Molecular Survey and Genetic Analysis of Ehrlichia canis in Rhipicephalus sanguineus Ticks Infesting Dogs in Northern Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Tick Collection and Species Identification
2.2. DNA Extraction from Tick Specimens
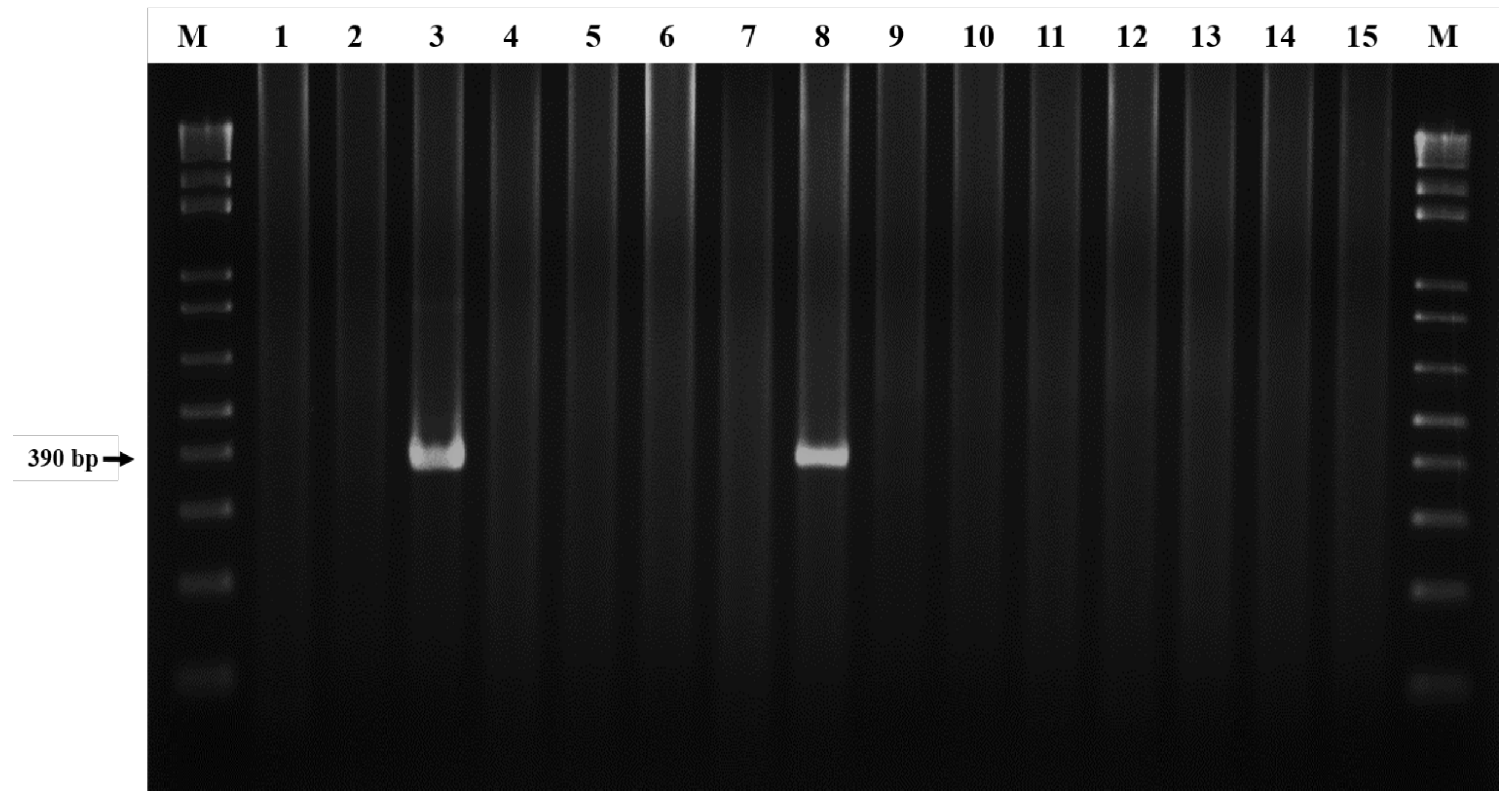
2.3. DNA Amplification by Nested Polymerase Chain Reaction
2.4. Gene Sequencing and Phylogenetic Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results
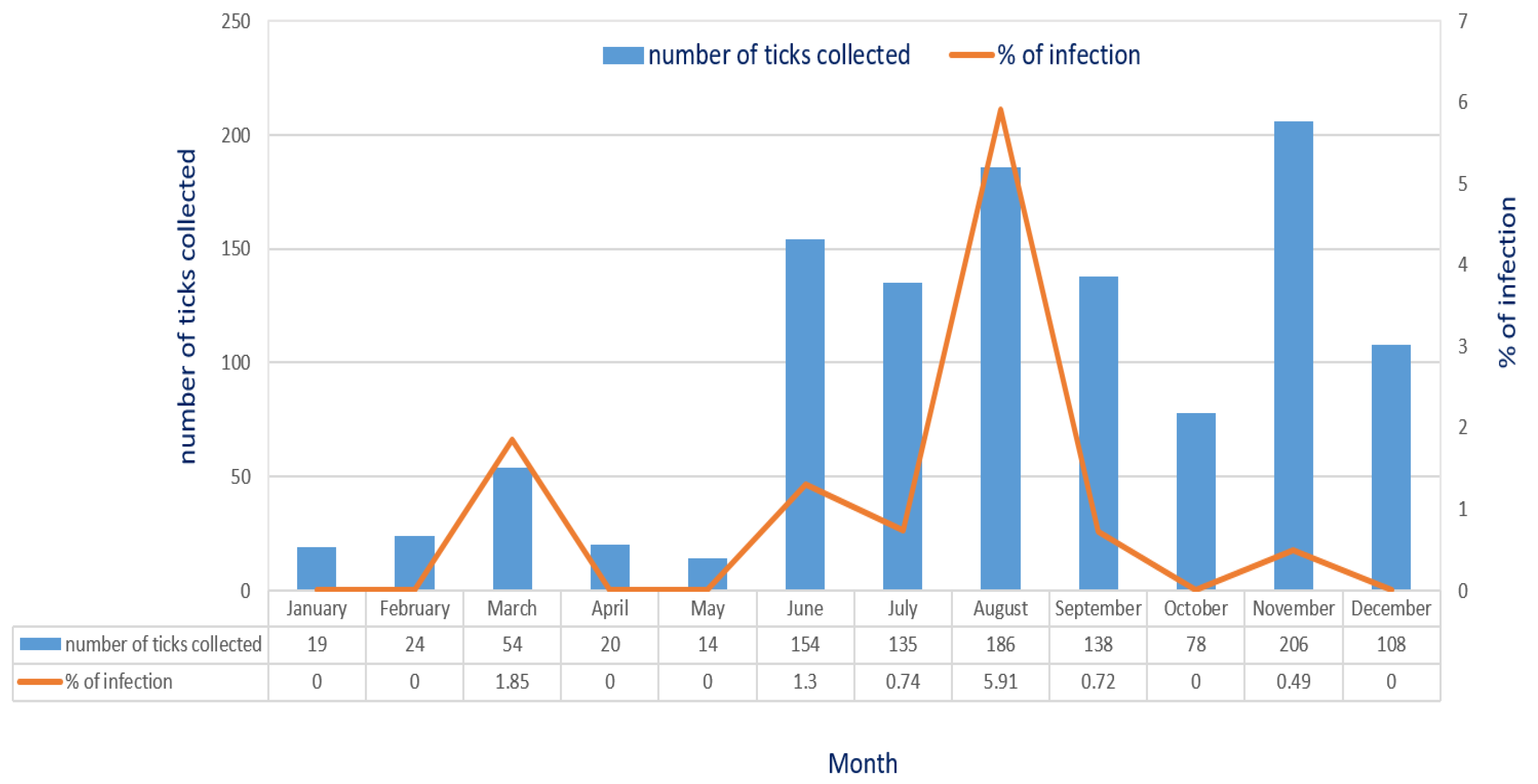
3.1. Detection of E. canis Infection in R. sanguineus Ticks of Taiwan
3.2. Genetic Analysis of E. canis Detected in R. sanguineus Ticks
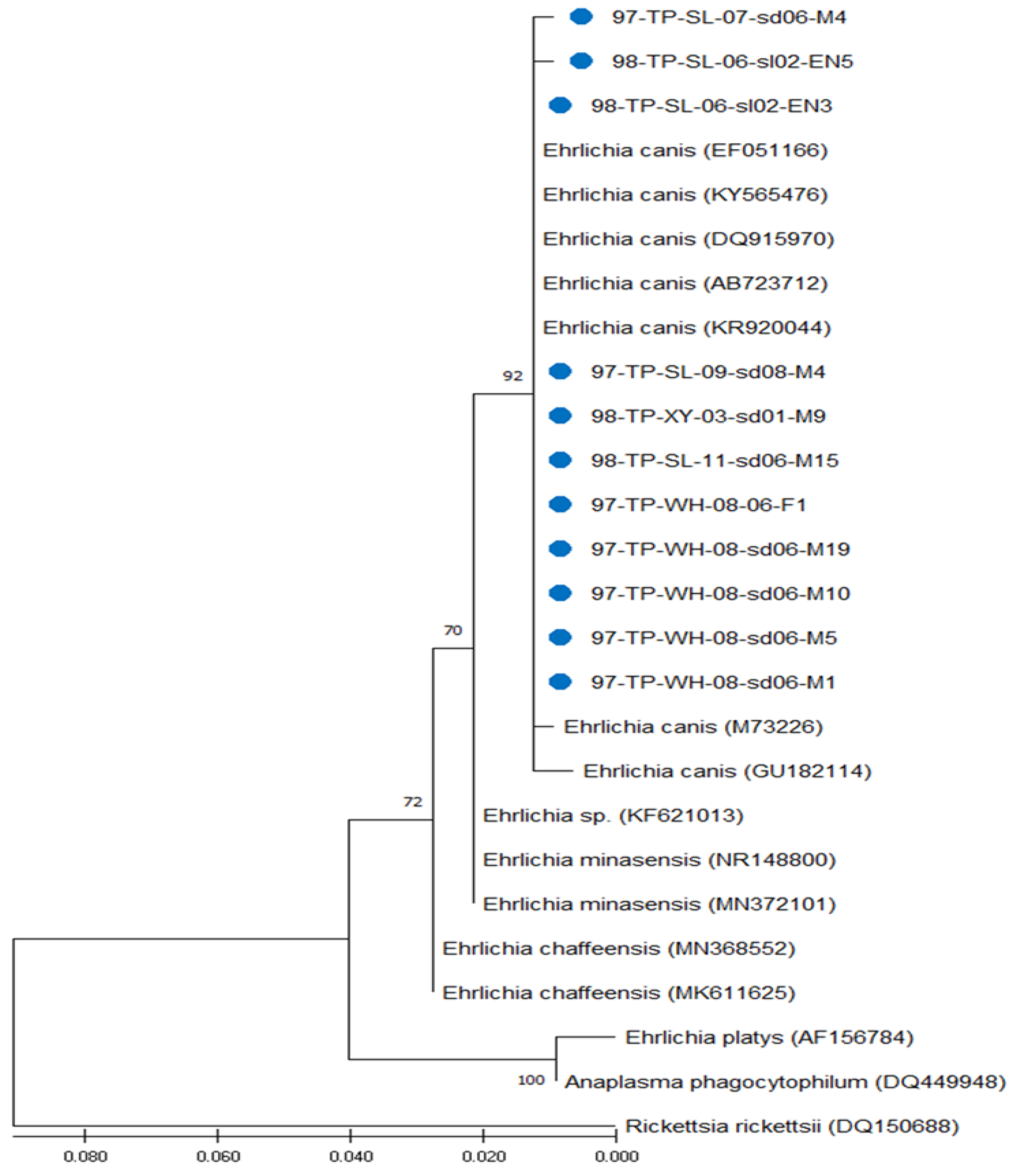
3.3. Phylogenetic Analysis of Ehrlichia Strains Detected in R. sanguineus Ticks of Taiwan
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rikihisa, Y. The tribe Ehrlichieae and ehrlichial diseases. Clin. Microbiol. Rev. 1991, 4, 286–308. [Google Scholar] [CrossRef]
- Rar, V.; Golovljova, I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: Pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infec. Genet. Evolu. 2011, 11, 1842–1861. [Google Scholar] [CrossRef] [PubMed]
- Donatien, A.; Lestoquard, F. Existence en Algerie d’une Rickettsia du chien. Bull. Soc. Pathol. Exot. 1935, 28, 418–419. [Google Scholar]
- Skotarczak, B. Canine ehrlichiosis. Ann. Agric. Environ. Med. 2003, 10, 137–141. [Google Scholar] [PubMed]
- Cardoso, L.; Tuna, J.; Vieira, L.; Yisaschar-Mekuzas, Y.; Baneth, G. Molecular detection of Anaplasma platys and Ehrlichia canis in dogs from the north of Portugal. Vet. J. 2010, 183, 232–233. [Google Scholar] [CrossRef]
- Yuasa, Y.; Tsai, Y.L.; Chang, C.C.; Hsu, T.H.; Chou, C.C. The prevalence of Anaplasma platys and a potential novel Anaplasma species exceed that of Ehrlichia canis in asymptomatic dogs and Rhipicephalus sanguineus in Taiwan. J. Vet. Med. Sci. 2017, 79, 1494–1502. [Google Scholar] [CrossRef]
- Dantas-Torres, F. Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasit. Vectors 2010, 3, 26. [Google Scholar] [CrossRef]
- Walker, J.B.; Keirans, J.E.; Horak, I.G. The Genus Rhipicephalus (Acari, Ixodidae): A Guide to the Brown Ticks of the World; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Shaw, S.E.; Day, M.J.; Birtles, R.J.; Breitschwerdt, E.B. Tick-borne infectious diseases of dogs. Trends Parasitol. 2001, 17, 74–80. [Google Scholar] [CrossRef]
- Otranto, D.; Dantas-Torres, F.; Breitschwerdt, E.B. Managing canine vector-borne diseases of zoonotic concern: Part one. Trends Parasitol. 2009, 25, 157–163. [Google Scholar] [CrossRef]
- Eremeeva, M.E.; Zambrano, M.L.; Anaya, L.; Beati, L.; Karpathy, S.E.; Santos-Silva, M.M.; Salceda, B.; MacBeth, D.; Olguin, H.; Dasch, G.A.; et al. Rickettsia rickettsii in Rhipicephalus ticks, Mexicali, Mexico. J. Med. Entomol. 2011, 48, 418–421. [Google Scholar] [CrossRef]
- Chao, L.L.; Shih, C.M. Molecular analysis of Rhipicephalus sanguineus (Acari: Ixodidae), an incriminated vector tick for Babesia vogeli in Taiwan. Exp. Appl. Acarol. 2016, 70, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.L.; Liao, H.T.; Ho, T.Y.; Shih, C.M. First detection and molecular identification of Babesia gibsoni from Rhipicephalus sanguineus ticks. Acta Trop. 2017, 166, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.L.; Liao, H.T.; Shih, C.M. First detection and genetic identification of Hepatozoon canis in Rhipicephalus sanguineus sensu lato ticks collected from dogs of Taiwan. Ticks Tick-Borne Dis. 2019, 10, 929–934. [Google Scholar] [CrossRef]
- Chao, L.L.; Ko, P.Y.; Shih, C.M. Molecular screening and genetic identification of Anaplasma platys in brown dog tick (Rhipicephalus sanguineus s. l.) infested on stray dogs in Taiwan. Microorganisms 2024, 12, 1779. [Google Scholar] [CrossRef]
- Shih, C.M.; Chao, L.L. Molecular Detection and Genetic Identification of Rickettsia Infection in Rhipicephalus sanguineus (Acari: Ixodidae) ticks collected from Southern Taiwan. Exp. Appl. Acarol. 2021, 85, 291–304. [Google Scholar] [CrossRef]
- Chao, L.L.; Robinson, M.; Liang, Y.F.; Shih, C.M. First detection and molecular identification of Rickettsia massiliae, a human pathogen, in Rhipicephalus sanguineus ticks collected from Southern Taiwan. PLoS Negl. Trop. Dis. 2022, 16, e0010917. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.E.; Summer, J.W.; Dawson, J.E.; Tzianabos, T.; Greene, C.R.; Olson, J.G.; Fishbein, D.B.; Olsen-Rasmuseen, M.; Holloway, B.P.; George, E.H.; et al. Detection of the etiological agent of human ehrlichiosis by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 775–780. [Google Scholar] [CrossRef]
- Dawson, J.E.; Biggie, K.L.; Warner, C.K.; Cookson, K.; Jenkins, S.; Levine, J.F.; Olson, J.G. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human disease, in dogs from southeast Virginia. Am. J. Vet. Res. 1996, 57, 1175–1179. [Google Scholar] [CrossRef]
- Murphy, G.L.; Ewing, S.A.; Whitworth, L.C.; Fox, J.C.; Kocan, A.A. A molecular and serologic survey of Ehrlichia canis, E. chaffeensis and E. ewingii in dogs and ticks from Oklahoma. Vet. Parasitol. 1998, 79, 325–339. [Google Scholar] [CrossRef]
- Kim, C.M.; Yi, Y.H.; Yu, D.H.; Lee, M.R.; Cho, M.R.; Desai, A.R.; Shringi, S.; Klein, T.A.; Kim, H.C.; Song, J.W.; et al. Tick-borne rickettsial pathogens in ticks and small mammals in Korea. App. Envirn. Microbiol. 2006, 72, 5766–5776. [Google Scholar] [CrossRef]
- Maekawa, N.; Konnai, S.; Balbin, M.M.; Mingala, C.N.; Gicana, K.R.B.; Bernando, F.A.E.M.; Murata, S.; Obashi, K. Molecular detection and phylogenetic analysis of Ehrlichia canis in a Philippine dog. Ticks Tick-Borne Dis. 2018, 9, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Leal, S.; Clemes, Y.S.; Lopes, M.G.; Acosta, I.C.L.; Serpa, M.C.A.; Mayorga, L.F.S.P.; Gennari, S.M.; Gonzalez-Acuna, D.; Labruna, M.B. Novel Ehrlichia sp. detected in magellanic prnguins (Sphenicus magellanicus) and in the seabird tick Ixodes uriae from Magdalena Island, southern Chile. Ticks Tick-Borne Dis. 2019, 10, 101256. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.; Hove, P.; Sharma, B.; Mattew-Belmar, V.; Karasek, I.; Lanza-Perea, M.; Werners, A.H.; Wilkerson, M.J.; Ganta, R.R. Molecular detection and characterization of Anaolasma platys and Ehrlichia canis in dogs from Caribbean. Ticks Tick-Borne Dis. 2021, 12, 101727. [Google Scholar] [CrossRef]
- Muraro, L.S.; Nogueira, M.F.; Borges, A.M.C.M.; de Oliveira Souza, A.; Vieira, T.S.W.J.; de Aguiar, D.M. Detection of Ehrlichia sp. in Amblyomma sculptum parasitizing horses from Brazilian Pantanal wetland. Ticks Tick-Borne Dis. 2021, 12, 101658. [Google Scholar] [CrossRef]
- Su, H.; Onoda, E.; Tai, H.; Fugita, H.; Sakabe, S.; Azuma, K.; Akachi, S.; Oishi, S.; Abe, F.; Ando, S.; et al. Diversity unearthed by the estimated molecular phylogeny and ecologically quantitative characteristics of uncultured Ehrlichia bacteria in Haemaphysalis ticks, Japan. Sci. Rep. 2022, 11, 687. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Bio. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 52, 1119–1134. [Google Scholar]
- Peres, M.; Rikihisa, Y.; Wen, B. Ehrlichia canis-like agent isolated from a man in Venezuela: Antigenic and genetic characterization. J. Clin. Microbiol. 1996, 34, 2133–2139. [Google Scholar] [CrossRef]
- Peres, M.; Bodor, M.; Zhang, C.; Xiong, Q.; Rikihisa, Y. Human infection with Ehrlichia canis accompanied by clinical signs in Venezuela. Ann. N. Y. Acad. Sci. 2006, 1078, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Unver, A.; Peres, M.; Orellana, N.; Huang, H.; Rikihisa, Y. Molecular and antigenic comparison of Ehrlichia canis isolated from dogs, ticks, and a human in Venezuela. J. Clin. Microbiol. 2001, 39, 2788–2793. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.L.; Hsieh, C.K.; Ho, T.Y.; Shih, C.M. First zootiological survey of hard ticks (Acari: Ixodidae) infesting dogs in northern Taiwan. Exp. Appl. Acarol. 2019, 77, 105–115. [Google Scholar] [CrossRef]
- Parola, P.; Socolovschi, C.; Jeanjean, L.; Bitam, I.; Fournier, P.E.; Sotto, A.; Labauge, P.; Raoult, D. Warmer weather linked to tick attack and emergence of severe rickettsioses. PLoS Negl. Trop. Dis. 2008, 2, e338. [Google Scholar] [CrossRef]
- Chang, A.C.H.; Chang, W.L.; Lin, C.T.; Pan, M.J.; Lee, S.C. Canine infectious cyclic thrombocytopenia found in Taiwan. J. Vet. Med. Sci. 1996, 58, 473–476. [Google Scholar] [CrossRef]
- Ewing, S.A. Canine ehrliciosis. Adv. Vet. Sci. Comp. Med. 1969, 13, 331–353. [Google Scholar]
- Groves, M.G.; Dennis, G.L.; Amys, H.L.; Huxsoll, D.L. Transmission of Ehrlichia canis to dogs by ticks (Rhipicephalus sanguineus). Am. J. Vet. Res. 1975, 36, 937–940. [Google Scholar] [CrossRef]
- Harvey, J.W.; Simpson, C.F.; Gaskin, J.M.; Sameck, J.H. Ehrlichiosis in wolves, dogs, and wolf-dog crosses. J. Am. Vet. Med. Assoc. 1979, 175, 901–905. [Google Scholar] [CrossRef]
- Lewis, G.E., Jr.; Ristic, M.; Smith, R.D.; Lincoln, T.; Stephenson, E.H. The brown dog tick Rhipicephalus sanguineus and the dog as experimental hosts of Ehrlichia canis. Am. J. Vet. Res. 1977, 38, 1953–1955. [Google Scholar] [CrossRef]
- Mathew, J.S.; Ewing, S.A.; Barker, R.W.; Fox, J.C.; Dawson, J.E.; Warner, C.K.; Murphy, G.L.; Kocan, K.M. Attempted transmission of Ehrlichia canis by Rhipicephalus sanguineus after passage in cell culture. Am. J. Vet. Res. 1996, 57, 1594–1598. [Google Scholar] [CrossRef]
- Johnson, E.M.; Ewing, S.A.; Barker, R.W.; Fox, J.C.; Crow, D.W.; Kocan, K.M. Experimental transmission of Ehrlichia canis (Rickettsiales: Ehrlichieae) by Dermacentor variabilis (Acari: Ixodidae). Vet. Parasitol. 1998, 74, 277–288. [Google Scholar] [CrossRef]
- Bremer, W.G.; Schaefer, J.J.; Wagner, E.R.; Ewing, S.A.; Rikihisa, Y.; Needham, G.R.; Jittapalapong, S.; Moore, D.L.; Stich, R.W. Transstadial and intrastadial experimental transmission of Ehrlichia canis by male Rhipicephalus sanguineus. Vet. Parasitol. 2005, 131, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Snellgrove, A.N.; Krapiunaya, I.; Ford, S.L.; Stanley, H.M.; Wickson, A.G.; Hartzer, K.L.; Levin, M.L. Vector competence of Rhipicephalus sanguineus sensu lato for Anaplasma platys. Ticks Tick-Borne Dis. 2020, 11, 101517. [Google Scholar] [CrossRef] [PubMed]
- Randolph, S.E.; Gern, L.; Nuttall, P.A. Co-feeding ticks epidemiological significance for tick-borne pathogen transmission. Parasitol. Today 1996, 12, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Laatamna, A.; Strube, C.; Bakkes, D.K.; Schaper, S.; Aziza, F.Z.; Chelef, H.B.; Amrane, N.E.H.; Bedraoui, R.; Dobler, G.; Chitimia Dobler, L. Molecular detection of tick-borne pathogens in Rhipicephalus sanguineus sensu stricto collected from dogs in the steppe and high plateau regions of Algeria. Acta Trop. 2022, 234, 106582. [Google Scholar] [CrossRef]
- Zemtova, G.E.; Apanaskevich, D.A.; Reeves, W.K.; Hahn, M.; Snellgrove, A.; Levin, M.L. Phylogeographical of Rhipicephalus sanguineus sensu lato and its relationships with climatic factors. Exp. Appl. Acarol. 2016, 69, 191–203. [Google Scholar] [CrossRef]
- EI-Sayed, A.; Kamel, M. Climate changes and their role in emergence and re-emergence of diseases. Environ Sci. Pollut. Res. 2020, 27, 22336–22352. [Google Scholar] [CrossRef]

| Strain | Origin of Bacterial Strain | 16S rRNA Gene Accession Number a | |
|---|---|---|---|
| Biological | Geographic | ||
| Taiwan strain | |||
| 97-TP-SL-07-sd06-M4 | Rhipicephalus sanguineus | Taiwan | OP389160 |
| 97-TP-SL-09-sd08-M4 | Rhipicephalus sanguineus | Taiwan | OP389165 |
| 98-TP-SL-06-sl02-EN3 | Rhipicephalus sanguineus | Taiwan | OP389212 |
| 98-TP-SL-06-sl02-EN5 | Rhipicephalus sanguineus | Taiwan | OP389213 |
| 97-TP-WH-08-sd06-M10 | Rhipicephalus sanguineus | Taiwan | OP392572 |
| 97-TP-WH-08-sd06-M19 | Rhipicephalus sanguineus | Taiwan | OP392573 |
| 97-TP-WH-08-sd06-M1 | Rhipicephalus sanguineus | Taiwan | OP392574 |
| 97-TP-WH-08-06-F1 | Rhipicephalus sanguineus | Taiwan | OP392575 |
| 97-TP-WH-08-sd06-M5 | Rhipicephalus sanguineus | Taiwan | OP392578 |
| 98-TP-XY-03-sd01-M9 | Rhipicephalus sanguineus | Taiwan | OP392580 |
| 98-TP-SL-11-sd06-M15 | Rhipicephalus sanguineus | Taiwan | OP392581 |
| Ehrlichia canis | Unknown | USA | M73226 |
| Ehrlichia canis | Dog blood | Malaysia | KR920044 |
| Ehrlichia canis | Dog blood | India | GU182114 |
| Ehrlichia canis | Leopard cat | Japan | AB723712 |
| Ehrlichia canis | Unknown | USA | DQ915970 |
| Ehrlichia canis | Dog blood | Taiwan | KY565476 |
| Ehrlichia canis | Dog | Portugal | EF051166 |
| Ehrlichia sp. | Cattle blood | Brazil | KF621013 |
| Ehrlichia minasensis | Rhipicephalus microplus | Czech Republic | NR148800 |
| Ehrlichia minasensis | Hyalomma tick | Egypt | MN372101 |
| Ehrlichia chaffeensis | Hyalomma tick | Egypt | MN368552 |
| Ehrlichia chaffeensis | Deer blood | USA | MK611625 |
| Ehrlichia platys | Unknown | China | AF156784 |
| Anaplasma phagocytophilum Rickettsia rickettsii | Dermacentor silvarum Dog | China USA | DQ449948 DQ150688 |
| Life Stage of Tick | E. canis Infection Detected by Nested PCR | % of E. canis Infection | |
|---|---|---|---|
| Number of Ticks Positive | Number of Ticks Examined | ||
| Nymph | 2 | 331 | 0.60 |
| Male | 8 | 610 | 1.31 |
| Female | 7 | 254 | 2.76 |
| Total | 17 | 1195 | 1.42 |
| Bacterial Strains b | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. 97-TP-WH-08-sd06-M1 (Taiwan) | – | ||||||||||||||||
| 2. 97-TP-WH-08-sd06-M5 (Taiwan) | 0.000 | – | |||||||||||||||
| 3. 97-TP-WH-08-sd06-M10 (Taiwan) | 0.000 | 0.000 | – | ||||||||||||||
| 4. 97-TP-WH-08-sd06-M19 (Taiwan) | 0.000 | 0.000 | 0.000 | – | |||||||||||||
| 5. 97-TP-WH-08-06-F1 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | – | ||||||||||||
| 6. 98-TP-SL-11-sd06-M15 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | |||||||||||
| 7. 98-TP-XY-03-sd01-M9 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | ||||||||||
| 8. 97-TP-SL-09-sd08-M4 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | |||||||||
| 9. 98-TP-SL-06-sl02-EN3 (Taiwan) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | ||||||||
| 10. Ehrlichia canis, Malaysia (KR920044) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | |||||||
| 11. Ehrlichia canis, Japan (AB723712) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | ||||||
| 12. Ehrlichia canis, Portugal (EF051166) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | – | |||||
| 13. Ehrlichia canis, USA (M73226) | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | 0.003 | – | ||||
| 14. Ehrlichia minasensis (MN372101) | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.009 | 0.010 | 0.012 | – | |||
| 15. Ehrlichia chafeensis (MN368552) | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.015 | 0.016 | 0.018 | 0.006 | – | ||
| 16. Anaplasma phagocytophilum (DQ449948) | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.054 | 0.053 | 0.057 | 0.050 | 0.044 | – | |
| 17. Rickettsia rickettsii (DQ150688) | 0.157 | 0.157 | 0.157 | 0.157 | 0.157 | 0.157 | 0.157 | 0.157 | 0.159 | 0.157 | 0.157 | 0.153 | 0.161 | 0.153 | 0.153 | 0.172 | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shih, C.-M.; Ko, P.-Y.; Chao, L.-L. Molecular Survey and Genetic Analysis of Ehrlichia canis in Rhipicephalus sanguineus Ticks Infesting Dogs in Northern Taiwan. Microorganisms 2025, 13, 1372. https://doi.org/10.3390/microorganisms13061372
Shih C-M, Ko P-Y, Chao L-L. Molecular Survey and Genetic Analysis of Ehrlichia canis in Rhipicephalus sanguineus Ticks Infesting Dogs in Northern Taiwan. Microorganisms. 2025; 13(6):1372. https://doi.org/10.3390/microorganisms13061372
Chicago/Turabian StyleShih, Chien-Ming, Pei-Yin Ko, and Li-Lian Chao. 2025. "Molecular Survey and Genetic Analysis of Ehrlichia canis in Rhipicephalus sanguineus Ticks Infesting Dogs in Northern Taiwan" Microorganisms 13, no. 6: 1372. https://doi.org/10.3390/microorganisms13061372
APA StyleShih, C.-M., Ko, P.-Y., & Chao, L.-L. (2025). Molecular Survey and Genetic Analysis of Ehrlichia canis in Rhipicephalus sanguineus Ticks Infesting Dogs in Northern Taiwan. Microorganisms, 13(6), 1372. https://doi.org/10.3390/microorganisms13061372







