Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterium Isolation and Characterization of Geobacillus sp. G4
2.2. DNA Extraction and Sequencing
2.3. Quality Analysis, Genome Assembly, and Annotation of Geobacillus sp. G4
2.4. Taxonomic Classification of Geobacillus sp. G4
2.4.1. Digital DNA Hybridization (dDDH) and Average Nucleotide Identity (ANI)
2.4.2. 16S rRNA Gene-Based Phylogenetic Tree Reconstruction
2.4.3. Phylogenetic Tree Reconstruction
3. Results
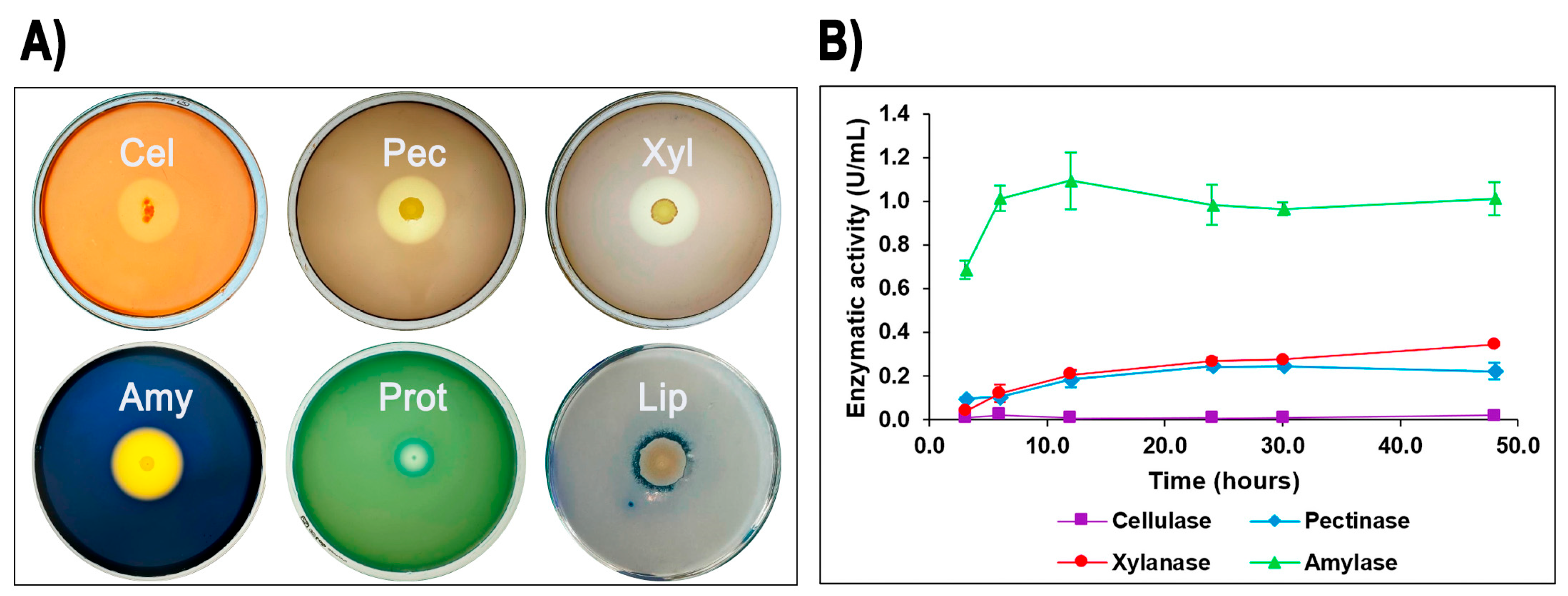
3.1. Characterization of Geobacillus sp. G4
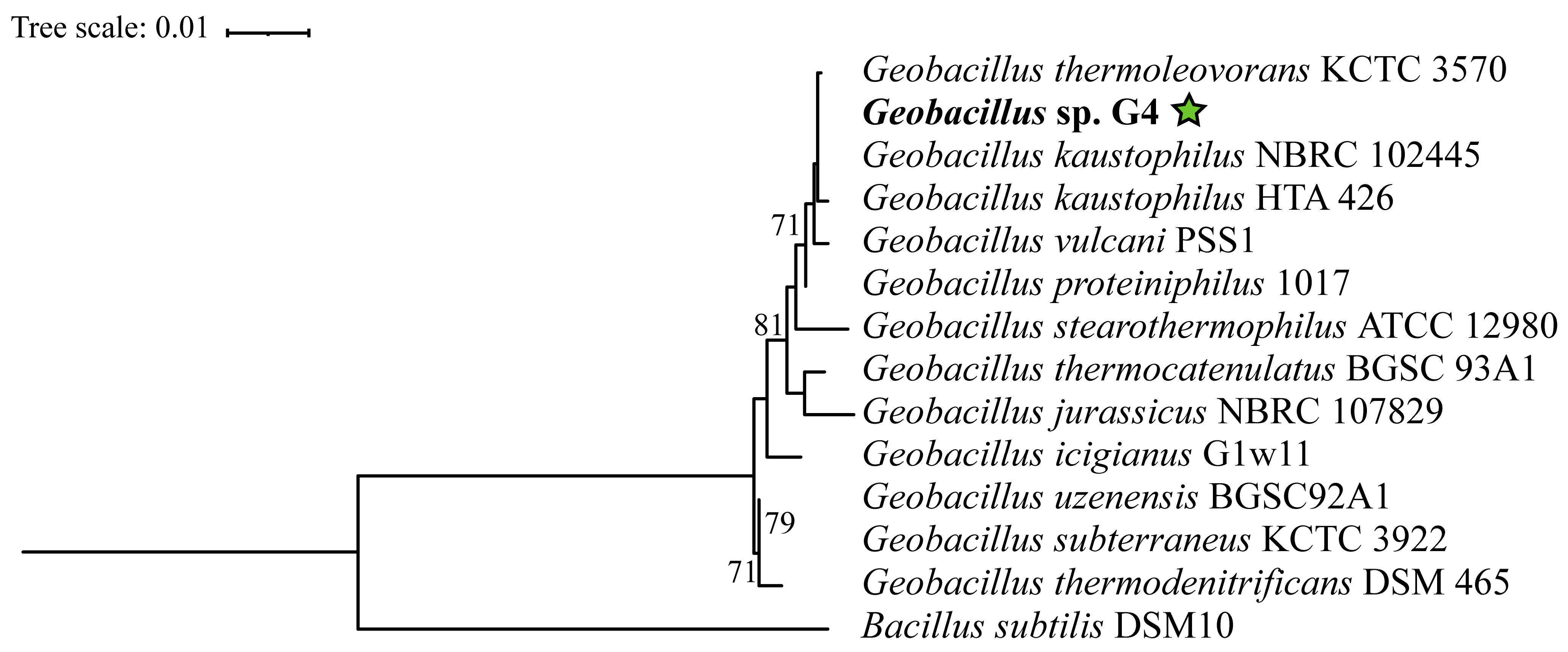
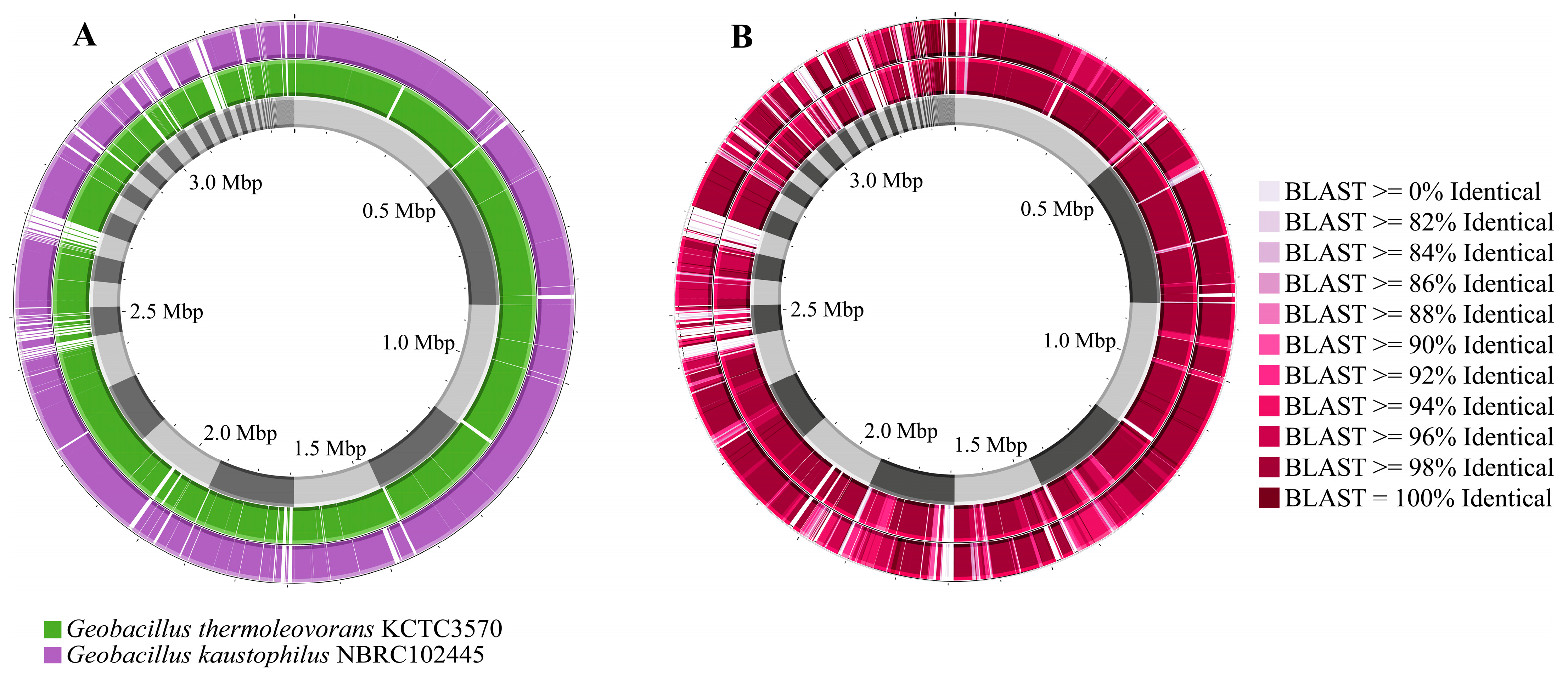
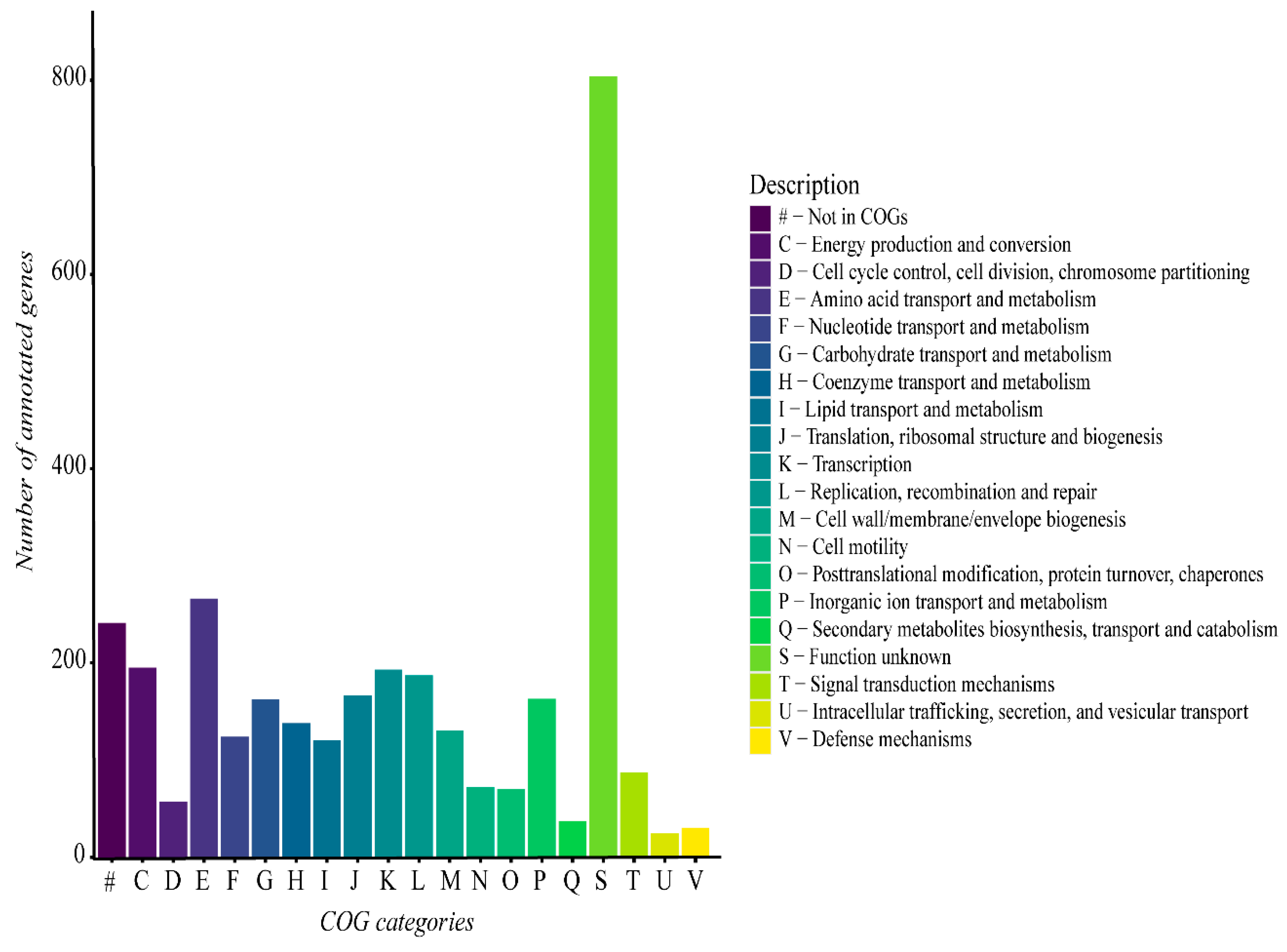
3.2. Genome Description and Taxonomic Classification of Geobacillus sp. G4
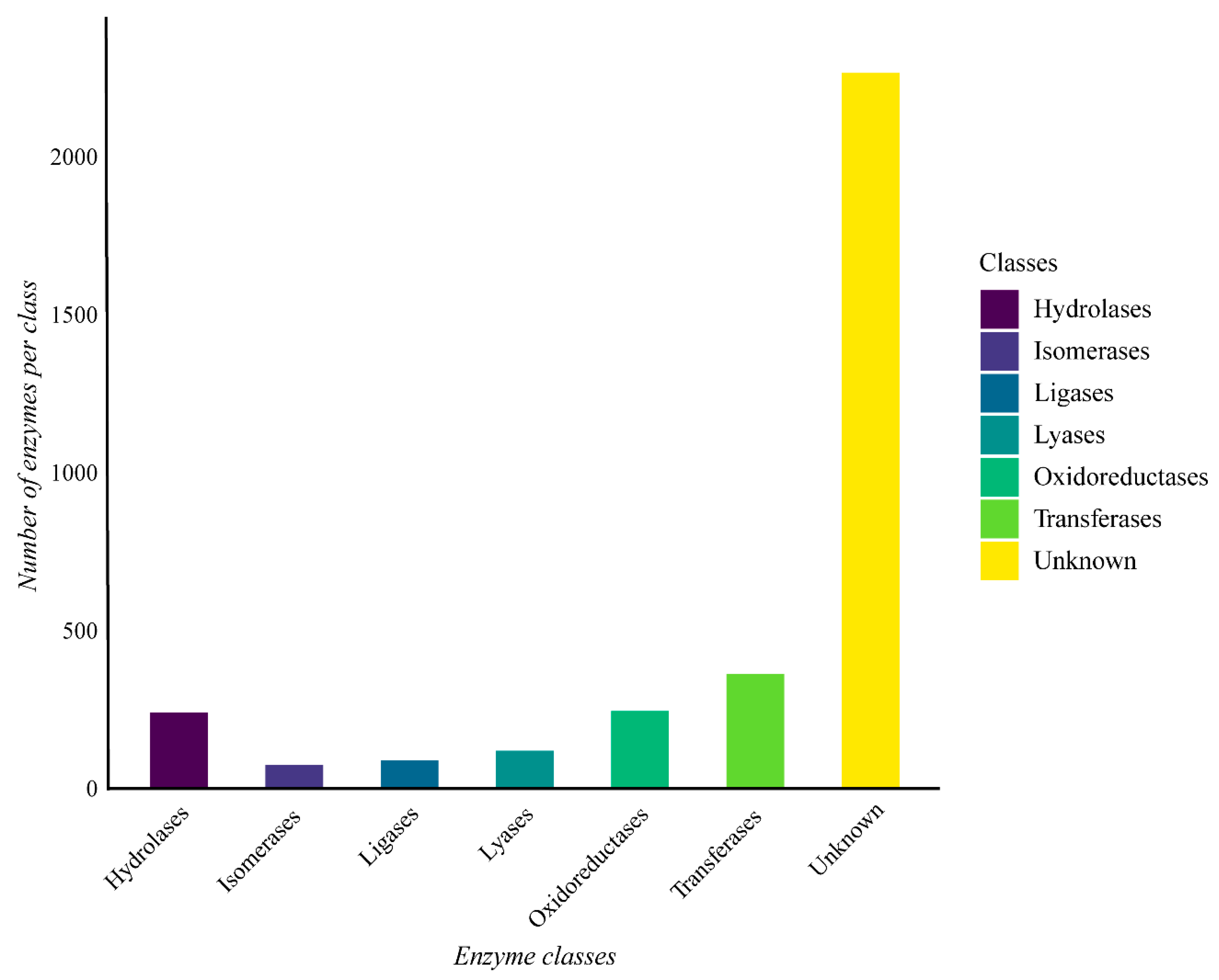
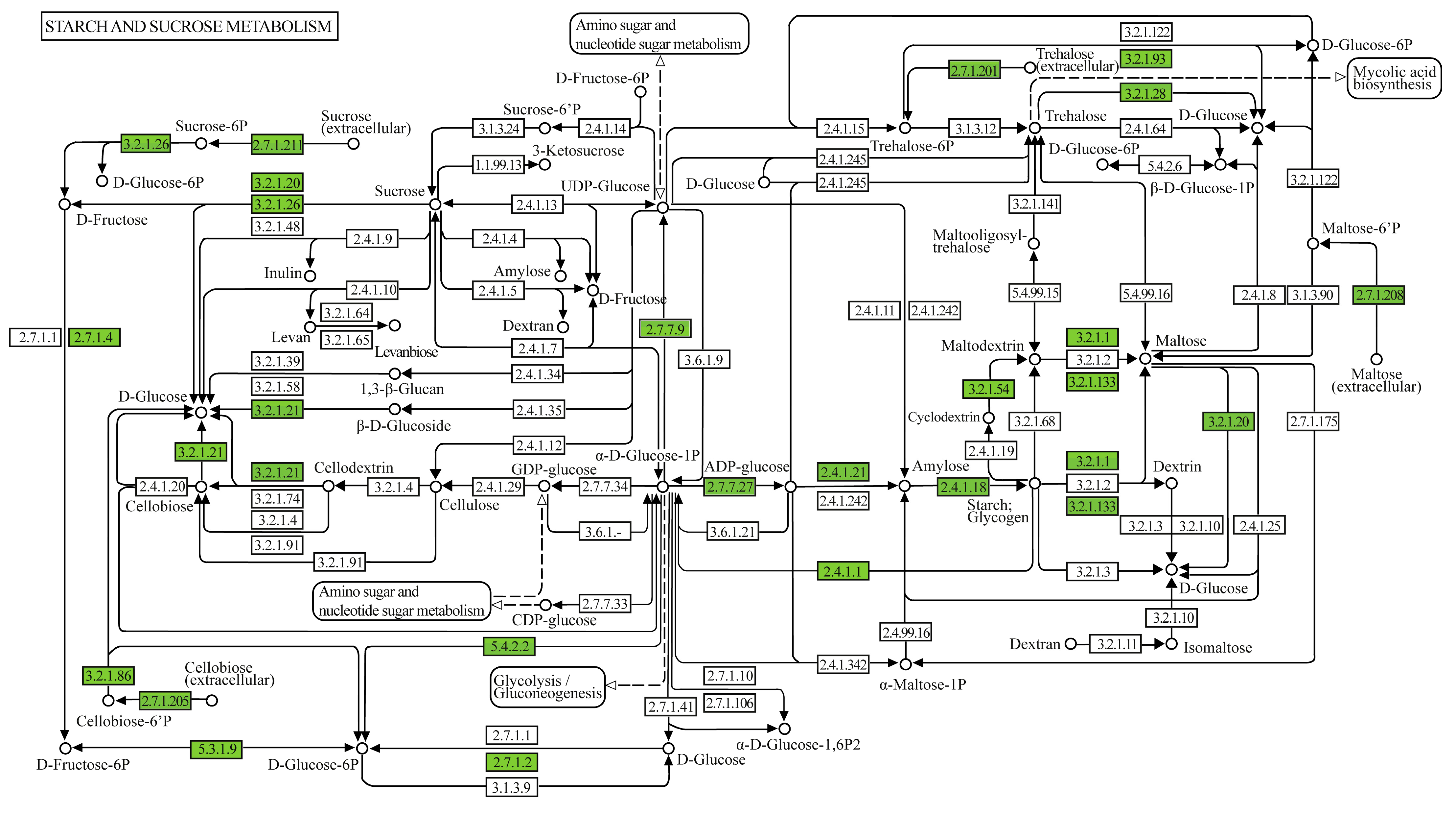
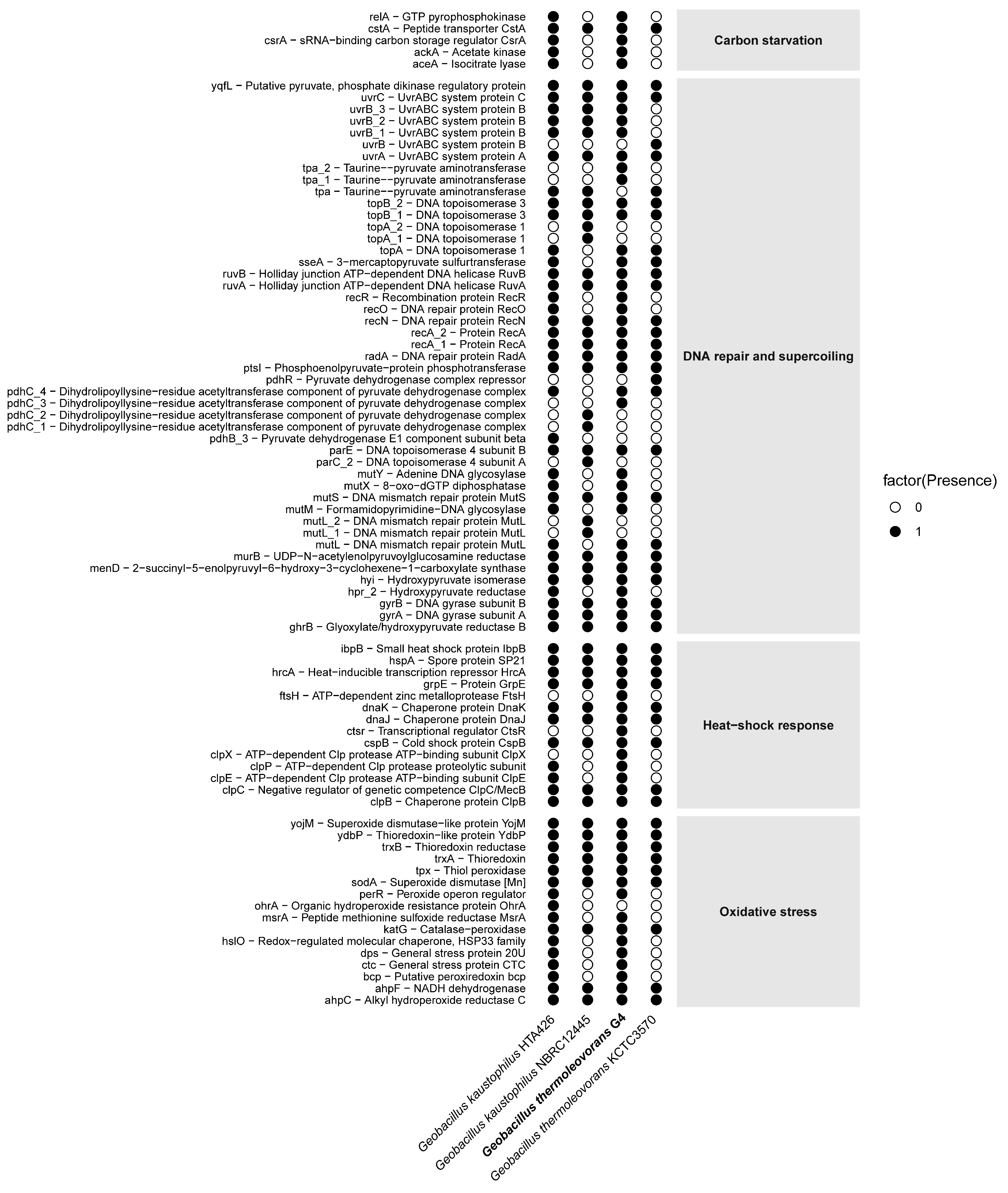
3.3. Functional Analysis of Geobacillus thermoleovorans G4 Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Najar, I.N.; Thakur, N. A Systematic Review of the Genera Geobacillus and Parageobacillus: Their Evolution, Current Taxonomic Status and Major Applications. Microbiology 2020, 166, 800–816. [Google Scholar] [CrossRef] [PubMed]
- Dumorné, K.; Córdova, D.C.; Astorga-Eló, M.; Renganathan, P. Extremozymes: A Potential Source for Industrial Applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic Study of Aerobic Thermophilic Bacilli: Descriptions of Geobacillus subterraneus Gen. Nov., Sp. Nov. and Geobacillus uzenensis Sp. Nov. from Petroleum Reservoirs and Transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef]
- LPSN—List of Prokaryotic Names with Standing in Nomenclature. Available online: https://lpsn.dsmz.de/ (accessed on 30 March 2025).
- Coorevits, A.; Dinsdale, A.E.; Halket, G.; Lebbe, L.; de Vos, P.; van Landschoot, A.; Logan, N.A. Taxonomic Revision of the Genus Geobacillus: Emendation of Geobacillus, G. stearothermophilus, G. jurassicus, G. toebii, G. thermodenitrificans and G. Thermoglucosidans (Nom. Corrig., Formerly ‘Thermoglucosidasius’); Transfer of Bacillus thermantarcticus to the genus as G. thermantarcticus comb. nov.; proposal of Caldibacillus debilis gen. nov., comb. nov.; transfer of G. tepidamans to Anoxybacillus as A. tepidamans comb. nov.; and proposal of Anoxybacillus caldiproteolyticus sp. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 1470–1485. [Google Scholar] [CrossRef]
- Kuisiene, N.; Raugalas, J.; Chitavichius, D. Geobacillus lituanicus Sp. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 1991–1995. [Google Scholar] [CrossRef]
- Meintanis, C.; Chalkou, K.I.; Kormas, K.A.; Lymperopoulou, D.S.; Katsifas, E.A.; Hatzinikolaou, D.G.; Karagouni, A.D. Application of RpoB Sequence Similarity Analysis, REP-PCR and BOX-PCR for the Differentiation of Species within the Genus Geobacillus. Lett. Appl. Microbiol. 2008, 46, 395–401. [Google Scholar] [CrossRef]
- Zeigler, D.R. The Geobacillus Paradox: Why Is a Thermophilic Bacterial Genus so Prevalent on a Mesophilic Planet? Microbiology 2014, 160, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J. Some (Bacilli) like It Hot: Genomics of Geobacillus Species. Microb. Biotechnol. 2015, 8, 40–48. [Google Scholar] [CrossRef]
- Nazina, T.N.; Lebedeva, E.V.; Poltaraus, A.B.; Tourova, T.P.; Grigoryan, A.A.; Sokolova, D.S.; Lysenko, A.M.; Osipov, G.A. Geobacillus gargensis sp. Nov., a Novel Thermophile from a Hot Spring, and the Reclassification of Bacillus vulcani as Geobacillus vulcani Comb. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2019–2024. [Google Scholar] [CrossRef]
- Nazina, T.N.; Sokolova, D.S.; Grigoryan, A.A.; Shestakova, N.M.; Mikhailova, E.M.; Poltaraus, A.B.; Tourova, T.P.; Lysenko, A.M.; Osipov, G.A.; Belyaev, S.S. Geobacillus jurassicus sp. Nov., a New Thermophilic bacterium Isolated from a High-Temperature Petroleum Reservoir, and the Validation of the Geobacillus Species. Syst. Appl. Microbiol. 2005, 28, 43–53. [Google Scholar] [CrossRef]
- Miñana-Galbis, D.; Pinzón, D.L.; Lorén, J.G.; Manresa, À.; Oliart-Ros, R.M. Reclassification of Geobacillus pallidus (Scholz et al. 1988) Banat et al. 2004 as Aeribacillus pallidus Gen. Nov., Comb. Nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1600–1604. [Google Scholar] [CrossRef] [PubMed]
- Valdez, S.; de la Vega, F.V.; Pairazaman, O.; Castellanos, R.; Esparza, M. Hyperthermophile Diversity Microbes in the Calientes Geothermal Field, Tacna, Peru. Braz. J. Microbiol. 2023, 54, 2927–2937. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, R.; Santos, K.C.R.; Ticona, A.R.P.; Ramirez-arua, H.E.; Bornás-acosta, S.A.; Silva, P.; Hamann, P.R.V.; Lopes, F.A.C. Draft Genome Sequence of Geobacillus sp. Strain G4 Isolated from the Calientes Geothermal Field in Tacna, Peru. Microbiol. Resour. Announc. 2024, 13, e0071724. [Google Scholar] [CrossRef] [PubMed]
- Cruz, V.; Vargas, V.; Matsuda, K. Geochemical Characterization of Thermal Waters in the Calientes Geothermal Field, Tacna, South of Peru. Proc. World Geotherm. Congr. 2010, 1, 1–7. [Google Scholar]
- Vargas, V.; Cruz Pauccara, V.; Antayhua Vera, Y.; Rivera Porras, M.; Cacya Dueñas, L. Estudio Geotérmico Del Campo Calientes; Instituto Geológico, Minero y Metalúrgico–INGEMMET: Lima, Peru, 2012; Volume 48. [Google Scholar]
- Cortez, Y.; Paul, S.; Villena, G.K.; Gutiérrez-Correa, M.; Rodríguez-Herrera, R.; Kharat, A.S. Isolation and Characterization of Cellulase Producing Bacterial Strains from an Amazonian Geothermal Spring in Peru. Microbiol. Res. J. Int. 2016, 15, 1–8. [Google Scholar] [CrossRef]
- Khaswal, A.; Chaturvedi, N.; Mishra, S.K.; Kumar, P.R.; Paul, P.K. Current Status and Applications of Genus Geobacillus in the Production of Industrially Important Products—A Review. Folia Microbiol. 2022, 67, 389–404. [Google Scholar] [CrossRef]
- Suzuki, H. Peculiarities and Biotechnological Potential of Environmental Adaptation by Geobacillus Species. Appl. Microbiol. Biotechnol. 2018, 102, 10425–10437. [Google Scholar] [CrossRef]
- Mol, M.; de Maayer, P. Elucidating the Biotechnological Potential of the Genera Parageobacillus and Saccharococcus through Comparative Genomic and Pan-Genome Analysis. BMC Genom. 2024, 25, 723. [Google Scholar] [CrossRef]
- van Staden, A.D.P.; van Zyl, W.F.; Trindade, M.; Dicks, L.M.T.; Smith, C. Therapeutic Application of Lantibiotics and Other Lanthipeptides: Old and New Findings. Appl. Environ. Microbiol. 2021, 87, e00186-21. [Google Scholar] [CrossRef]
- Brumm, P.J.; De Maayer, P.; Mead, D.A.; Cowan, D.A. Genomic Analysis of Six New Geobacillus Strains Reveals Highly Conserved Carbohydrate Degradation Architectures and Strategies. Front. Microbiol. 2015, 6, 137987. [Google Scholar] [CrossRef]
- De Maayer, P.; Brumm, P.J.; Mead, D.A.; Cowan, D.A. Comparative Analysis of the Geobacillus Hemicellulose Utilization Locus Reveals a Highly Variable Target for Improved Hemicellulolysis. BMC Genom. 2014, 15, 836. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Mac Faddin, J.F. Biochemical Tests for Identification of Medical Bacteria, 3rd ed.; Williams and Wilkins: Baltimore, MD, USA, 2000; ISBN 0683053183. [Google Scholar]
- Guta, M.; Abebe, G.; Bacha, K.; Cools, P. Screening and Characterization of Thermostable Enzyme-Producing Bacteria from Selected Hot Springs of Ethiopia. Microbiol. Spectr. 2024, 12, e0371023. [Google Scholar] [CrossRef]
- Ticona, A.R.P.; Ullah, S.F.; Hamann, P.R.V.; Lopes, F.A.C.; Noronha, E.F. Paenibacillus barengoltzii A1_50L2 as a Source of Plant Cell Wall Degrading Enzymes and Its Use on Lignocellulosic Biomass Hydrolysis. Waste Biomass Valorization 2021, 12, 393–405. [Google Scholar] [CrossRef]
- Oluoch, K.R.; Okanta, P.W.; Hatti-Kaul, R.; Mattiasson, B.; Mulaa, F.J. Protease-, Pectinase- and Amylase- Producing Bacteria from a Kenyan Soda Lake. Open Biotechnol. J. 2018, 12, 33–45. [Google Scholar] [CrossRef]
- Sharma, P.; Sharma, N.; Pathania, S.; Handa, S. Purification and Characterization of Lipase by Bacillus methylotrophicus PS3 under Submerged Fermentation and Its Application in Detergent Industry. J. Genet. Eng. Biotechnol. 2017, 15, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 2006, 34, 32–36. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; Cukura, A.; et al. The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Saitou, N. On the Maximum Likelihood Method in Molecular Phylogenetics. J. Mol. Evol. 1991, 32, 443–445. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Wu, M.; Scott, A.J. Phylogenomic Analysis of Bacterial and Archaeal Sequences with AMPHORA2. Bioinformatics 2012, 28, 1033–1034. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. The Genus Geobacillus and Their Biotechnological Potential. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 92, pp. 1–48. [Google Scholar] [CrossRef]
- Meslé, M.M.; Mueller, R.C.; Peach, J.; Eilers, B.; Tripet, B.P. Isolation and Characterization of Lignocellulose-Degrading Geobacillus thermoleovorans from Yellowstone National Park. Appl. Environ. Microbiol. 2022, 88, e00958-21. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, S.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Chor Leow, T.; Oslan, S.N. Expression and Characterization of Geobacillus stearothermophilus Sr74 Recombinant α-Amylase in Pichia Pastoris. Biomed. Res. Int. 2015, 2015, 529059. [Google Scholar] [CrossRef] [PubMed]
- Burhanoğlu, T.; Sürmeli, Y.; Şanlı-Mohamed, G. Identification and Characterization of Novel Thermostable α-Amylase from Geobacillus sp. GS33. Int. J. Biol. Macromol. 2020, 164, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Bischoff, K.M.; Sani, R.K. Highly Thermostable Xylanase Production from A Thermophilic Geobacillus sp. Strain WSUCF1 Utilizing Lignocellulosic Biomass. Front. Bioeng. Biotechnol. 2015, 3, 84. [Google Scholar] [CrossRef]
- Bibra, M.; Kunreddy, V.R.; Sani, R.K. Thermostable Xylanase Production by Geobacillus sp. Strain Duselr13, and Its Application in Ethanol Production with Lignocellulosic Biomass. Microorganisms 2018, 6, 93. [Google Scholar] [CrossRef]
- Shaheen, M.; Shah, A.A.; Hameed, A.; Hasan, F. Influence of Culture Conditions on Production and Activity of Protease from Bacillus subtilis Bs1. Pak. J. Bot. 2008, 40, 2161–2169. [Google Scholar]
- Morabandza, C.J.; Dibangou, V.; Mabika, F.A.S.; Gatse, E.V.; Ngoulou, T.B.; Itsouhou, N.; Nguimbi, E. Optimization of Culture Conditions for Protease Production Using Three Strains of Bacillus. J. Pure Appl. Microbiol. 2021, 15, 621–629. [Google Scholar] [CrossRef]
- Sung, J.; Ganbat, D.; Kim, S.B.; Lee, S.; Lee, D. Complete Genome Sequences of Geobacillus stearothermophilus Strains EF60045 and SJEF4-2 from Korean Hot Springs. Microbiol. Resour. Announc. 2024, 13, e0057324. [Google Scholar] [CrossRef]
- Miyazaki, K.; Hase, E.; Tokito, N. Complete Genome Sequence of Geobacillus sp. Strain E55-1, Isolated from Mine Geyser in Japan. Microbiol. Resour. Announc. 2020, 9, 10–1128. [Google Scholar] [CrossRef]
- Esin, A.; Ellis, T.; Warnecke, T. Horizontal Gene Flow into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation. bioRxiv 2018, 381442. [Google Scholar] [CrossRef]
- Aliyu, H.; Lebre, P.; Blom, J.; Cowan, D.; De Maayer, P. Phylogenomic Re-Assessment of the Thermophilic Genus Geobacillus. Syst. Appl. Microbiol. 2016, 39, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Sunna, A.; Tokajian, S.; Burghardt, J.; Rainey, F.; Antranikian, G.; Hashwa, F. Identification of Bacillus kaustophilus, Bacillus thermocatenulatus and Bacillus Strain HSR as Members of Bacillus thermoleovorans. Syst. Appl. Microbiol. 1997, 20, 232–237. [Google Scholar] [CrossRef]
- GArcía-Aparicio, M.P.; Ballesteros, I.; González, A.; Oliva, J.; Ballesteros, M.; Negro, M.J. Effect of Inhibitors Released During Steam-Explosion Pretreatment of Barley Straw on Enzymatic Hydrolysis. Appl. Biochem. Biotechnol. 2006, 129, 278–288. [Google Scholar] [CrossRef]
- Duarte, G.C.; Moreira, L.R.S.; Jaramillo, P.M.D.; Filho, E.X.F. Biomass-Derived Inhibitors of Holocellulases. Bioenergy Res. 2012, 5, 768–777. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in Industrially Important Thermostable Enzymes: A Review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Dutta, T.; Sahoo, R.; Sengupta, R.; Ray, S.S.; Bhattacharjee, A.; Ghosh, S. Novel Cellulases from an Extremophilic Filamentous Fungi Penicillium citrinum: Production and Characterization. J. Ind. Microbiol. Biotechnol. 2008, 35, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, R.; Yang, X.; Wu, H.; Xu, D.; Tang, Z.; Shen, Q. Thermostable Cellulase Production of Aspergillus fumigatus Z5 under Solid-State Fermentation and Its Application in Degradation of Agricultural Wastes. Int. Biodeterior. Biodegrad. 2011, 65, 717–725. [Google Scholar] [CrossRef]
- Waheeb, M.S.; Elkhatib, W.F.; Yassien, M.A.; Hassouna, N.A. Optimized Production and Characterization of a Thermostable Cellulase from Streptomyces thermodiastaticus Strain. AMB Express 2024, 14, 129. [Google Scholar] [CrossRef]
- Jayani, R.S.; Saxena, S.; Gupta, R. Microbial Pectinolytic Enzymes: A Review. Process Biochem. 2005, 40, 2931–2944. [Google Scholar] [CrossRef]
- Gummadi, S.N.; Panda, T. Purification and Biochemical Properties of Microbial Pectinases—A Review. Process Biochem. 2003, 38, 987–996. [Google Scholar] [CrossRef]
- Demir, N.; Nadaroglu, H.; Tasgin, E.; Adiguzel, A.; Gulluce, M. Purification and Characterization of a Pectin Lyase Produced by Geobacillus stearothermophilus Ah22 and Its Application in Fruit Juice Production. Ann. Microbiol. 2011, 61, 939–946. [Google Scholar] [CrossRef]
- Uma, J.L.; Rao, M.; Satyanarayana, T. Purification and Characterization of a Hyperthermostable and High Maltogenic α-Amylase of an Extreme Thermophile Geobacillus thermoleovorans. Appl. Biochem. Biotechnol. 2007, 142, 179–193. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, H.; Kong, Z.; Li, T.; Ma, L.; Liu, D.; Shen, Q. Pan-Genome Analyses of Geobacillus spp. Reveal Genetic Characteristics and Composting Potential. Int. J. Mol. Sci. 2020, 21, 3393. [Google Scholar] [CrossRef]
- Najar, I.N.; Sharma, P.; Das, R.; Mondal, K.; Singh, A.K.; Radha, A.; Sharma, V.; Sharma, S.; Thakur, N.; Gandhi, S.G.; et al. Exploring Probiotic Potential: A Comparative Genomics and In Silico Assessment of Genes within the Genus Geobacillus. bioRxiv 2024, 1, 2024.05.15.594408. [Google Scholar] [CrossRef]
- Evers, S.; Courvalin, P. Regulation of VanB-Type Vancomycin Resistance Gene Expression by the VanS B-VanR B Two-Component Regulatory System in Enterococcus Faecalis V583. J. Bacteriol. 1996, 178, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Z.; Zheng, Q.W.; Wei, T.; Zhang, Z.Q.; Zhao, C.F.; Zhong, H.; Xu, Q.Y.; Lin, J.F.; Guo, L.Q. Isolation and Characterization of Fengycins Produced by Bacillus amyloliquefaciens JFL21 and Its Broad-Spectrum Antimicrobial Potential Against Multidrug-Resistant Foodborne Pathogens. Front. Microbiol. 2020, 11, 579621. [Google Scholar] [CrossRef] [PubMed]
- Markelova, N.; Chumak, A. Antimicrobial Activity of Bacillus Cyclic Lipopeptides and Their Role in the Host Adaptive Response to Changes in Environmental Conditions. Int. J. Mol. Sci. 2025, 26, 336. [Google Scholar] [CrossRef]
- Matin, A. The Molecular Basis of Carbon-Starvation-Induced General Resistance in Escherichia coli. Mol. Microbiol. 1991, 5, 5–10. [Google Scholar] [CrossRef]
- Dubey, A.K.; Baker, C.S.; Suzuki, K.; Jones, A.D.; Pandit, P.; Romeo, T.; Babitzke, P. CsrA Regulates Translation of the Escherichia coli Carbon Starvation Gene, CstA, by Blocking Ribosome Access to the CstA Transcript. J. Bacteriol. 2003, 185, 4450–4460. [Google Scholar] [CrossRef]
- Flardh, K.; Axberg, T.; Albertson, N.H.; Kjelleberg, S. Stringent Control during Carbon Starvation of Marine Vibrio sp. Strain S14: Molecular Cloning, Nucleotide Sequence, and Deletion of the RelA Gene. J. Bacteriol. 1994, 176, 5949–5957. [Google Scholar] [CrossRef]
- Yeung, A.T.; Mattes, W.B.; Oh, E.Y.; Grossman, L. Enzymatic Properties of Purified Escherichia coli UvrABC Proteins. Proc. Natl. Acad. Sci. USA 1983, 80, 6157–6161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhang, L.; Xu, Q.; Zhang, L.; Yu, Y.; Hua, X. The Mismatch Repair System (MutS and MutL) in Acinetobacter baylyi ADP1. BMC Microbiol. 2020, 20, 40. [Google Scholar] [CrossRef] [PubMed]
- Zuo, F.L.; Yu, R.; Khaskheli, G.B.; Ma, H.Q.; Chen, L.L.; Zeng, Z.; Mao, A.J.; Chen, S.W. Homologous Overexpression of Alkyl Hydroperoxide Reductase Subunit C (AhpC) Protects Bifidobacterium Longum Strain NCC2705 from Oxidative Stress. Res. Microbiol. 2014, 165, 581–589. [Google Scholar] [CrossRef]
- Versace, G.; Palombo, M.; Menon, A.; Scarlato, V.; Roncarati, D. Feeling the Heat: The Campylobacter jejuni HrcA Transcriptional Repressor Is an Intrinsic Protein Thermosensor. Biomolecules 2021, 11, 1413. [Google Scholar] [CrossRef] [PubMed]


| Phenotypic Characteristics | Geobacillus sp. G4 * | G. kaustophilus ATCC 8005 ** | G. thermoleovorans ATCC 43513 ** |
|---|---|---|---|
| Gram Staining | Positive | Positive | Positive |
| Motility | Non-motile | Non-motile | Non-motile |
| Morphology | Rod shape | Rod shape | Rod shape |
| Oxygen Requirement | Aerobic | Aerobic | Aerobic |
| G+C (%) | 52 | 51–58 | 52–58 |
| Endospore Formation | + | + | + |
| Voges-Proskauer | - | - | - |
| Methyl red | + | + | no data |
| Catalase | + | + | + |
| Oxidase | + | + | + |
| Hydrolysis of: | |||
| Casein | + | + | Variable |
| Starch | + | + | + |
| Acid from: | |||
| Lactose | + | - | + |
| Cellobiose | + | + | Variable |
| Glycerol | + | + | + |
| Sucrose | + | + | Variable |
| Xylose | + | + | Variable |
| Growth at/in: | |||
| pH range | 6.0–7.5 | 6.2–7.5 | 5.0–9.0 |
| NaCl (%, w/v) | 0–2.5 | 0–2 | 0–4 |
| Temperature range (°C) | 50–70 | 40–75 | 37–70 |
| Acetate | + | + | + |
| Citrate | - | + | - |
| General Data | Assembly De Novo Geobacillus sp. G4 | Reference Genome G. kaustophilus HTA426 | Reference Genome G. thermoleovorans KCTC 3570 | Reference Genome G. kaustophilus NBRC102445 |
|---|---|---|---|---|
| Contigs | 164 | 2 | 2 | 1 |
| Total length | 3,362,411 | 3,544,776 | 3,450,609 | 3,670,957 |
| N50 (KB) | 236,119 | 3,716,831 | 1,192,538 | 750,728 |
| L50 | 5 | 1 | 2 | 1 |
| GC (%) | 52.59 | 52.35 | 52.50 | 51.73 |
| Coverage | 1224.95 | - | 224.2 | 150.5 |
| Completeness (%) | 99.45 | 99.31 | 99.44 | 93.81 |
| Contamination (%) | 0.95 | 0.22 | 30.89 | 0.24 |
| CDS | 2047 | 5668 | 4714 | 5548 |
| rRNA | 5 | 37 | 32 | 35 |
| tRNA | 88 | 129 | 100 | 111 |
| tmRNA | 1 | 2 | 1 | 3 |
| Genes | 2047 | 3655 | 3622 | 3734 |
| Putative protein | 1349 | 3386 | 3369 | 3401 |
| Isolated Strain | Type Reference Strains | ANIb (%) | ANIm (%) |
|---|---|---|---|
| G. thermoleovorans KCTC3570 | 97.65 | 98.30 | |
| G. kaustophilus HTA426 | 97.19 | 97.91 | |
| G. kaustophilus NBRC102445 | 97.11 | 97.85 | |
| Geobacillus sp. G4 | G. proteiniphilus 1017 G. thermocatenulatus BGSC93A1 G. jurassicus NBRC107829 Geobacillus vulcani PSS1 | 94.66 93.15 90.57 90.41 | 95.51 94.06 91.52 91.54 |
| G. stearothermophilus ATCC12980 G. uzenensis BGSC92A1 | 88.93 85.05 | 90.65 87.16 | |
| G. subterraneus KCTC3922 G. icigianus G1w11 | 85.04 84.64 | 87.12 87.28 | |
| G. thermodenitrificans DSM465 | 82.64 | 85.04 |
| Geobacillus sp. G4 | dDDH Formula 4-d4 (Similarity Based on Sequence Identity) | ||
|---|---|---|---|
| Reference Genomes | dDDH (d4%) | Model C.I. (%) | G+C Difference (%) |
| G. thermoleovorans KCTC3570 G. kaustophilus HTA426 | 83.2 79.1 | [80.4–85.7] [76.2–81.8] | 0.31 0.61 |
| G. kaustophilus NBRC102445 | 78.5 | [75.6–81.2] | 0.74 |
| G. proteiniphilus 1017 G. thermocatenulatus BGSC93A1 | 61.5 53.4 | [58.6–64.3] [50.7–56.1] | 0.84 0.82 |
| G. vulcani PSS1 | 43.3 | [40.8–45.9] | 0.19 |
| G. jurassicus NBRC107829 | 42.8 | [40.3–45.4] | 0.38 |
| G. stearothermophilus ATCC12980 | 38.7 | [36.2–41.2] | 0.18 |
| G. uzenensis BGSC92A1 | 30.4 | [28.1–32.9] | 0.37 |
| G. subterraneus KCTC3922 | 30.6 | [28.2–33.1] | 0.4 |
| G. icigianus G1w11 | 30.4 | [28.0–32.9] | 1.13 |
| G. thermodenitrificans DSM465 | 26.3 | [24.0–28.8] | 3.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ticona, A.R.P.; Santos, K.C.R.; Ramirez-Arua, H.E.; Castellanos, R.; Silva, J.P.; Hamann, P.R.V.; Noronha, E.F.; Lopes, F.A.C. Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru. Microorganisms 2025, 13, 1374. https://doi.org/10.3390/microorganisms13061374
Ticona ARP, Santos KCR, Ramirez-Arua HE, Castellanos R, Silva JP, Hamann PRV, Noronha EF, Lopes FAC. Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru. Microorganisms. 2025; 13(6):1374. https://doi.org/10.3390/microorganisms13061374
Chicago/Turabian StyleTicona, Alonso R. Poma, Karita C. R. Santos, Heber E. Ramirez-Arua, Roberto Castellanos, Jéssica Pinheiro Silva, Pedro R. Vieira Hamann, Eliane F. Noronha, and Fabyano A. C. Lopes. 2025. "Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru" Microorganisms 13, no. 6: 1374. https://doi.org/10.3390/microorganisms13061374
APA StyleTicona, A. R. P., Santos, K. C. R., Ramirez-Arua, H. E., Castellanos, R., Silva, J. P., Hamann, P. R. V., Noronha, E. F., & Lopes, F. A. C. (2025). Functional Characterization, Genome Assembly, and Annotation of Geobacillus sp. G4 Isolated from a Geothermal Field in Tacna, Peru. Microorganisms, 13(6), 1374. https://doi.org/10.3390/microorganisms13061374






