In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Lactic Acid Bacteria (LAB) from the Gastrointestinal Tract of Broiler Chickens
2.3. Molecular Identification of LAB by 16S rDNA Sequencing
2.4. Preliminary Screening of LAB
2.4.1. Assessment of Acid Tolerance in LAB Isolates
2.4.2. Assessment of Bile Salt Tolerance in LAB Isolates
2.5. Safety Assessment of LAB
2.5.1. Assessment of Haemolysis Activity in LAB Isolates
2.5.2. Determination of Antibiotic Resistance Profiles
2.6. Functional Characterization of LAB
2.6.1. Antagonistic Activity
2.6.2. Assessment of Extracellular Enzymatic Activity in LAB Isolates
2.6.3. Auto-Aggregation and Co-Aggregation
2.6.4. Cell Surface Hydrophobicity
2.7. Statistical Analysis
3. Results and Discussion
3.1. Isolation of Bacterial Strains and Identification of Lab Isolates by 16s rDNA Gene Sequencing
3.2. In Vitro Characterization of LAB
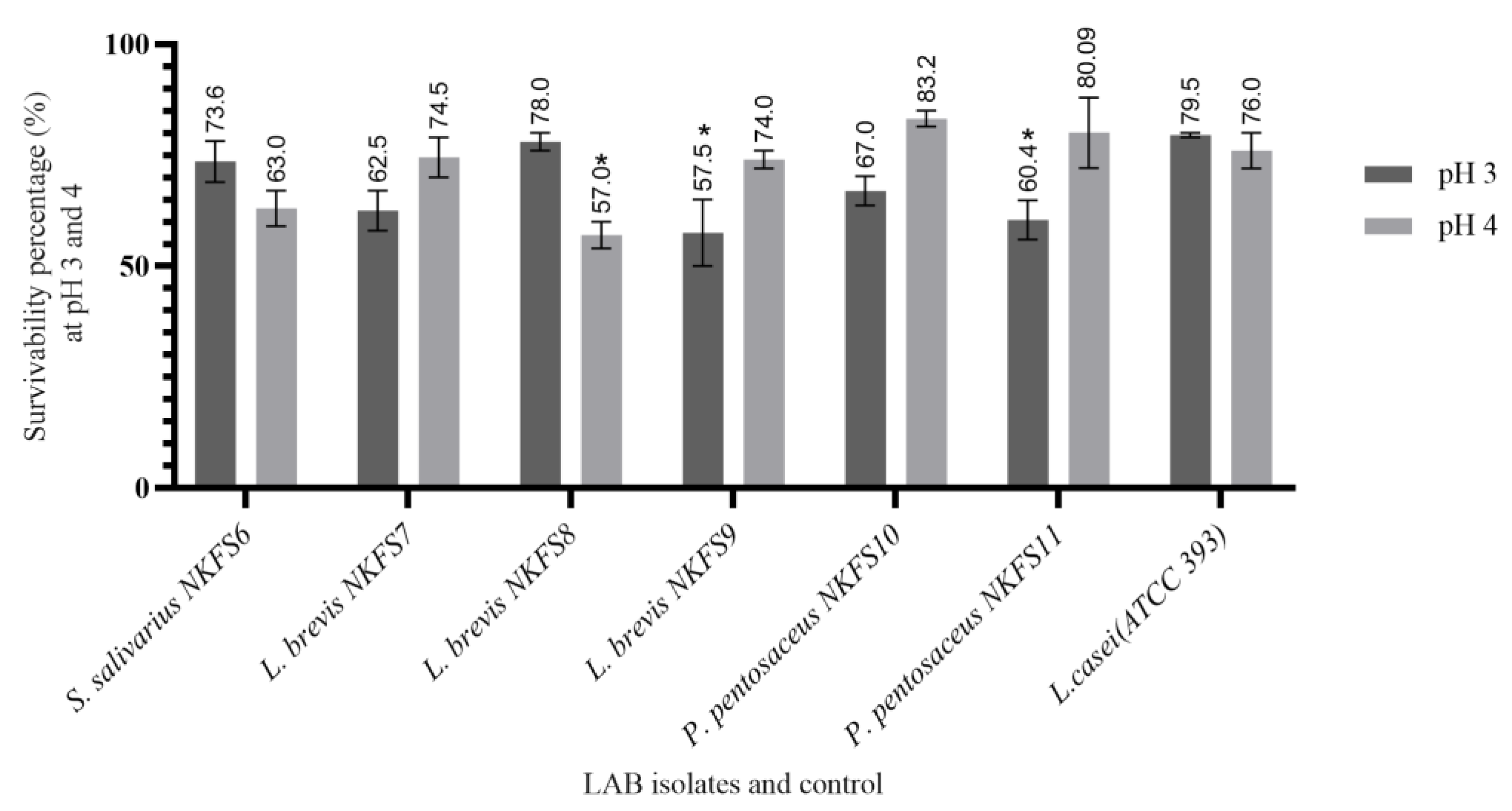
3.2.1. Acid Tolerance
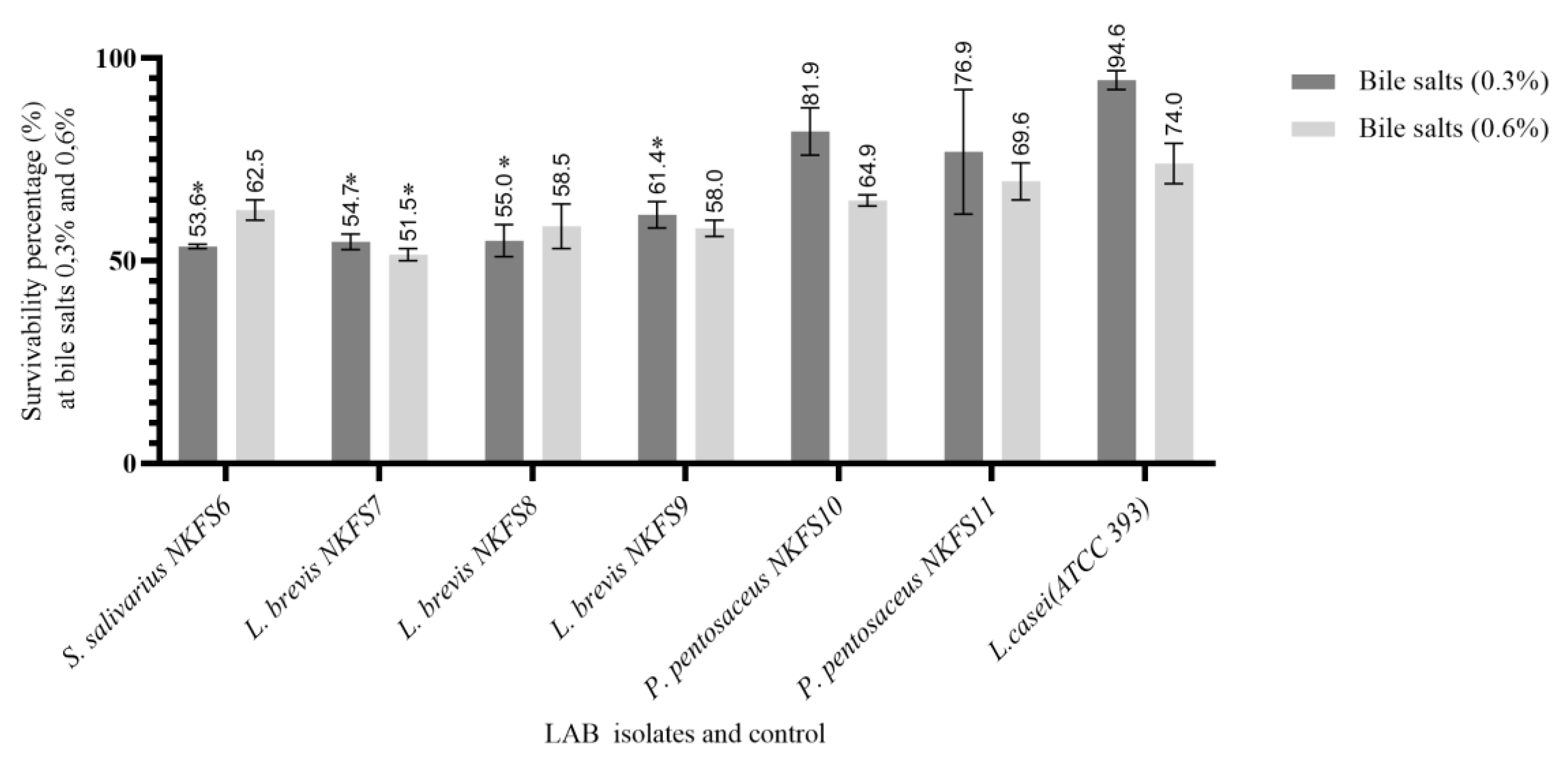
3.2.2. Bile Salt Tolerance
3.3. Safety Assessment
3.3.1. Haemolysis Activity
3.3.2. Antibiotic Susceptibility Testing
3.4. Functional Characteristics
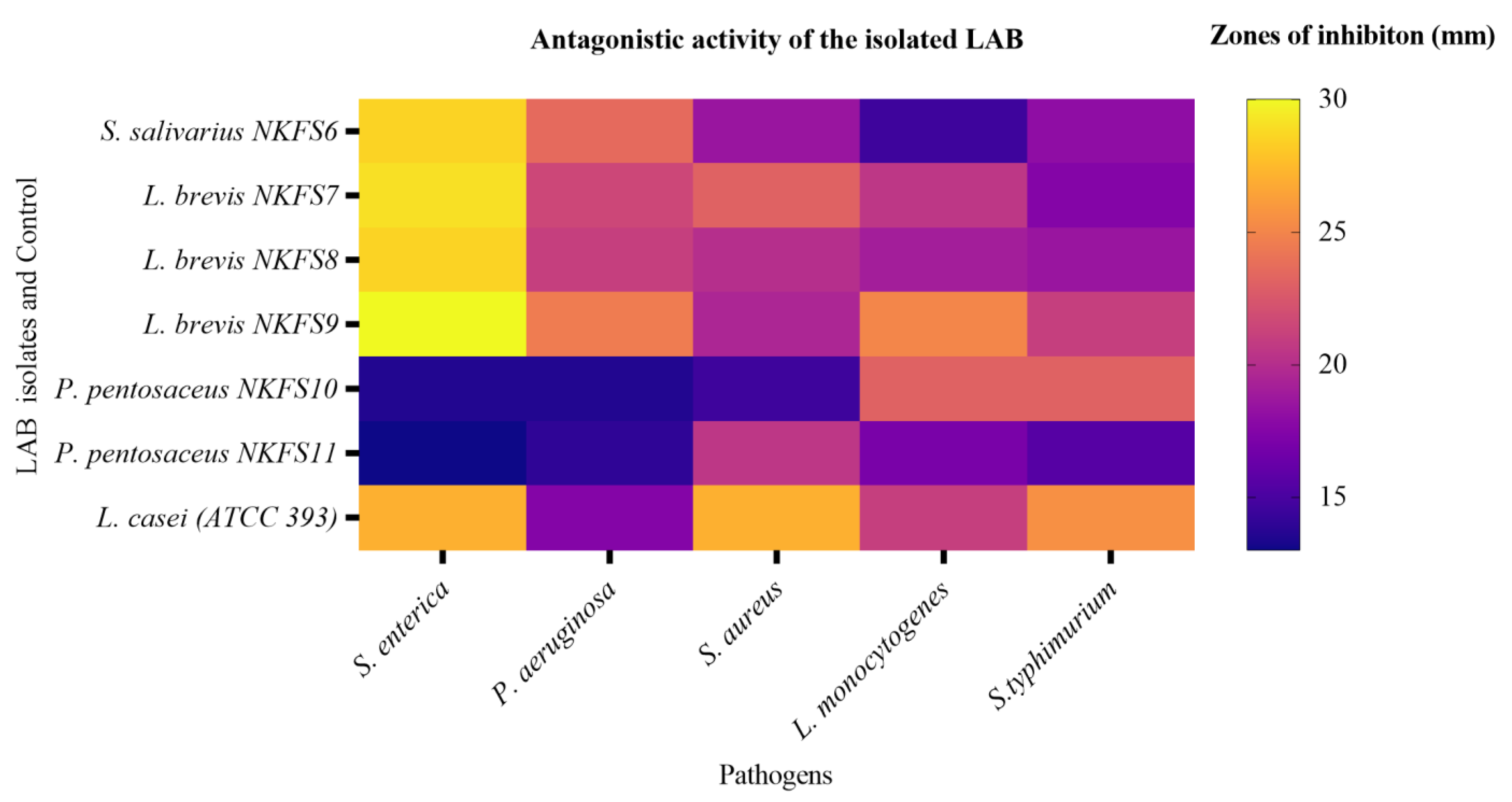
3.4.1. Antagonistic Activity of LAB
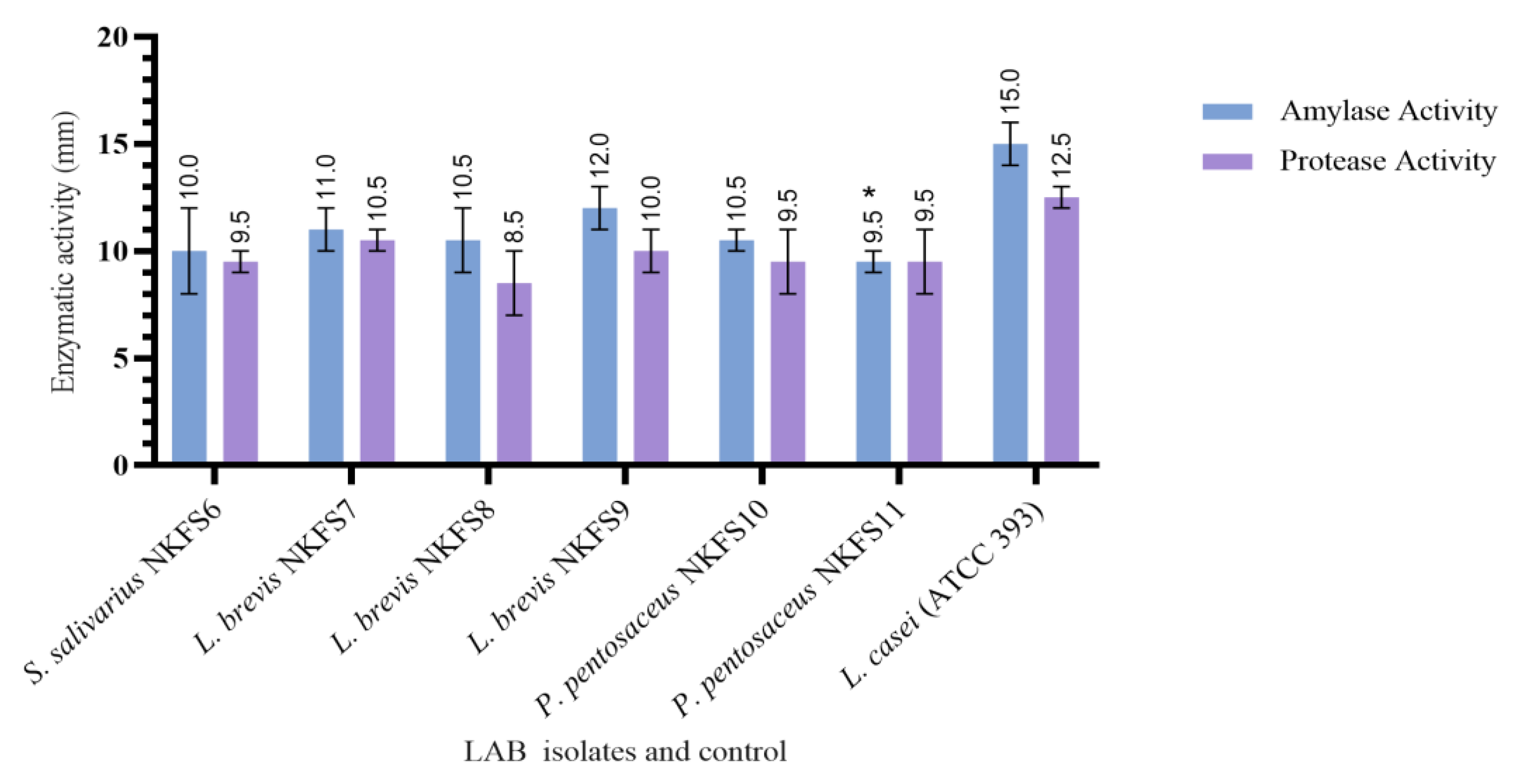
3.4.2. Extracellular Enzymatic Activity
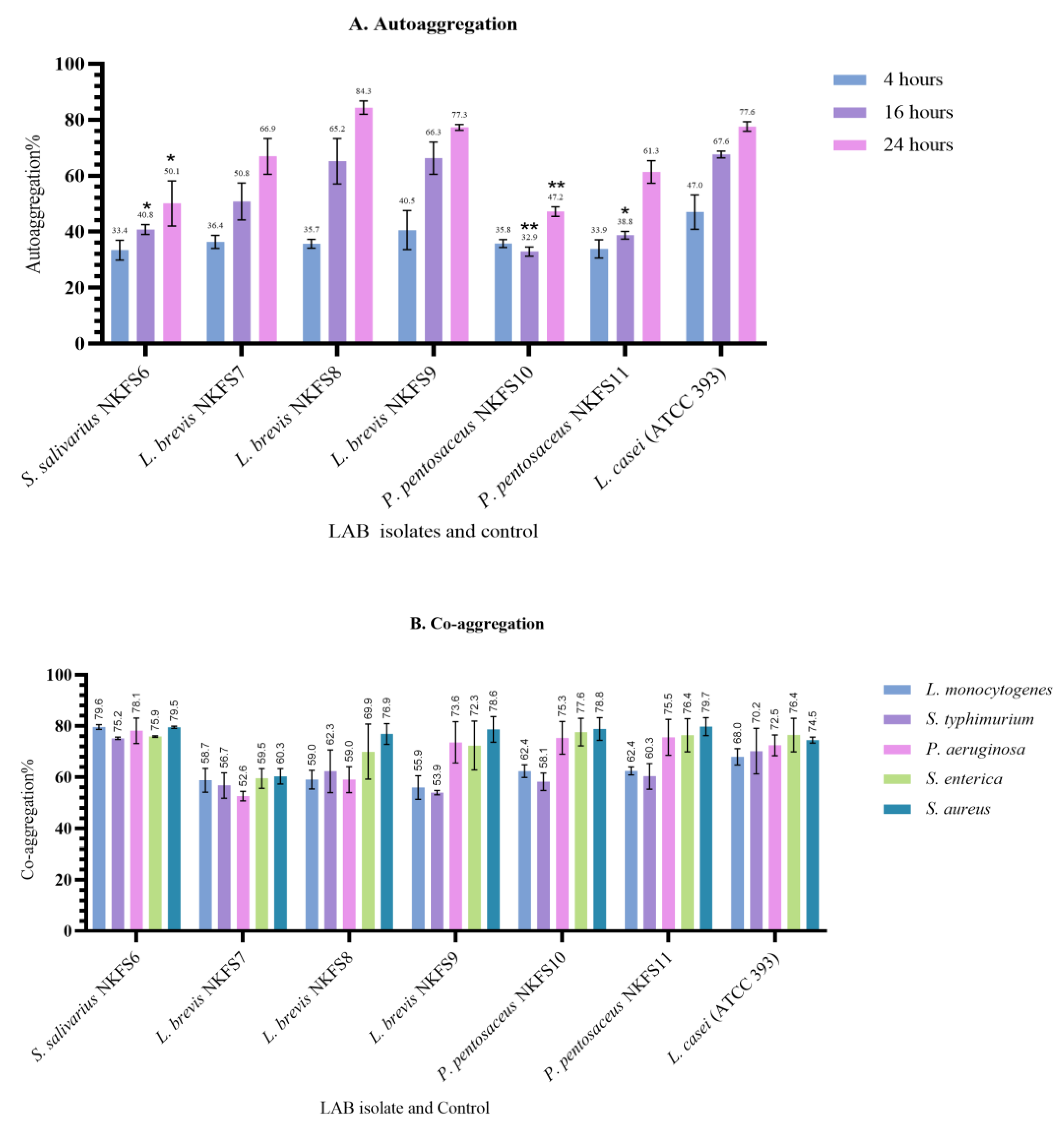
3.4.3. Auto-Aggregation and Co-Aggregation
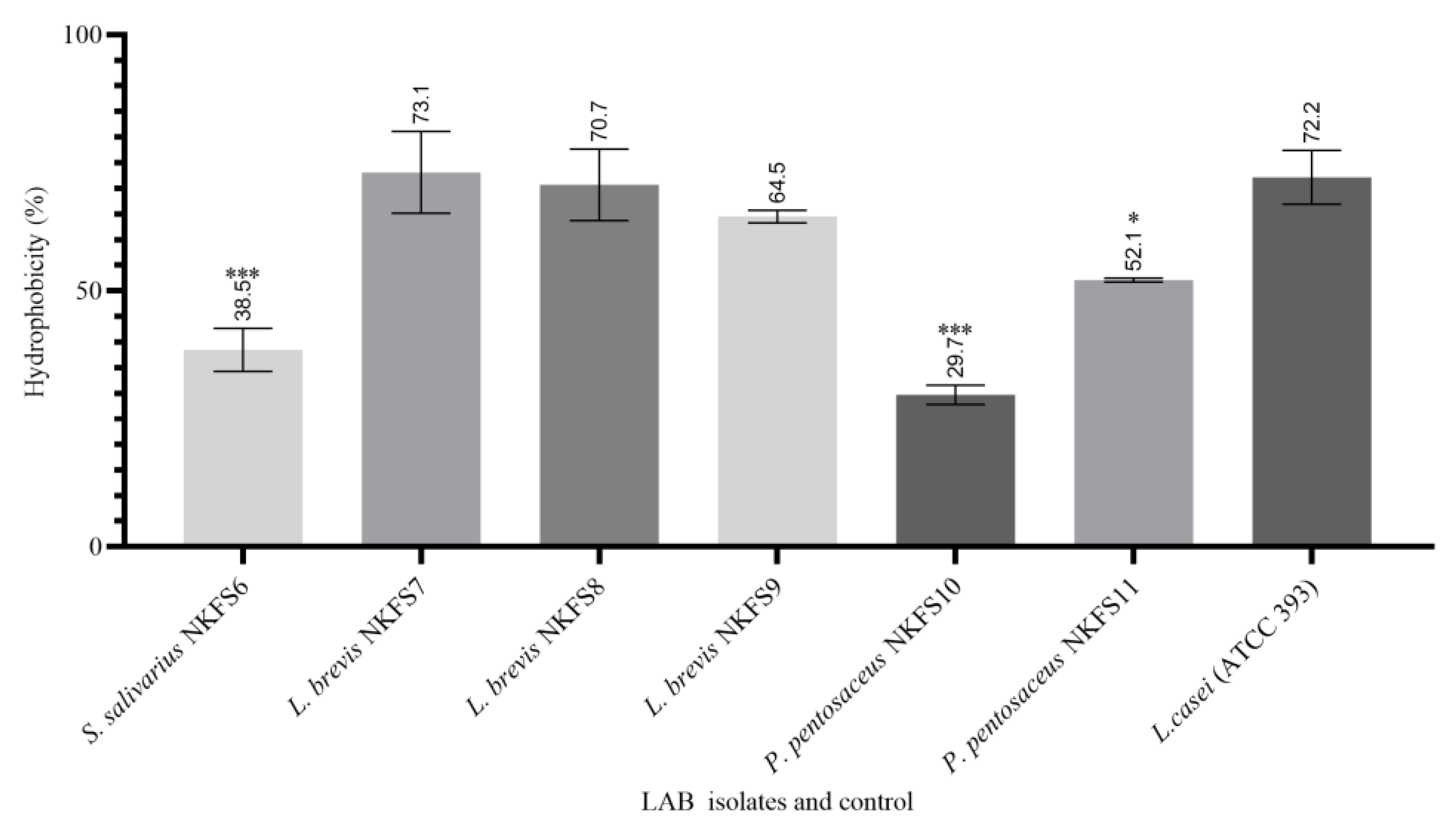
3.4.4. Cell Surface Hydrophobicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Idowu, P.A.; Zishiri, O.; Nephawe, K.A.; Mtileni, B. Current status and intervention of South Africa chicken production–A review. World’s Poult. Sci. J. 2021, 77, 115–133. [Google Scholar] [CrossRef]
- South African Poultry Association (SAPA). State of the SA Poultry Industry. Available online: https://www.sapoultry.co.za/state-of-the-sa-poultry-industry/ (accessed on 25 March 2025).
- Rajoka, M.S.R.; Hayat, H.F.; Sarwar, S.; Mehwish, H.M.; Ahmad, F.; Hussain, N.; Shah, S.Z.H.; Khurshid, M.; Siddiqu, M.; Shi, J. Isolation and evaluation of probiotic potential of lactic acid bacteria isolated from poultry intestine. Microbiology 2018, 87, 116–126. [Google Scholar] [CrossRef]
- Sirisopapong, M.; Shimosato, T.; Okrathok, S.; Khempaka, S. Assessment of lactic acid bacteria isolated from the chicken digestive tract for potential use as poultry probiotics. Anim. Biosci. 2023, 36, 1209–1220. [Google Scholar] [CrossRef]
- Blajman, J.; Gaziano, C.; Zbrun, M.V.; Soto, L.; Astesana, D.; Berisvil, A.; Scharpen, A.R.; Signorini, M.; Frizzo, L. In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res. Vet. Sci. 2015, 101, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Elzeini, H.M.; Ali, A.; Nasr, N.F.; Hassan, M.; Hassan, A.A.M.; Elenany, Y.E. Probiotic capability of novel lactic acid bacteria isolated from worker honey bees gut microbiota. FEMS Microbiol. Lett. 2021, 368, fnab030. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Hidayat, M.; Malaka, R.; Agustina, L.; Pakiding, W. Characteristics isolate bacteria lactic acid of origin digestive tract of broiler as a probiotic candidate for poultry. Int. J. Sci. Eng. Res. 2018, 9, 1787. [Google Scholar]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.; Ali, M.; Zhu, X.; Zhang, Y.; Li, S.; Li, K.; Kebzhai, F.; Li, J. Effects of lactic acid bacteria isolated from Tibetan chickens on the growth performance and gut microbiota of broiler. Front. Microbiol. 2023, 14, 1171074. [Google Scholar] [CrossRef]
- Kumar, M.; Kala, A.; Chaudhary, L.C.; Agarwal, N.; Kochewad, S.A. Microencapsulated and lyophilized Lactobacillus acidophilus improved gut health and immune status of predominant calves. Probiotics Antimicrob. Proteins 2022, 14, 523–534. [Google Scholar] [CrossRef]
- Amarantini, C.; Budarso, T.; Antika, Y.; Prakasita, V.J. Characterisation of Lactobacillus plantarum isolated from pickled cucumber, and its antagonist effect on pathogenic bacteria. Int. Food Res. J. 2020, 27, 805–813. [Google Scholar]
- Dejene, F.; Regasa Dadi, B.; Tadesse, D. In vitro antagonistic effect of lactic acid bacteria isolated from fermented beverage and finfish on pathogenic and foodborne pathogenic microorganism in Ethiopia. Int. J. Microbiol. 2021, 2021, 5370556. [Google Scholar] [CrossRef] [PubMed]
- Mufandaedza, J.; Viljoen, B.C.; Feresu, S.B.; Gadaga, T.H. Antimicrobial properties of lactic acid bacteria and yeast-lab cultures isolated from traditional fermented milk against pathogenic Escherichia coli and Salmonella enteritidis strains. Int. J. Food Microbiol. 2006, 108, 147–152. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, W.; Hu, Z.; Liu, Z.; Du, T.; Dai, Y.; Xiong, T. Isolation, characterization and selection of potential probiotic lactic acid bacteria from feces of wild boar, native pig and commercial pig. Livest. Sci. 2020, 237, 104036. [Google Scholar] [CrossRef]
- Musikasang, H.; Tani, A.; H-kittikun, A.; Maneerat, S. Probiotic potential of lactic acid bacteria isolated from the chicken gastrointestinal digestive tract. World J. Microbiol. Biotechnol. 2009, 25, 1337–1345. [Google Scholar] [CrossRef]
- Dahri, A.S.; Patrick, A.; Shaikh, N.; Mangi, J.; Bhatti, A.A.; Simair, A.A. Evaluation of the lactic acid bacteria in different types of yogurt consumed in Pakistan. Int. J. Agric. Food Sci. 2020, 17, 149–153. [Google Scholar]
- Shamsudin, W.N.F.; Loo, S.S.; Ho, Y.W.; Abdullah, N.; Saad, W.Z.; Wan, K.L. Probiotic properties of lactobacillus isolates from chicken intestines. Microbiology 2019, 7, 8–13. [Google Scholar] [CrossRef]
- Shazali, N.; Foo, H.L.; Loh, T.C.; Choe, D.W.; Abdul Rahim, R. Prevalence of antibiotic resistance in lactic acid bacteria isolated from the faeces of broiler chicken in Malaysia. Gut Pathog. 2014, 6, 1. [Google Scholar] [CrossRef]
- Salehizadeh, M.; Modarressi, M.H.; Mousavi, S.N.; Ebrahimi, M.T. Evaluation of lactic acid bacteria isolated from poultry feces as potential probiotic and its in vitro competitive activity against Salmonella typhimurium. Vet. Res. Forum 2020, 11, 67. [Google Scholar]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.-J. Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef]
- Armas, F.; Camperio, C.; Marianelli, C. In vitro assessment of the probiotic potential of Lactococcus lactis lmg 7930 against ruminant mastitis-causing pathogens. PLoS ONE 2017, 12, e0169543. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Tan, J.S.; Bashokouh, F.; Ibrahim, T.A.T.; Mustafa, S.; Vakhshiteh, F.; Sivasamboo, S.; Ariff, A.B. In vitro assessment of Pediococcus acidilactici kp10 for its potential use in the food industry. BMC Microbiol. 2017, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Marchwińska, K.; Gwiazdowska, D. Isolation and probiotic potential of lactic acid bacteria from swine feces for feed additive composition. Arch. Microbiol. 2022, 204, 61. [Google Scholar] [CrossRef]
- Casarotti, S.N.; Carneiro, B.M.; Todorov, S.D.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In vitro assessment of safety and probiotic potential characteristics of Lactobacillus strains isolated from water buffalo mozzarella cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef]
- Nurcahyo, H.; Dale, A.; Furqon, F.Y.A. Isolation and characterization of lactic acid bacteria (lab) from small intestine content of duck (Anas sp.) as a probiotic candidate. J. Phys. Conf. Ser. 2019, 1397, 012043. [Google Scholar] [CrossRef]
- Noohi, N.; Papizadeh, M.; Rohani, M.; Talebi, M.; Pourshafie, M.R. Screening for probiotic characters in Lactobacilli isolated from chickens revealed the intra-species diversity of Lactobacillus brevis. Anim. Nutr. 2021, 7, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Torres-Maravilla, E.; Boucard, A.-S.; Al Azzaz, J.; Gontier, S.; Kulakauskas, S.; Langella, P.; Bermúdez-Humarán, L. Assessment of the safety of Levilactobacillus brevis cncm i-5321, a probiotic candidate strain isolated from pulque with anti-proliferative activities. J. Benef. Microbes 2023, 14, 335–348. [Google Scholar] [CrossRef]
- Kaci, G.; Goudercourt, D.; Dennin, V.; Pot, B.; Doré, J.; Ehrlich, S.D.; Renault, P.; Blottière, H.M.; Daniel, C.; Delorme, C. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl. Environ. Microbiol. 2014, 80, 928–934. [Google Scholar] [CrossRef]
- García-Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.L.; Nicoli, J.R.; Moreira-Silva, J.; Rodríguez, Z.; Fuertes, H.; Nuñez, O.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Poultry 2016, 108, 125–132. [Google Scholar] [CrossRef]
- Shahbaz, F.; Muccee, F.; Shahab, A.; Safi, S.Z.; Alomar, S.Y.; Qadeer, A. Isolation and in vitro assessment of chicken gut microbes for probiotic potential. Front. Microbiol. 2024, 15, 1278439. [Google Scholar] [CrossRef]
- Yang, S.J.; Kim, K.-T.; Kim, T.Y.; Paik, H.-D. Probiotic properties and antioxidant activities of Pediococcus pentosaceus sc28 and Levilactobacillus brevis ku15151 in fermented black gamju. Foods 2020, 9, 1154. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhang, G.J.L. Synbiotic encapsulation of probiotic Lactobacillus plantarum by alginate-arabinoxylan composite microspheres. LWT 2018, 93, 135–141. [Google Scholar] [CrossRef]
- Guan, N.; Liu, L. Microbial response to acid stress: Mechanisms and applications. Appl. Microbiol. Biotechnol. 2020, 104, 51–65. [Google Scholar] [CrossRef]
- Reis, N.A.; Saraiva, M.A.; Duarte, E.A.; de Carvalho, E.A.; Vieira, B.B.; Evangelista-Barreto, N.S. Probiotic properties of lactic acid bacteria isolated from human milk. J. Appl. Microbiol. 2016, 121, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Topçu, K.C.; Kaya, M.; Kaban, G.J.L. Probiotic properties of lactic acid bacteria strains isolated from pastırma. LWT 2020, 134, 110216. [Google Scholar] [CrossRef]
- Harnentis, H.; Marlida, Y.; Nur, Y.S.; Wizna, W.; Santi, M.A.; Septiani, N.; Adzitey, F.; Huda, N. Novel probiotic lactic acid bacteria isolated from Indigenous fermented foods from West Sumatra, Indonesia. Vet. World 2020, 13, 1922–1927. [Google Scholar] [CrossRef]
- Ahmed, A.-S.I.; El Moghazy, G.M.; Elsayed, T.R.; Goda, H.A.L.; Khalafalla, G.M. Molecular identification and in vitro evaluation of probiotic functional properties of some Egyptian lactic acid bacteria and yeasts. J. Genet. Eng. Biotechnol. 2021, 19, 114. [Google Scholar] [CrossRef]
- Lee, M.-G.; Kang, M.-J.; Kim, S.; Jeong, H.; Kang, D.; Paik, H.; Park, Y.-S. Safety assessment of Levilactobacillus brevis ku15006: A comprehensive analysis of its phenotypic and genotypic properties. Probiotics Antimicrob. Proteins 2024, 17, 1117–1131. [Google Scholar] [CrossRef]
- Keresztény, T.; Libisch, B.; Orbe, S.C.; Nagy, T.; Kerényi, Z.; Kocsis, R.; Posta, K.; Papp, P.P.; Olasz, F. Isolation and characterization of lactic acid bacteria with probiotic attributes from different parts of the gastrointestinal tract of free-living wild boars in Hungary. Probiotics Antimicrob. Proteins 2024, 16, 1221–1239. [Google Scholar] [CrossRef]
- Li, X.; Li, W.; Zhao, L.; Li, Y.; He, W.; Ding, K.; Cao, P. Characterization and assessment of native lactic acid bacteria from broiler intestines for potential probiotic properties. Microorganisms 2024, 12, 749. [Google Scholar] [CrossRef]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. Aims Microbiol. 2018, 4, 482. [Google Scholar] [CrossRef] [PubMed]
- Sirichoat, A.; Flórez, A.B.; Vázquez, L.; Buppasiri, P.; Panya, M.; Lulitanond, V.; Mayo, B. Antibiotic susceptibility profiles of lactic acid bacteria from the human vagina and genetic basis of acquired resistances. Int. J. Mol. Sci. 2020, 21, 2594. [Google Scholar] [CrossRef]
- Zhang, Q.; Pan, Y.; Wang, M.; Sun, L.; Xi, Y.; Li, M.; Zeng, Q. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from the vagina of yak (Bos grunniens). Microbiology 2022, 10, e13177. [Google Scholar] [CrossRef] [PubMed]
- El-Deeb, W.M.; Fayez, M.; Elsohaby, I.; Ghoneim, I.; Al-Marri, T.; Kandeel, M.; ElGioushy, M. Isolation and characterization of vaginal Lactobacillus spp. In dromedary camels (Camelus dromedarius): In vitro evaluation of probiotic potential of selected isolates. Microbiology 2020, 8, e8500. [Google Scholar] [CrossRef]
- Dowarah, R.; Verma, A.K.; Agarwal, N.; Singh, P.; Singh, B.R. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE 2018, 13, e0192978. [Google Scholar] [CrossRef]
- Wu, Y.; Yue, S.; Yu, J.; Bian, F.; Chen, G.; Zhang, Y. Probiotic characterization of lactic acid bacteria from donkey feces in China. Animals 2025, 15, 207. [Google Scholar] [CrossRef]
- Abdel Tawab, F.I.; Abd Elkadr, M.H.; Sultan, A.M.; Hamed, E.O.; El-Zayat, A.S.; Ahmed, M.N. Probiotic potentials of lactic acid bacteria isolated from Egyptian fermented food. Sci. Rep. 2023, 13, 16601. [Google Scholar] [CrossRef] [PubMed]
- Adeniyi, B.; Adesuyi, A.; Ayeni, F.; Ogunbanwo, T.; Agidigbi, T.J. Poultry gastrointestinal-derived lactic acid bacteria (pGIT-d-LAB) inhibit multiple antibiotic-resistant bacterial and fungal pathogens. Avicenna J. Med. Biotechnol. 2024, 16, 111. [Google Scholar]
- Reuben, R.; Roy, P.; Sarkar, S.; Alam, A.R.U.; Jahid, I. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef]
- De Almeida Júnior, W.L.G.; da Silva Ferrari, Í.; de Souza, J.V.; da Silva, C.D.A.; da Costa, M.M.; Dias, F.S. Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 2015, 53, 96–103. [Google Scholar] [CrossRef]
- Padmavathi, T.; Bhargavi, R.; Priyanka, P.R.; Niranjan, N.R.; Pavitra, P.V. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J. Genet. Eng. Biotechnol. 2018, 16, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Nallala, V.S.; Sadishkumar, V.; Jeevaratnam, K. Molecular characterization of antimicrobial Lactobacillus isolates and evaluation of their probiotic characteristics in vitro for use in poultry. Food Biotechnol. 2017, 31, 20–41. [Google Scholar] [CrossRef]
- Makzum, S.; Ghadam, P.; Ramezani, M. Isolation, functional evaluation of probiotic properties and molecular identification of strains isolated from Iranian poultry’s gut. Iran J. Microbiol. 2023, 15, 267–277. [Google Scholar] [CrossRef]
- Dlamini, Z.C.; Langa, R.L.S.; Aiyegoro, O.A.; Okoh, A.I. Safety evaluation and colonisation abilities of four lactic acid bacteria as future probiotics. Probiotics Antimicrob. Proteins 2019, 11, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Kathiriya, M.; Hati, S.; Prajapati, J.; Vekariya, Y. Assessment of in vitro probiotic potential of lactic acid bacteria. J. Dairy Sci. Technol. 2018, 5, 17–30. [Google Scholar]
- Toledo-Arana, A.; Valle, J.; Solano, C.; Arrizubieta, M.J.; Cucarella, C.; Lamata, M.; Amorena, B.; Leiva, J.; Penadés, J.R.; Lasa, I. The enterococcal surface protein, esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 2001, 67, 4538–4545. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penadés, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Boris, S.; Suárez, J.E.; Barbés, C. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J. Appl. Microbiol. 1997, 83, 413–420. [Google Scholar] [CrossRef]
- Gupta, S.; Mohanty, U.; Majumdar, R.K. Isolation and characterization of lactic acid bacteria from traditional fermented fish product shidal of India with reference to their probiotic potential. LWT 2021, 146, 111641. [Google Scholar] [CrossRef]
- Falah, F.; Vasiee, A.; Behbahani, B.A.; Yazdi, F.T.; Moradi, S.; Mortazavi, S.A.; Roshanak, S. Evaluation of adherence and anti-infective properties of probiotic Lactobacillus fermentum strain 4-17 against Escherichia coli causing urinary tract infection in humans. Microb. Pathog. 2019, 131, 246–253. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef]
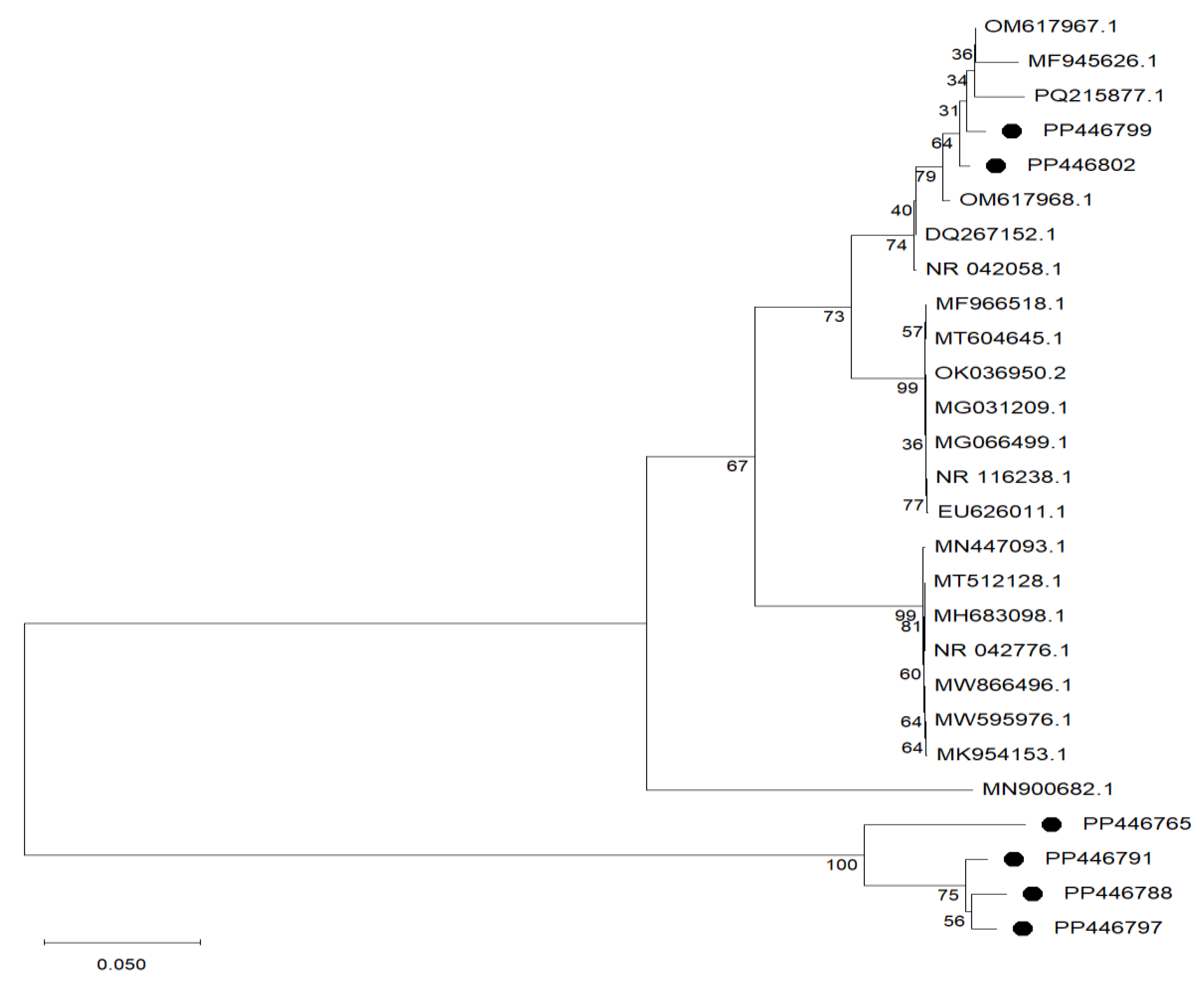
| Isolate ID | Morphology | Gram-Stain | Catalase Test | Phylogenetic Affiliation | GenBank Accession No. | Similarity |
|---|---|---|---|---|---|---|
| SI4 | Cocci | + | − | Streptococcus salivarius NR042776.1 | PP446765 | 100% |
| SI6 | Bacilli | + | − | Levilactobacillus brevis NR116238.1 | PP446788 | 99.85% |
| SI8 | Bacilli | + | − | Levilactobacillus brevis NR116238.1 | PP446791 | 99.85% |
| SI9 | Bacilli | + | − | Levilactobacillus brevis NR116238.1 | PP446797 | 99.85% |
| SI23 | Cocci | + | − | Pediococcus pentosaceus NR042058.1 | PP446799 | 99.58% |
| SI38 | Cocci | + | − | Pediococcus pentosaceus NR042058.1 | PP446802 | 99.51% |
| Isolate ID | Haemolytic Activity | GM | S | C | CRO | CIP | VA | E | AMP | P | OX | TE | NV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. salivarius NKFS6 | - | I | I | S | S | R | R | S | S | I | R | S | R |
| L. brevis NKFS7 | - | R | R | S | S | R | R | S | S | I | R | S | R |
| L. brevis NKFS8 | - | S | S | S | S | R | R | S | S | I | R | S | R |
| L. brevis NKFS9 | - | R | I | S | S | R | R | S | S | I | R | S | R |
| P. pentosaceus NKFS10 | - | R | R | R | R | R | R | R | R | R | R | R | R |
| P. pentosaceus NKFS11 | - | R | R | S | S | R | R | S | S | R | I | S | R |
| L. casei (ATCC 393) | - | R | R | S | R | R | R | S | S | S | R | R | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokwe, N.H.; Tshabuse, F.; Swalaha, F.M. In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders. Microorganisms 2025, 13, 1231. https://doi.org/10.3390/microorganisms13061231
Kokwe NH, Tshabuse F, Swalaha FM. In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders. Microorganisms. 2025; 13(6):1231. https://doi.org/10.3390/microorganisms13061231
Chicago/Turabian StyleKokwe, Nwabisa Happiness, Freedom Tshabuse, and Feroz Mahomed Swalaha. 2025. "In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders" Microorganisms 13, no. 6: 1231. https://doi.org/10.3390/microorganisms13061231
APA StyleKokwe, N. H., Tshabuse, F., & Swalaha, F. M. (2025). In Vitro Characterization and Safety Assessment of Streptococcus salivarius, Levilactobacillus brevis and Pediococcus pentosaceus Isolated from the Small Intestine of Broiler Breeders. Microorganisms, 13(6), 1231. https://doi.org/10.3390/microorganisms13061231








