Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration and Study Areas
2.2. Sample Collection
2.3. Enrichment and Isolation of Diarrhoeagenic Pathogens
2.4. Bacterial Identification
2.4.1. Mass Spectrometry Technology (MALDI-TOF)
2.4.2. Conventional PCR for Identification of Target Pathogens
- a.
- DNA extraction
- b.
- DNA Amplification by conventional multiplex PCR
2.4.3. Library Preparation and Sequencing
2.4.4. Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Prevalence of Potential Diarrhoeagenic Pathogens by Culture-Based Methods
3.2. Isolates Confirmed as Diarrhoeagenic Pathogens
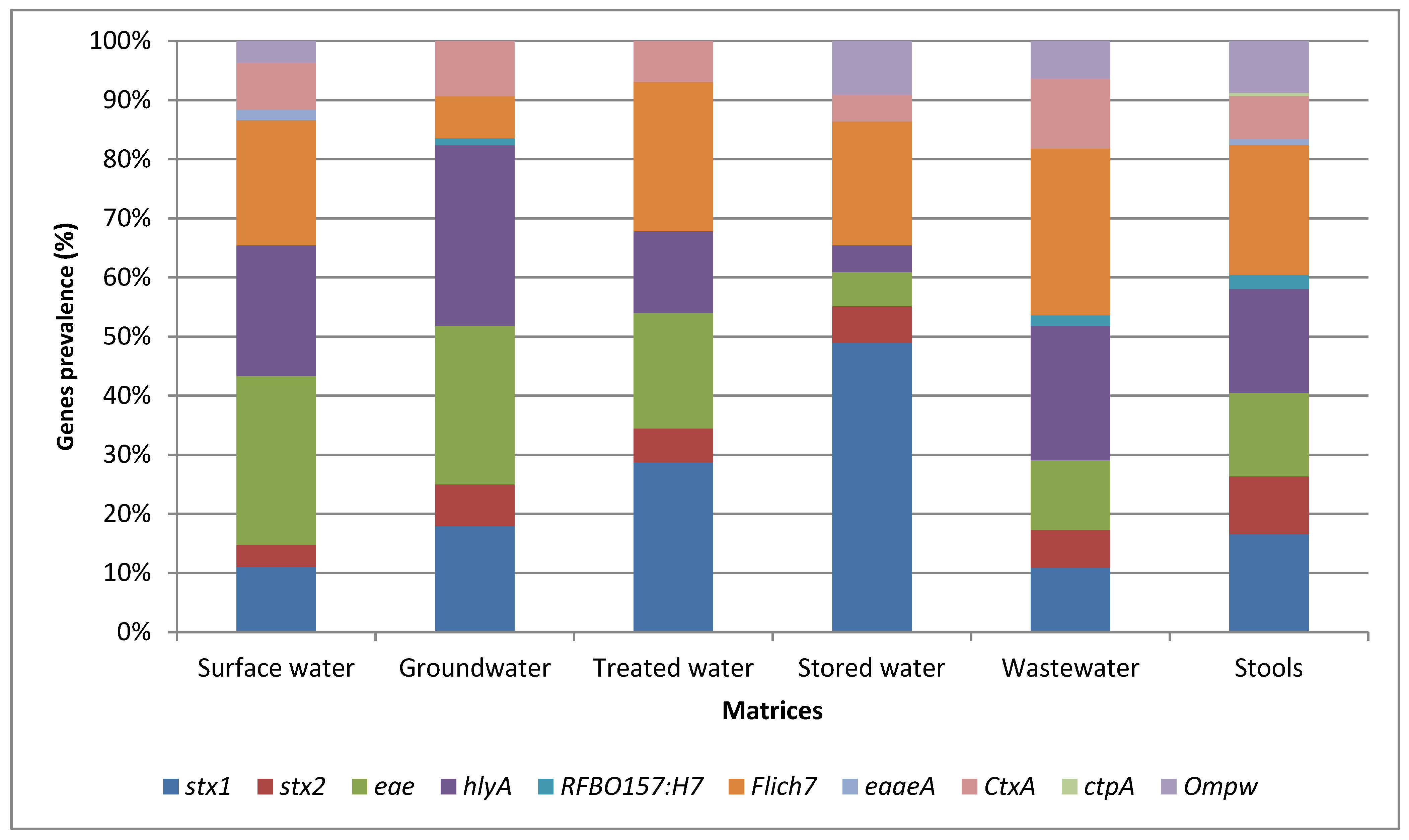
3.3. Abundance of Virulence-Associated Genes Identified from Various Matrices by PCR
3.4. Bioinformatic Analysis
3.4.1. Identification
3.4.2. Most Abundant Virulence Factors (VFs)
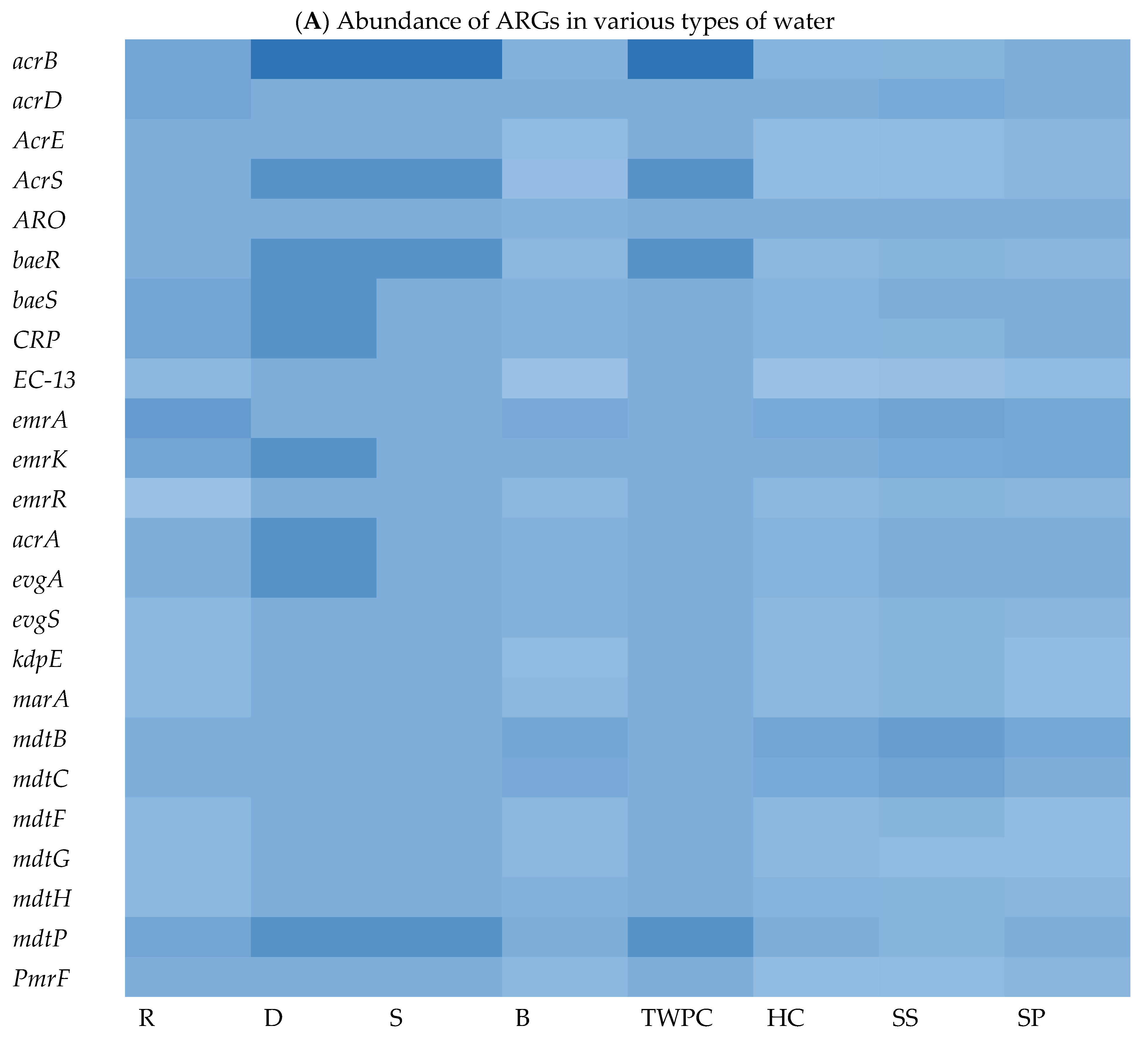
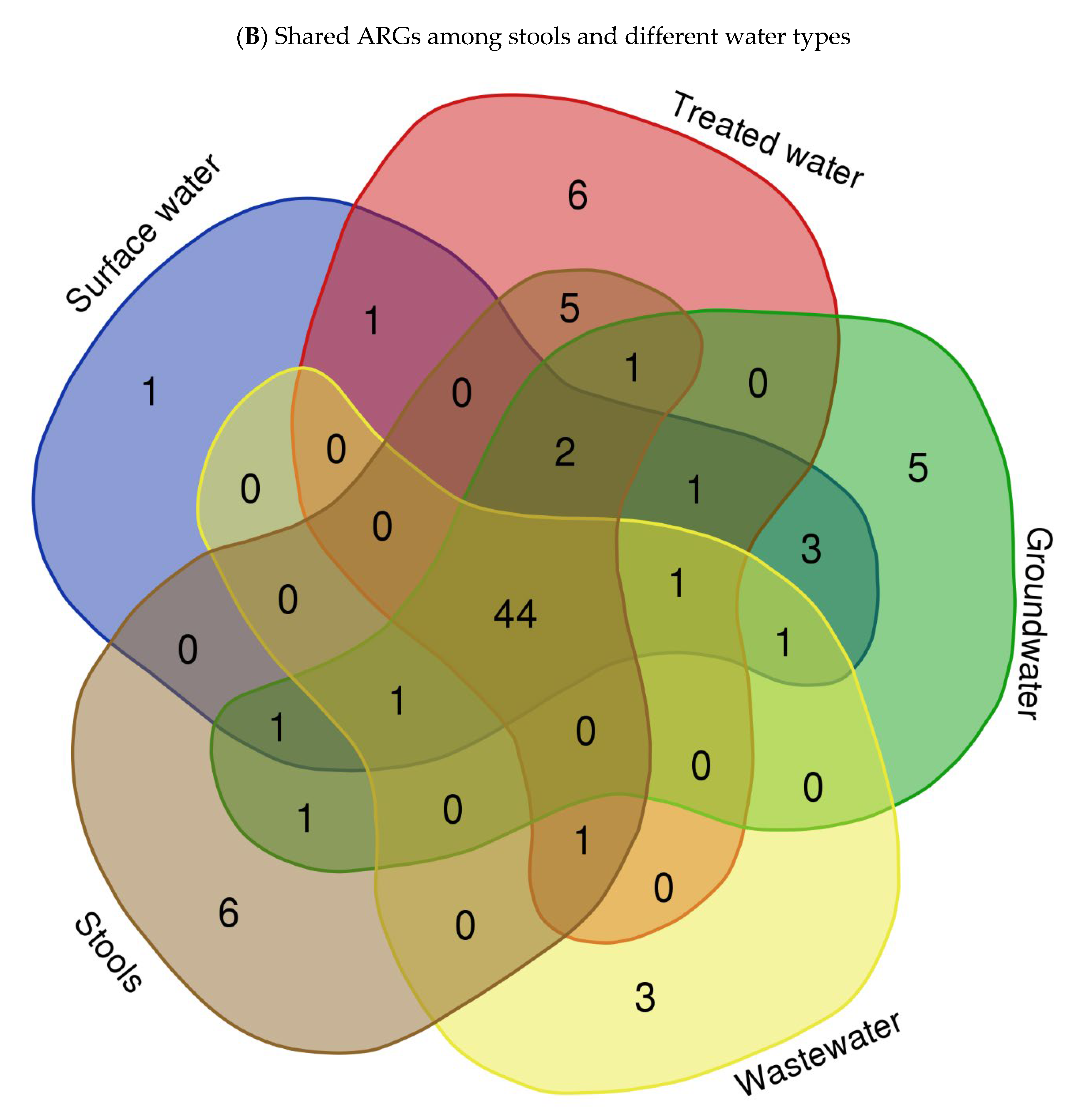
3.4.3. Most Abundant Antibiotic Resistance Genes (ARGs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omarova, A.; Tussupova, K.; Berndtsson, R.; Kalishev, M.; Sharapatova, K. Protozoan parasites in drinking water: A system approach for improved water, sanitation and hygiene in developing countries. Int. J. Environ. Res. Public. Health 2018, 15, 495. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for Drinking Water Quality: Fourth Edition, Incoporating the First and Second Addenda; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Edokpayi, J.N.; Rogawski, E.T.; Kahler, D.M.; Hill, C.L.; Reynolds, C.; Nyathi, E.; Smith, J.A.; Odiyo, J.O.; Samie, A.; Bessong, P.; et al. Challenges to sustainable safe drinking water: A case study of water quality and use across seasons in rural communities in Limpopo Province, South Africa. Water 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Amatobi, D.A.; Agunwamba, J.C. Improved quantitative microbial risk assessment (QMRA) for drinking water sources in developing countries. Appl. Water Sci. 2022, 12, 49. [Google Scholar] [CrossRef]
- Karama, M.; Mainga, A.O.; Cenci-Goga, B.T.; Malahlela, M.; El-Ashram, S.; Kalake, A. Molecular profiling and antimicrobial resistance of Shiga toxin-producing Escherichia coli O26, O45, O103, O121, O145 and O157 isolates from cattle on cow-calf operations in South Africa. Sci. Rep. 2019, 9, 11930. [Google Scholar] [CrossRef]
- Liu, Y.; Li, H.; Chen, X.; Tong, P.; Zhang, Y.; Zhu, M.; Su, Z.; Yao, G.; Li, G.; Cai, W. Characterization of Shiga toxin-producing Escherichia coli isolated from cattle and sheep in Xinjiang Province, China, using whole-genome sequencing. Transbound. Emerg. Dis. 2022, 69, 413–422. [Google Scholar] [CrossRef]
- Greig, D.R.; Hickey, T.J.; Boxall, M.D.; Begum, H.; Gentle, A.; Jenkins, C.; Chattaway, M.A. A real-time multiplex PCR for the identification and typing of Vibrio cholerae, Diagn. Microbiol. Infect. Dis. 2018, 90, 171–176. [Google Scholar] [CrossRef]
- Pasquali, F.; Palma, F.; Trevisani, M.; Parisi, A.; Lucchi, A.; De Cesare, A.; Manfreda, G. Whole genome sequencing based typing and characterisation of Shiga-toxin producing Escherichia coli strains belonging to O157 and O26 serotypes and isolated in dairy farms. Ital. J. Food Saf. 2018, 7, 181–188. [Google Scholar] [CrossRef]
- Bénard, A.H.; Guenou, E.; Fookes, M.; Ateudjieu, J.; Kasambara, W.; Siever, M.; Debes, A.K. Whole genome sequence of Vibrio cholerae directly from dried spotted filter paper. PLoS Negl. Trop. Dis. 2019, 13, e0007330. [Google Scholar] [CrossRef]
- Delannoy, S.; Mariani-Kurkdjian, P.; Webb, H.E.; Bonacorsi, S.; Fach, P. The mobilome; A major contributor to Escherichia coli stx2-Positive O26: H11 strains intra-serotype diversity. Front. Microbiol. 2017, 8, 1625. [Google Scholar] [CrossRef]
- Amézquita-López, B.A.; Soto-Beltrán, M.; Lee, B.G.; Yambao, J.C.; Quiñones, B. Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 2018, 51, 425–434. [Google Scholar] [CrossRef]
- Rumore, J.; Tschetter, L.; Kearney, A.; Kandar, R.; McCormick, R.; Walker, M.; Peterson, C.L.; Reimer, A.; Nadon, C. Evaluation of whole-genome sequencing for outbreak detection of Verotoxigenic Escherichia coli O157:H7 from the Canadian perspective. BMC Genom. 2018, 19, 870. [Google Scholar] [CrossRef] [PubMed]
- Shridhar, P.B.; Worley, J.N.; Gao, X.; Yang, X.; Noll, L.W.; Shi, X.; Bai, J.; Meng, J.; Nagaraja, T.G. Analysis of virulence potential of Escherichia coli O145 isolated from cattle feces and hide samples based on whole genome sequencing. PLoS ONE 2019, 14, e0225057. [Google Scholar] [CrossRef] [PubMed]
- Alhamlan, F.S.; Al-Qahtani, A.A.; Al-Ahdal, M.N. Recommended advanced techniques for waterborne pathogen detection in developing countries. J. Infect. Dev. Ctries. 2015, 9, 128–135. [Google Scholar] [CrossRef] [PubMed]
- García-Aljaro, C.; Blanch, A.R.; Campos, C.; Jofre, J.; Lucena, F. Pathogens, faecal indicators and human-specific microbial source-tracking markers in sewage. J. Appl. Microbiol. 2019, 126, 701–717. [Google Scholar] [CrossRef]
- Cuénod, A.; Foucault, F.; Pflüger, V.; Egli, A. Factors associated with MALDI-TOF mass spectral quality of species identification in clinical routine diagnostics. Front. Cell. Infect. Microbiol. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Ramaite, K.; Ekwanzala, M.D.; Dewar, J.B.; Momba, M.N.B. Human-associated methicillin-resistant Staphylococcus aureus Clonal Complex 80 isolated from cattle and aquatic environments. Antibiotics 2021, 10, 1038. [Google Scholar] [CrossRef]
- Bannon, J.; Melebari, M.; Jordao, C., Jr.; Leon-Velarde, C.G.; Warriner, K. Incidence of Top 6 Shiga Toxigenic Escherichia Coli within Two Ontario Beef Processing Facilities: Challenges in Screening and Confirmation Testing. AIMS Microbiol. 2016, 2, 278–291. [Google Scholar] [CrossRef]
- Fayemi, O.E.; Akanni, G.B.; Elegbeleye, J.A.; Aboaba, O.O.; Njage, P.M. Prevalence, characterization and antibiotic resistance of Shiga toxigenic Escherichia coli serogroups isolated from fresh beef and locally processed ready-to-eat meat products in Lagos, Nigeria. Food Microbiol. 2021, 347, 109191. [Google Scholar] [CrossRef]
- Mehrabadi, J.F.; Morsali, P.; Nejad, H.R.; Fooladi, A.A.I. Detection of toxigenic Vibrio cholerae with new multiplex PCR. J. Infect. Public. Health 2012, 5, 263–267. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 16 April 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Inform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Javaid, M.; Qasim, H.; Zia, H.Z.; Bashir, M.A.; Samiullah, K.; Hashem, M.; Morsy, K.; Dajem, S.B.; Muhammad, T.; Shaheen, M.; et al. Bacteriological composition of groundwater and its role in human health. J. King Saud Univ. Sci. 2022, 34, 102128. [Google Scholar] [CrossRef]
- Gwimbi, P.; George, M.; Ramphalile, M. Bacterial contamination of drinking water sources in rural villages of Mohale Basin, Lesotho: Exposures through neighbourhood sanitation and hygiene practices. Environ. Health Prev. Med. 2019, 24, 33. [Google Scholar] [CrossRef]
- Sila, O.N.A. Physico-chemical and bacteriological quality of water sources in rural settings, a case study of Kenya, Africa. Sci. Afr. 2019, 2, e00018. [Google Scholar] [CrossRef]
- Pang, X.; Qiu, Y.; Gao, T.; Zurawell, R.; Neumann, N.F.; Craik, S.; Lee, B.E. Prevalence, levels and seasonal variations of human enteric viruses in six major rivers in Alberta, Canada. Water Res. 2019, 153, 349–356. [Google Scholar] [CrossRef]
- Genter, F.; Putri, G.L.; Pratama, M.A.; Priadi, C.; Willetts, J.; Foster, T. Microbial contamination of groundwater self-supply in urban Indonesia: Assessment of sanitary and socio-economic risk factors. Water Resour. Res. 2022, 58, e2021WR031843. [Google Scholar] [CrossRef]
- Obanor, O.; Afegbua, S.L.; Ameh, J.B. Sanitary status and water quality of some drinking water sources and antibiogram of Shiga toxin-producing Escherichia coli O157:H7 isolated from Shika, Zaria, Nigeria. Int. J. Environ. Health Res. 2022, 33, 1604–1616. [Google Scholar] [CrossRef]
- Hada, H.S.; Stemmler, J.; Grossbard, M.L.; West, P.A.; Potrikus, C.J.; Hastings, J.W. Characterization of non-O1 serovar Vibrio cholerae (Vibrio albensis). Syst. Appl. Microbiol. 1985, 6, 203–209. [Google Scholar] [CrossRef]
- Gronthoud, F.A. (Ed.) Practical Clinical Microbiology and Infectious Diseases: A Hands-On Guide; CRC Press: Boca Raton, FL, USA, 2020; p. 484. [Google Scholar]
- Kurwadkar, S. Occurrence and distribution of organic and inorganic pollutants in groundwater. Water Environ. Res. 2019, 91, 1001–1008. [Google Scholar] [CrossRef]
- Hakim, A.L.; Saputra, D.D.; Tanika, L.; Kusumawati, I.A.; Sari, R.R.; Andreotti, F.; Bagbohouna, M.; Abdurrahim, A.Y.; Wamucii, C.; Lagneaux, E.G.; et al. Protected spring and sacred forest institutions at the instrumental—relational value interface. Curr. Opin. Environ. Sustain. 2023, 62, 101292. [Google Scholar] [CrossRef]
- Tayh, G.; Boubaker, S.M.; Khedher, R.B.; Jbeli, M.; Chehida, F.B.; Mamlouk, A.; Messadi, L. Prevalence, virulence genes, and antimicrobial profiles of Escherichia coli O157:H7 isolated from healthy cattle in Tunisia. J. Infect. Dev. Ctries 2022, 16, 1308–1316. [Google Scholar] [CrossRef]
- Nhu, N.T.K.; Phan, M.D.; Forde, B.M.; Murthy, A.M.; Peters, K.M.; Day, C.J.; Schembri, M.A. Complex multilevel control of hemolysin production by uropathogenic Escherichia coli. mBio 2019, 10, e02248-19. [Google Scholar] [CrossRef]
- Awad, W.S.; El-Sayed, A.A.; Mohammed, F.F.; Bakry, N.M.; Abdou, N.E.M.; Kamel, M.S. Molecular characterisation of pathogenic Escherichia coli isolated from diarrheic and in-contact cattle and buffalo calves. Trop. Anim. Health Prod. 2020, 52, 3173–3185. [Google Scholar] [CrossRef]
- Lou, R.N.; Jacobs, A.; Wilder, A.P.; Therkildsen, N.O. A beginner’s guide to low-coverage whole genome sequencing for population genomics. Mol. Ecol. 2021, 30, 5966–5993. [Google Scholar] [CrossRef]
- Traoré, A.N.; Mulaudzi, K.; Chari, G.J.; Foord, S.H.; Mudau, L.S.; Barnard, T.G.; Potgieter, N. The impact of human activities on microbial quality of rivers in the Vhembe District, South Africa. Int. J. Environ. Res. Public Health 2016, 13, 817. [Google Scholar] [CrossRef]
- Totsika, M.; Wells, T.J.; Beloin, C.; Valle, J.; Allsopp, L.P.; King, N.P.; Ghigo, J.M.; Schembri, M.A. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 2179–2189. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, J.; Ambikan, A.; Jernberg, C.; Ehricht, R.; Scheutz, F.; Xiong, Y.; Matussek, A. Molecular characterization and comparative genomics of clinical hybrid Shiga toxin-producing and enterotoxigenic Escherichia coli (STEC/ETEC) strains in Sweden. Sci. Rep. 2019, 9, 5619. [Google Scholar] [CrossRef] [PubMed]
- Ateba, C.N.; Mbewe, M. Detection of Escherichia coli O157:H7 virulence genes in isolates from beef, pork, water, human and animal species in the Northwest Province, South Africa: Public health implications. Res. Microbiol. 2011, 162, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, M.; Nabi, B.; Daswani, M.; Viquar, I.; Pal, N.; Sharma, P.; Tiwari, S.; Sarma, D.K.; Shubham, S.; Kumar, M. Role of bacterial efflux pump proteins in antibiotic resistance across microbial species. Microb. Pathog. 2023, 181, 106182. [Google Scholar] [CrossRef] [PubMed]
- Pulingam, T.; Parumasivam, T.; Gazzali, A.M.; Sulaiman, A.M.; Chee, J.Y.; Lakshmanan, M.; Chin, C.F.; Sudesh, K. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 2022, 170, 106103. [Google Scholar] [CrossRef]
- Yang, J.; Wang, H.; Roberts, D.J.; Du, H.N.; Yu, X.F.; Zhu, N.Z.; Meng, X.Z. Persistence of antibiotic resistance genes from river water to tap water in the Yangtze River Delta. Sci. Total Environ. 2020, 742, 140592. [Google Scholar] [CrossRef]
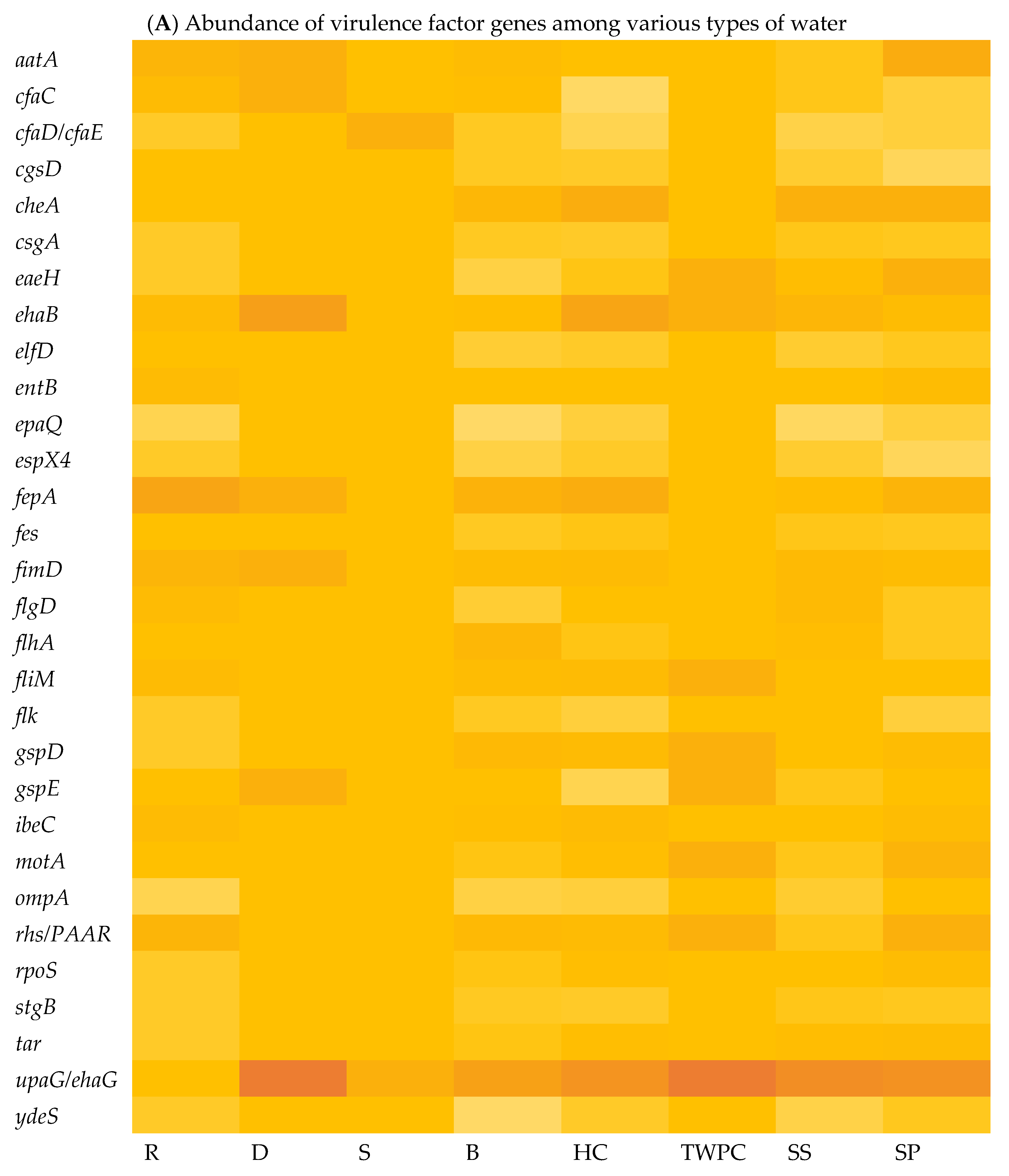

| Organism | Genes | Primer Sequence (F: Forward, R: Reverse) 5′ to 3′ | Product Size (bp) | Annealing Temp (°C) | References |
|---|---|---|---|---|---|
| STEC | stx1 | F-ATAAATCGCCATTCGTTGACTAC | 180 | 55 | [18] |
| R-AGAACGCCCACTGAGATCATC | |||||
| stx2 | F-GGCACTGTCTGAAACTGCTCC | 255 | 55 | ||
| R-TCGCCAGTTATCTGACATTCTG | |||||
| Eae | F-GACCCGGCACAAGCATAAGC | 384 | 55 | ||
| R-CCACCTGCAGCAACAAGAGG | |||||
| hlyA | F-GCATCATCAAGCGTACGTTCC | 534 | 55 | ||
| R-AATGAGCCAAGCTGGTTAAGCT | |||||
| E. coli O157:H7 | rfbO157:H7 | F-CGGACATCCATGTGATATGG | 259 | 56 | [19] |
| R-TTGCCTATGTACAGCTAATCC | |||||
| fliCH7 | F-TACCATCGCAAAAGCAACTCC | 247 | 56 | ||
| R-GTCGGCAACGTTAGTGATACC | |||||
| eaeA | F-AAG CGA CTG AGG TCA CT | 450 | 56 | ||
| R-ACG CTG CTC ACT AGA TGT | |||||
| V. cholerae | ctxA | F-GGTCTTATGCCAGAGGACAG | 219 | 50 | [20] |
| R-GTTGGGTGCAGTGGCTATAAC | |||||
| tcpA | F-ATTCTTGGTGATCTCATGATAAGG | 295 | 50 | ||
| R-TTAATTCACCACAAATATCTGCC | |||||
| ompW | F-CACCAAGAAGGTGACTTTATTGTG | 588 | 50 | ||
| R-GAACTTATAACCACCCGCG | |||||
| R-ACCCAGTTTGCAGTTCCGAATGT |
| Matrices | Total Sample | Total Number of Samples Testing Positive (%) | |||||
|---|---|---|---|---|---|---|---|
| STEC | E. coli O157:H7 | V. Cholerae | |||||
| Surface water | 136 | 32 | (23.5%) | 8 | (5.9%) | 31 | (22.8%) |
| Groundwater | 664 | 25 | (3.8%) | 20 | (3.0%) | 9 | (1.4%) |
| Treated water * | 640 | 17 | (2.7%) | 14 | (2.2%) | 15 | (2.3%) |
| Stored water | 1624 | 96 | (5.9%) | 30 | (1.85%) | 88 | (5.4%) |
| Wastewater | 104 | 9 | (8.7%) | 13 | (12.5%) | 18 | (17.3%) |
| Stool | 135 | 44 | (32.6%) | 41 | (30.4%) | 45 | (33.3%) |
| Total | 3303 | 223 | (6.8%) | 126 | (3.8%) | 206 | 6.2%) |
| Matrices | Total Sample | Total Number of Samples Testing Positive (%) | |||||
|---|---|---|---|---|---|---|---|
| STEC | E. coli O157:H7 | V. Cholerae | |||||
| Surface water | 71 | 10 | (14.1%) | 5 | (0.7%) | 7 | (9.9%) |
| Groundwater | 68 | 25 | (36.8%) | 19 | (2.8%) | 10 | (14.7%) |
| Treated water | 37 | 20 | (54.1%) | 6 | (1.6%) | 8 | (21.6%) |
| Stored water * | 209 | 89 | (42.6%) | 34 | (1.6%) | 23 | (11.0%) |
| Wastewater | 40 | 24 | (60.0%) | 9 | (2.3%) | 7 | (17.5%) |
| Stools | 130 | 36 | (27.7%) | 14 | (1.1%) | 21 | (16.2%) |
| Total | 555 | 204 | (36.8%) | 87 | (1.6%) | 76 | (13.7%) |
| Sample ID | Species | No. of Contigs | Total Length (bp) | No. of Sequence | N50 | GC (%) | No. of Genes | GenBank Accession No. |
|---|---|---|---|---|---|---|---|---|
| B,MK1 | Escherichia coli | 1584 | 1,255,779 | 4243 | 801 | 51.81 | 3094 | JASBBF000000000 |
| B,MK2 | Escherichia coli | 1703 | 1,326,996 | 4829 | 774 | 52.55 | 3445 | JASBBG000000000 |
| B,MK3 | Escherichia coli | 2863 | 2,971,202 | 4815 | 1122 | 52.97 | 3838 | JASBBH000000000 |
| B,TM | Escherichia coli | 1586 | 1,178,354 | 5527 | 738 | 50.71 | 3695 | JASBBI000000000 |
| B,TM1 | Escherichia coli | 1455 | 1,088,444 | 4993 | 741 | 52.11 | 3429 | JASBBJ000000000 |
| B,TM2 | Escherichia coli | 3507 | 3,968,282 | 5004 | 1224 | 52.07 | 4278 | JASBBK000000000 |
| B,TM3 | Escherichia coli | 2989 | 3,280,046 | 4779 | 1203 | 52.78 | 3925 | JASBBL000000000 |
| D,MK | Escherichia coli | 3588 | 3,745,404 | 6211 | 1119 | 51.32 | 4860 | JASBBM000000000 |
| HC,CC4 | Escherichia coli | 1895 | 1,569,200 | 4783 | 852 | 52.71 | 3506 | JASBBN000000000 |
| HC,CC5 | Escherichia coli | 3385 | 3,542,931 | 6013 | 1134 | 52.11 | 4564 | JASBBO000000000 |
| HC,MK1 | Escherichia coli | 1663 | 1,241,949 | 5009 | 738 | 50.88 | 3557 | JASBBP000000000 |
| HC,MK3 | Escherichia coli | 1535 | 1,148,875 | 5144 | 741 | 52.18 | 3604 | JASBBQ000000000 |
| HC,MK | Escherichia coli | 1588 | 1,195,056 | 5661 | 756 | 52.92 | 3824 | JASBBR000000000 |
| HC,TM4 | Escherichia coli | 1728 | 1,338,484 | 5087 | 774 | 52.48 | 3637 | JASBBS000000000 |
| R,CC1 | Escherichia coli | 2819 | 2,790,526 | 5045 | 1047 | 52.36 | 4022 | JASBBT000000000 |
| R,TM1 | Escherichia coli | 1638 | 1,175,496 | 6354 | 705 | 49.75 | 3996 | JASBBU000000000 |
| R,TM3 | Escherichia coli | 1611 | 1,280,091 | 4593 | 798 | 52.2 | 3270 | JASBBV000000000 |
| S,TM1 | Escherichia coli | 1883 | 1,512,031 | 5449 | 813 | 52.45 | 3573 | JASBBW000000000 |
| SP,CC1 | Escherichia coli | 1429 | 1,069,863 | 5260 | 753 | 51.81 | 3617 | JASBBX000000000 |
| SP,MK1 | Escherichia coli | 1787 | 1,476,801 | 4787 | 837 | 52.49 | 3370 | JASBBY000000000 |
| SP,MK2 | Escherichia coli | 3026 | 3,153,597 | 4333 | 1104 | 52.79 | 3205 | JASBBZ000000000 |
| SP,MK3 | Escherichia coli | 1624 | 1,247,824 | 4963 | 771 | 52.39 | 4016 | JASBCA000000000 |
| SS,CC1 | Escherichia coli | 2532 | 2,348,190 | 4842 | 966 | 52.71 | 3405 | JASBCB000000000 |
| SS,CC2 | Escherichia coli | 2454 | 2,372,235 | 4924 | 1011 | 52.3 | 3774 | JASBCC000000000 |
| SS,MK | Escherichia coli | 3048 | 3,199,724 | 4544 | 1125 | 52.68 | 3462 | JASBCD000000000 |
| SS,MK2 | Escherichia coli | 1776 | 1,443,500 | 4962 | 831 | 52.32 | 3981 | JASBCE000000000 |
| SS,MK3 | Escherichia coli | 1461 | 1,060,329 | 4769 | 717 | 52.02 | 3493 | JASBCF000000000 |
| TWPC,CC1 | Escherichia coli | 1826 | 1,527,059 | 4486 | 846 | 53.03 | 3290 | JASBCG000000000 |
| Matrices | Total Sample | Total Number of Samples Testing Positive (%) | |||||
|---|---|---|---|---|---|---|---|
| STEC | E. coli O157:H7 | V. Cholerae | |||||
| Surface water | 18 | 9 | (50.0%) | 5 | (27.8%) | 4 | (22.2%) |
| Groundwater | 66 | 23 | (34.8%) | 18 | (27.3%) | 8 | (12.1%) |
| Treated water | 35 | 19 | (54.3%) | 6 | (17.1%) | 6 | (17.1%) |
| Stored water * | 158 | 76 | (48.1%) | 43 | (27.2%) | 22 | (13.9%) |
| Wastewater | 40 | 20 | (50.0%) | 11 | (27.5%) | 7 | (17.5%) |
| Stools | 50 | 33 | (66.0%) | 12 | (24.0%) | 18 | (36.0%) |
| Total | 367 | 180 | (49.0%) | 95 | (25.9%) | 65 | (17.7%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murei, A.; Momba, M.N.B. Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing. Microorganisms 2025, 13, 1373. https://doi.org/10.3390/microorganisms13061373
Murei A, Momba MNB. Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing. Microorganisms. 2025; 13(6):1373. https://doi.org/10.3390/microorganisms13061373
Chicago/Turabian StyleMurei, Arinao, and Maggy Ndombo Benteke Momba. 2025. "Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing" Microorganisms 13, no. 6: 1373. https://doi.org/10.3390/microorganisms13061373
APA StyleMurei, A., & Momba, M. N. B. (2025). Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing. Microorganisms, 13(6), 1373. https://doi.org/10.3390/microorganisms13061373







