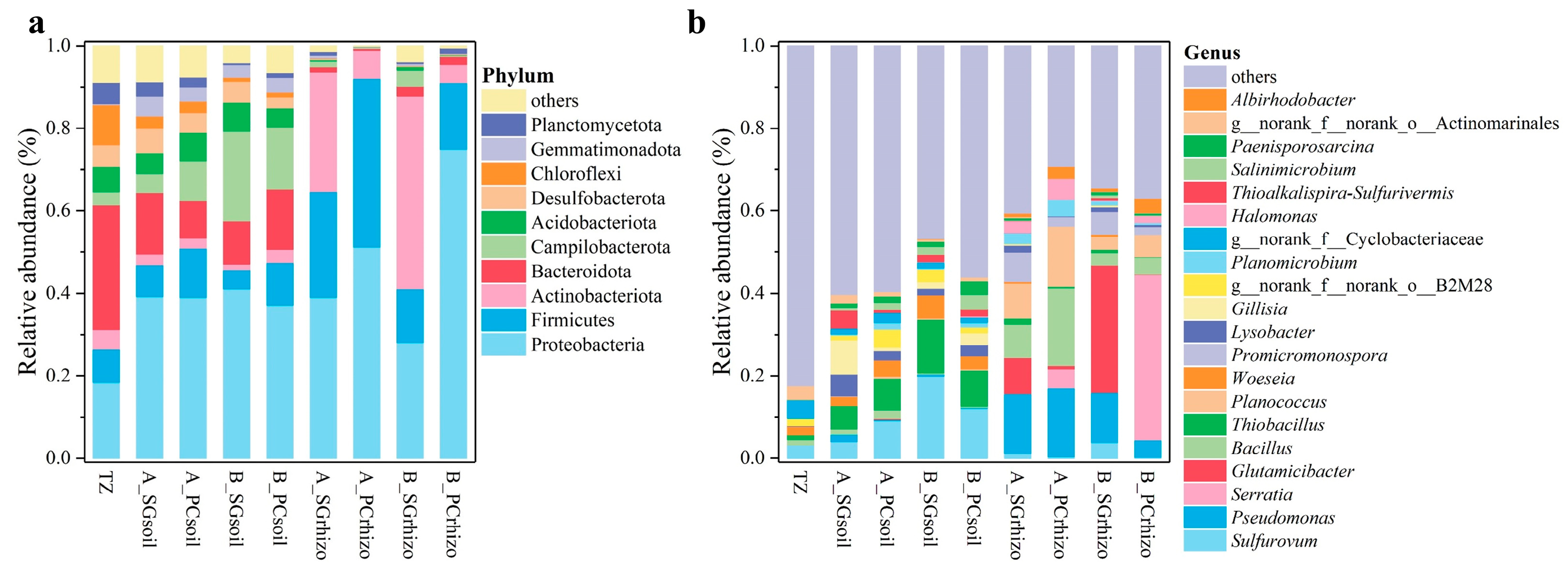
3.3. Analysis of Bacterial Community Composition
3.4. Analysis of Bacterial Community Composition and Environmental Factors
3.5. Predictive Analysis of Bacterial Community Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TZ | tidal zone area |
| Plot A | Suaeda glauca-dominant area |
| Plot B | Phragmites communis-dominant area |
| SG | Suaeda glauca |
| PC | Phragmites communis |
| A_SGsoil | peri root soil of Suaeda glauca in plot A |
| A_PCsoil | peri root soil of Phragmites communis in plot A |
| A_SGrhizo | rhizosphere soil of Suaeda glauca in plot A |
| A_PCrhizo | rhizosphere soil of Phragmites communis in plot A |
| B_SGsoil | peri root soil of Suaeda glauca in plot B |
| B_PCsoil | peri root soil of Phragmites communis in plot B |
| B_SGrhizo | rhizosphere soil of Suaeda glauca in plot B |
| B_PCrhizo | rhizosphere soil of Phragmites communis in plot B |
References
- He, Q.; Li, Z.; Daleo, P.; Lefcheck, J.S.; Thomsen, M.S.; Adams, J.B.; Bouma, T.J. Coastal wetland resilience through local, regional and global conservation. Nat. Rev. Biodivers. 2025, 1, 50–67. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, X.; Xu, X.; Zou, Z.; Chen, B.; Qin, Y.; Zhang, X.; Dong, J.; Liu, D.; Pan, L.; et al. Rebound in China’s coastal wetlands following conservation and restoration. Nat. Sustain. 2021, 4, 1076–1083. [Google Scholar] [CrossRef]
- Wang, Y.-S.; Gu, J.-D. Ecological responses, adaptation and mechanisms of mangrove wetland ecosystem to global climate change and anthropogenic activities. Int. Biodeterior. Biodegrad. 2021, 162, 105248. [Google Scholar] [CrossRef]
- An, X.; Wang, Z.; Jiao, K.; Teng, X.; Zhou, R.; Xu, M.; Lian, B. Bacterial community characteristics in the rhizosphere of Suaeda glauca versus bulk soil in coastal silt soil modified by sea-sand and their implications. Front. Mar. Sci. 2023, 9, 1001449. [Google Scholar] [CrossRef]
- An, X.; Wang, Z.; Teng, X.; Zhou, R.; Wang, X.; Xu, M.; Lian, B. Rhizosphere bacterial diversity and environmental function prediction of wild salt-tolerant plants in coastal silt soil. Ecol. Indic. 2022, 134, 108503. [Google Scholar] [CrossRef]
- Ortiz-Álvarez, R.; Fierer, N.; de los Ríos, A.; Casamayor, E.O.; Barberán, A. Consistent changes in the taxonomic structure and functional attributes of bacterial communities during primary succession. ISME J. 2018, 12, 1658–1667. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Gao, Y.; Zhang, Y.; Tong, S.; Liu, X. Early establishment of Suaeda salsa population as affected by soil moisture and salinity: Implications for pioneer species introduction in saline-sodic wetlands in Songnen Plain, China. Ecol. Indic. 2019, 107, 105654. [Google Scholar] [CrossRef]
- Lambertini, C.; Guo, W.-Y.; Ye, S.; Eller, F.; Guo, X.; Li, X.-Z.; Sorrell, B.K.; Speranza, M.; Brix, H. Phylogenetic diversity shapes salt tolerance in Phragmites australis estuarine populations in East China. Sci. Rep. 2020, 10, 17645. [Google Scholar] [CrossRef]
- Liu, X.; Feng, T.; Zhang, Y.; Liu, Y.; Wang, P. Vegetation restoration affects soil hydrological processes in typical natural and planted forests on the Loess Plateau. J. Hydrol. 2025, 650, 132465. [Google Scholar] [CrossRef]
- Lever, M.A.; Torti, A.; Eickenbusch, P.; Michaud, A.B.; Šantl-Temkiv, T.; Jørgensen, B.B. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 2015, 6, 476. [Google Scholar] [CrossRef] [PubMed]
- Lueders, T.; Manefield, M.; Friedrich, M.W. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 2004, 6, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Ma, Y.; Wang, X.; Zhang, B.; Zhang, G.; Bahadur, A.; Chen, T.; Liu, G.; Zhang, W.; et al. Research progress regarding the role of halophilic and halotolerant microorganisms in the eco-environmental sustainability and conservation. J. Clean. Prod. 2023, 418, 138054. [Google Scholar] [CrossRef]
- de Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef]
- Luo, C.; He, Y.; Chen, Y. Rhizosphere microbiome regulation: Unlocking the potential for plant growth. Curr. Res. Microb. Sci. 2025, 8, 100322. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Liu, F.; Mo, X.; Kong, W.; Song, Y. Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total. Environ. 2020, 740, 140144. [Google Scholar] [CrossRef]
- Yin, X.; Song, Y.; Shen, J.; Sun, L.; Fan, K.; Chen, H.; Sun, K.; Ding, Z.; Wang, Y. The role of rhizosphere microbial community structure in the growth and development of different tea cultivars. Appl. Soil. Ecol. 2025, 206, 105817. [Google Scholar] [CrossRef]
- Ma, J.; Ibekwe, A.M.; Yang, C.-H.; Crowley, D.E. Bacterial diversity and composition in major fresh produce growing soils affected by physiochemical properties and geographic locations. Sci. Total. Environ. 2016, 563–564, 199–209. [Google Scholar] [CrossRef]
- Li, W.; Dong, X.; Lu, R.; Zhou, Y.; Zheng, P.; Feng, D.; Wang, Y. Microbial ecology of sulfur cycling near the sulfate-methane transition of deep-sea cold seep sediments. Environ. Microbiol. 2021, 23, 6844–6858. [Google Scholar] [CrossRef]
- Ranadev, P.; Revanna, A.; Bagyaraj, D.J.; Shinde, A.H. Sulfur oxidizing bacteria in agro ecosystem and its role in plant productivity-a review. J. Appl. Microbiol. 2023, 134, lxad161. [Google Scholar] [CrossRef] [PubMed]
- Rolando, J.L.; Kolton, M.; Song, T.; Kostka, J.E. The core root microbiome of Spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes, USA. Microbiome 2022, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Xavier, J.F.; Costa, D.P.d.; Gonçalves, J.V.d.S.; Pinto, M.C.F.; Goulart, R.S.; Zonta, E.; Coelho, I.d.S. Different halophytes orchestrate microbial diversity in the rhizosphere of salinity-impacted soils. Appl. Soil. Ecol. 2024, 202, 105588. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Plant growth-promoting soil bacteria: Nitrogen fixation, phosphate solubilization, siderophore production, and other biological activities. Plants 2023, 12, 4074. [Google Scholar] [CrossRef]
- Patani, A.; Patel, M.; Islam, S.; Yadav, V.K.; Prajapati, D.; Yadav, A.N.; Sahoo, D.K.; Patel, A. Recent advances in Bacillus-mediated plant growth enhancement: A paradigm shift in redefining crop resilience. World J. Microbiol. Biotechnol. 2024, 40, 77. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Jabborova, D.; Räsänen, L.A.; Liao, H. Coordination between Bradyrhizobium and Pseudomonas alleviates salt stress in soybean through altering root system architecture. J. Plant Interact. 2017, 12, 100–107. [Google Scholar] [CrossRef]
- Ahmad, M.; Zahir, Z.A.; Khalid, M.; Nazli, F.; Arshad, M. Efficacy of Rhizobium and Pseudomonas strains to improve physiology, ionic balance and quality of mung bean under salt-affected conditions on farmer’s fields. Plant Physiol. Biochem. 2013, 63, 170–176. [Google Scholar] [CrossRef]
- Peng, X.; Wang, Q.; Lang, D.; Li, Y.; Zhang, W.; Zhang, X. Bacillus cereus G2 facilitates N cycle in soil, further improves N uptake and assimilation, and accelerates proline and glycine betaine metabolisms of Glycyrrhiza uralensis subjected to salt stress. J. Agric. Food Chem. 2023, 71, 15485–15496. [Google Scholar] [CrossRef]
- Feng, L.; Li, Q.; Zhou, D.; Jia, M.; Liu, Z.; Hou, Z.; Ren, Q.; Ji, S.; Sang, S.; Lu, S.; et al. B. subtilis CNBG-PGPR-1 induces methionine to regulate ethylene pathway and ROS scavenging for improving salt tolerance of tomato. Plant J. 2024, 117, 193–211. [Google Scholar] [CrossRef]
- Lu, S.; Feng, L.; Zhou, D.; Jia, M.; Liu, Z.; Hou, Z.; Li, Q.; Yu, J. Complete genome sequence of Bacillus subtilis CNBG-PGPR-1 for studying the promotion of plant growth. Mol. Plant Microbe Interact. 2022, 35, 1115–1119. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Z.; Wang, Y.; Wang, X.; Liu, P.; Han, M.; Zhou, W. Improving soil phosphorus availability in saline areas by marine bacterium Bacillus paramycoides. Environ. Sci. Pollut. Res. 2023, 30, 112385–112396. [Google Scholar] [CrossRef] [PubMed]
- Orhan, F.; Demirci, A. Salt stress mitigating potential of halotolerant/halophilic plant growth promoting. Geomicrobiol. J. 2020, 37, 663–669. [Google Scholar] [CrossRef]
- Zhou, N.; Zhao, S.; Tian, C.-Y. Effect of halotolerant rhizobacteria isolated from halophytes on the growth of sugar beet (Beta vulgaris L.) under salt stress. FEMS Microbiol. Lett. 2017, 364, fnx091. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.H.A.; Alkhalifah, D.H.M.; Al Yousef, S.A.; Beemster, G.T.S.; Mousa, A.S.M.; Hozzein, W.N.; AbdElgawad, H. Salinity stress enhances the antioxidant capacity of Bacillus and Planococcus species isolated from saline lake environment. Front. Microbiol. 2020, 11, 561816. [Google Scholar] [CrossRef]
- Kang, S.-M.; Khan, A.L.; Hamayun, M.; Hussain, J.; Joo, G.-J.; You, Y.-H.; Kim, J.-G.; Lee, I.-J. Gibberellin-producing Promicromonospora sp. SE188 improves Solanum lycopersicum plant growth and influences endogenous plant hormones. J. Microbiol. 2012, 50, 902–909. [Google Scholar] [CrossRef]
- Siddikee, M.A.; Chauhan, P.S.; Anandham, R.; Han, G.-H.; Sa, T. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 2010, 20, 1577–1584. [Google Scholar] [CrossRef]
- Najafi Zilaie, M.; Mosleh Arani, A.; Etesami, H.; Dinarvand, M. Improved salinity and dust stress tolerance in the desert halophyte Haloxylon aphyllum by halotolerant plant growth-promoting rhizobacteria. Front. Plant Sci. 2022, 13, 948260. [Google Scholar] [CrossRef] [PubMed]
- Zilaie, M.N.; Arani, A.M.; Etesami, H.; Dinarvand, M.; Dolati, A. Halotolerant plant growth-promoting rhizobacteria-mediated alleviation of salinity and dust stress and improvement of forage yield in the desert halophyte Seidlitzia rosmarinus. Environ. Exp. Bot. 2022, 201, 104952. [Google Scholar] [CrossRef]
- de Carvalho Neta, S.J.; Araújo, V.L.V.P.; Fracetto, F.J.C.; da Silva, C.C.G.; de Souza, E.R.; Silva, W.R.; Lumini, E.; Fracetto, G.G.M. Growth-promoting bacteria and arbuscular mycorrhizal fungus enhance maize tolerance to saline stress. Microbiol. Res. 2024, 284, 127708. [Google Scholar] [CrossRef]
- Behera, B.C.; Yadav, H.; Singh, S.K.; Mishra, R.R.; Sethi, B.K.; Dutta, S.K.; Thatoi, H.N. Phosphate solubilization and acid phosphatase activity of Serratia sp. isolated from mangrove soil of Mahanadi river delta, Odisha, India. J. Genet. Eng. Biotechnol. 2017, 15, 169–178. [Google Scholar] [CrossRef]
- Lucero, C.T.; Lorda, G.S.; Anzuay, M.S.; Ludueña, L.M.; Taurian, T. Peanut endophytic phosphate solubilizing bacteria increase growth and P content of soybean and maize plants. Curr. Microbiol. 2021, 78, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Pei, H.; Xu, Z. Low nitrogen stress stimulating the indole-3-acetic acid biosynthesis of Serratia sp. ZM is vital for the survival of the bacterium and its plant growth-promoting characteristic. Arch. Microbiol. 2017, 199, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.A.; Soliman, E.R.S. Characterization and genomics identification of key genes involved in denitrification-DNRA-nitrification pathway of plant growth-promoting rhizobacteria (Serratia marcescens OK482790). BMC Microbiol. 2023, 23, 210. [Google Scholar] [CrossRef]
- Kulkova, I.; Wróbel, B.; Dobrzyński, J. Serratia spp. as plant growth-promoting bacteria alleviating salinity, drought, and nutrient imbalance stresses. Front. Microbiol. 2024, 15, 1342331. [Google Scholar] [CrossRef]
- Jung, H.S.; Lee, J.; Hyeon, J.W.; Hyun, S.; Jeon, C.O. Albirhodobacter confluentis sp. nov., isolated from an estuary. Int. J. Syst. Evol. Microbiol. 2018, 68, 289–293. [Google Scholar] [CrossRef]
- León-Barrios, M.; Ramírez-Bahena, M.H.; Igual, J.M.; Peix, Á.; Velázquez, E. Phyllobacterium salinisoli sp. nov., isolated from a Lotus lancerottensis root nodule in saline soil from Lanzarote. Int. J. Syst. Evol. Microbiol. 2018, 68, 1085–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Ouyang, L.; Xu, Z.; Zhang, L. Rhizobacteria of Populus euphratica promoting plant growth against heavy metals. Int. J. Phytoremediat. 2015, 17, 973–980. [Google Scholar] [CrossRef]
- Bresson, J.; Varoquaux, F.; Bontpart, T.; Touraine, B.; Vile, D. The PGPR strain Phyllobacterium brassicacearum STM196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis. New Phytol. 2013, 200, 558–569. [Google Scholar] [CrossRef]
- Li, C.-M.; Wu, H.-Z.; Wang, Y.-X.; Zhu, S.; Wei, C.-H. Enhancement of phenol biodegradation: Metabolic division of labor in co-culture of Stenotrophomonas sp. N5 and Advenella sp. B9. J. Hazard. Mater. 2020, 400, 123214. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.-X.; Li, Y.-H.; Ding, J.-Z.; Du, H.-M.; Zhao, Z.; Zhou, L.-N.; Liu, C.; Gao, S.-B.; Cao, M.-J.; et al. Overexpression of the Suaeda salsa SsNHX1 gene confers enhanced salt and drought tolerance to transgenic Zea may. J. Integr. Agric. 2018, 17, 2612–2623. [Google Scholar] [CrossRef]
- Kraft, B.; Strous, M.; Tegetmeyer, H.E. Microbial nitrate respiration—Genes, enzymes and environmental distribution. J. Biotechnol. 2011, 155, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Yang, L.; Wang, S.; Liu, J.; Yang, H. Physiological and metabolic profiles of common reed provide insights into plant adaptation to low nitrogen conditions. Biochem. Syst. Ecol. 2017, 73, 3–10. [Google Scholar] [CrossRef]
- Nieder, R.; Benbi, D.K.; Scherer, H.W. Fixation and defixation of ammonium in soils: A review. Biol. Fertil. Soils. 2011, 47, 1–14. [Google Scholar] [CrossRef]



| Properties | Sampling Sites | ||
|---|---|---|---|
| TZ | Plot A | Plot B | |
| pH | 8.33 ± 0.04 a | 8.37 ± 0.08 a | 8.43 ± 0.11 a |
| EC (mS/cm) | 3.08 ± 0.15 b | 4.31 ± 0.28 a | 3.69 ± 0.56 b |
| TN (g/kg) | 1.20 ± 0.03 a | 1.06 ± 0.09 b | 1.07 ± 0.03 b |
| TP (g/kg) | 0.75 ± 0.01 a | 0.70 ± 0.01 b | 0.71 ± 0.02 b |
| TK (g/kg) | 19.81 ± 0.90 a | 18.13 ± 1.46 b | 20.31 ± 0.41 a |
| AN (mg/kg) | 73.03 ± 2.52 a | 59.27 ± 9.89 b | 57.04 ± 0.35 b |
| AP (mg/kg) | 8.77 ± 1.21 a | 8.60 ± 0.19 a | 8.83 ± 0.26 a |
| AK (mg/kg) | 438.40 ± 33.91 b | 514.00 ± 40.03 a | 502.20 ± 38.01 a |
| SOC (g/kg) | 16.51 ± 0.92 a | 11.70 ± 1.12 b | 12.84 ± 0.44 b |
| Avail. K (mg/kg) | 142.05 ± 14.38 a | 156.85 ± 16.70 a | 147.90 ± 15.17 a |
| Avail. Ca (mg/kg) | 150.20 ± 23.01 a | 103.30 ± 42.72 ab | 84.85 ± 25.66 b |
| Avail. Na (mg/kg) | 2730.00 ± 122.77 b | 3780.50 ± 327.50 a | 3200.00 ± 478.00 b |
| Avail. Mg (mg/kg) | 156.55 ± 25.04 a | 140.35 ± 38.63 a | 125.80 ± 36.50 a |
| Avail. HCO3 (mg/kg) | 42.58 ± 1.58 a | 35.50 ± 2.30 b | 36.60 ± 3.34 b |
| Avail. Cl (mg/kg) | 3538.23 ± 136.52 b | 5698.92 ± 753.95 a | 4400.05 ± 743.97 b |
| Avail. SO4 (mg/kg) | 1729.93 ± 170.86 a | 1128.11 ± 247.87 b | 1136.71 ± 194.57 b |
| SAR | 220.45 ± 7.28 b | 348.80 ± 29.73 a | 315.08 ± 18.34 a |
| Environmental Factors | Suaeda glauca | Phragmites communis | ||
|---|---|---|---|---|
| r2 | p | r2 | p | |
| TK | 0.7061 | 0.001 *** | 0.6255 | 0.002 ** |
| TP | 0.2108 | 0.136 | 0.4399 | 0.008 ** |
| TN | 0.1138 | 0.369 | 0.0174 | 0.848 |
| AP | 0.1324 | 0.31 | 0.2356 | 0.112 |
| AK | 0.0392 | 0.727 | 0.1103 | 0.375 |
| AN | 0.2439 | 0.079 | 0.1117 | 0.338 |
| SOC | 0.0427 | 0.721 | 0.2755 | 0.053 |
| Avail. HCO3 | 0.2924 | 0.065 | 0.2908 | 0.068 |
| Avail. Cl | 0.2163 | 0.141 | 0.4686 | 0.003 ** |
| Avail. SO4 | 0.0117 | 0.91 | 0.0145 | 0.866 |
| Avail. K | 0.2127 | 0.126 | 0.114 | 0.378 |
| Avail. Ca | 0.1559 | 0.227 | 0.1562 | 0.234 |
| Avail. Na | 0.2613 | 0.09 | 0.3794 | 0.023 * |
| Avail. Mg | 0.0368 | 0.699 | 0.089 | 0.46 |
| pH | 0.0794 | 0.502 | 0.2091 | 0.158 |
| EC | 0.4195 | 0.013 * | 0.6676 | 0.001 *** |
| SAR | 0.5492 | 0.004 ** | 0.5731 | 0.002 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, H.; Jia, M.; Xue, J.; Liu, Z.; Zhou, D.; Hou, Z.; Yu, J.; Lu, S. Complementary Rhizosphere Microbial Strategies Drive Functional Specialization in Coastal Halophyte Succession: Differential Adaptation of Suaeda glauca and Phragmites communis to Saline–Alkali Stress. Microorganisms 2025, 13, 1399. https://doi.org/10.3390/microorganisms13061399
Dai H, Jia M, Xue J, Liu Z, Zhou D, Hou Z, Yu J, Lu S. Complementary Rhizosphere Microbial Strategies Drive Functional Specialization in Coastal Halophyte Succession: Differential Adaptation of Suaeda glauca and Phragmites communis to Saline–Alkali Stress. Microorganisms. 2025; 13(6):1399. https://doi.org/10.3390/microorganisms13061399
Chicago/Turabian StyleDai, Hao, Mingyun Jia, Jianhui Xue, Zhuangzhuang Liu, Dongqin Zhou, Zhaoqi Hou, Jinping Yu, and Shipeng Lu. 2025. "Complementary Rhizosphere Microbial Strategies Drive Functional Specialization in Coastal Halophyte Succession: Differential Adaptation of Suaeda glauca and Phragmites communis to Saline–Alkali Stress" Microorganisms 13, no. 6: 1399. https://doi.org/10.3390/microorganisms13061399
APA StyleDai, H., Jia, M., Xue, J., Liu, Z., Zhou, D., Hou, Z., Yu, J., & Lu, S. (2025). Complementary Rhizosphere Microbial Strategies Drive Functional Specialization in Coastal Halophyte Succession: Differential Adaptation of Suaeda glauca and Phragmites communis to Saline–Alkali Stress. Microorganisms, 13(6), 1399. https://doi.org/10.3390/microorganisms13061399






