Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Accessions
2.2. Phylogenetic Analysis
2.3. Orthology and Protein Family Analysis
2.4. Identification of Genes and Pathways Across Acidithiobacillia Genomes
2.5. Recovery of Transcriptional Units and Upstream Regions
2.6. Identification and Phylogeny of Sigma Factors in Acidithiobacillia Representatives
2.7. Identification of Sigma54-Dependent Two-Component Systems
2.8. Identification and Analysis of Putative DNA Binding Sites
2.9. Matrix Scanning of Binding Sites
3. Results
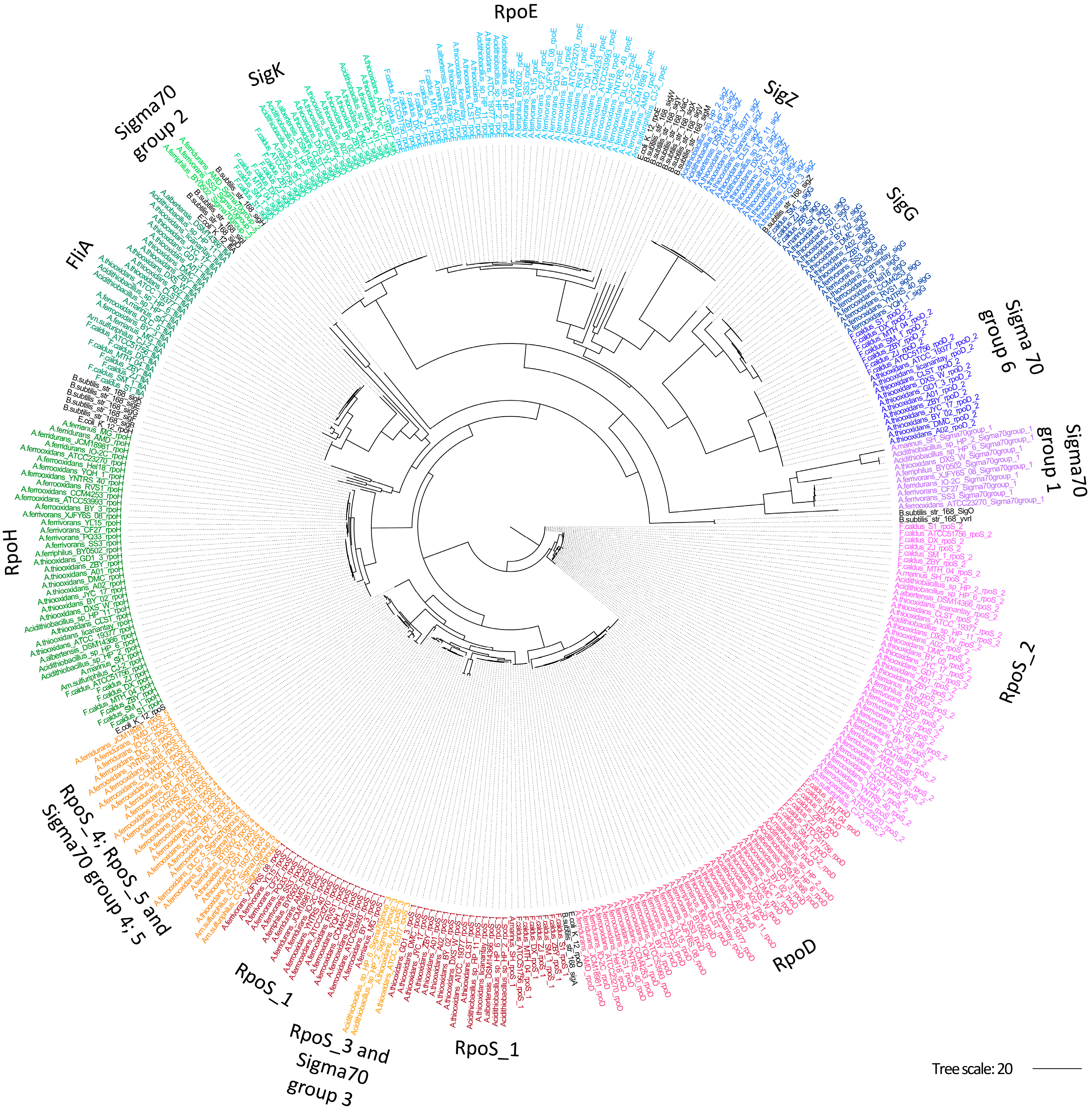
3.1. Acidithiobacillia Display a Diverse Sigma Factor Repertoire Providing Clues on Extreme Acidophile Adaptability
3.2. Sigma Factors Phylogeny Reflects Divergence Signatures of Acidithiobacillia Phenotypes
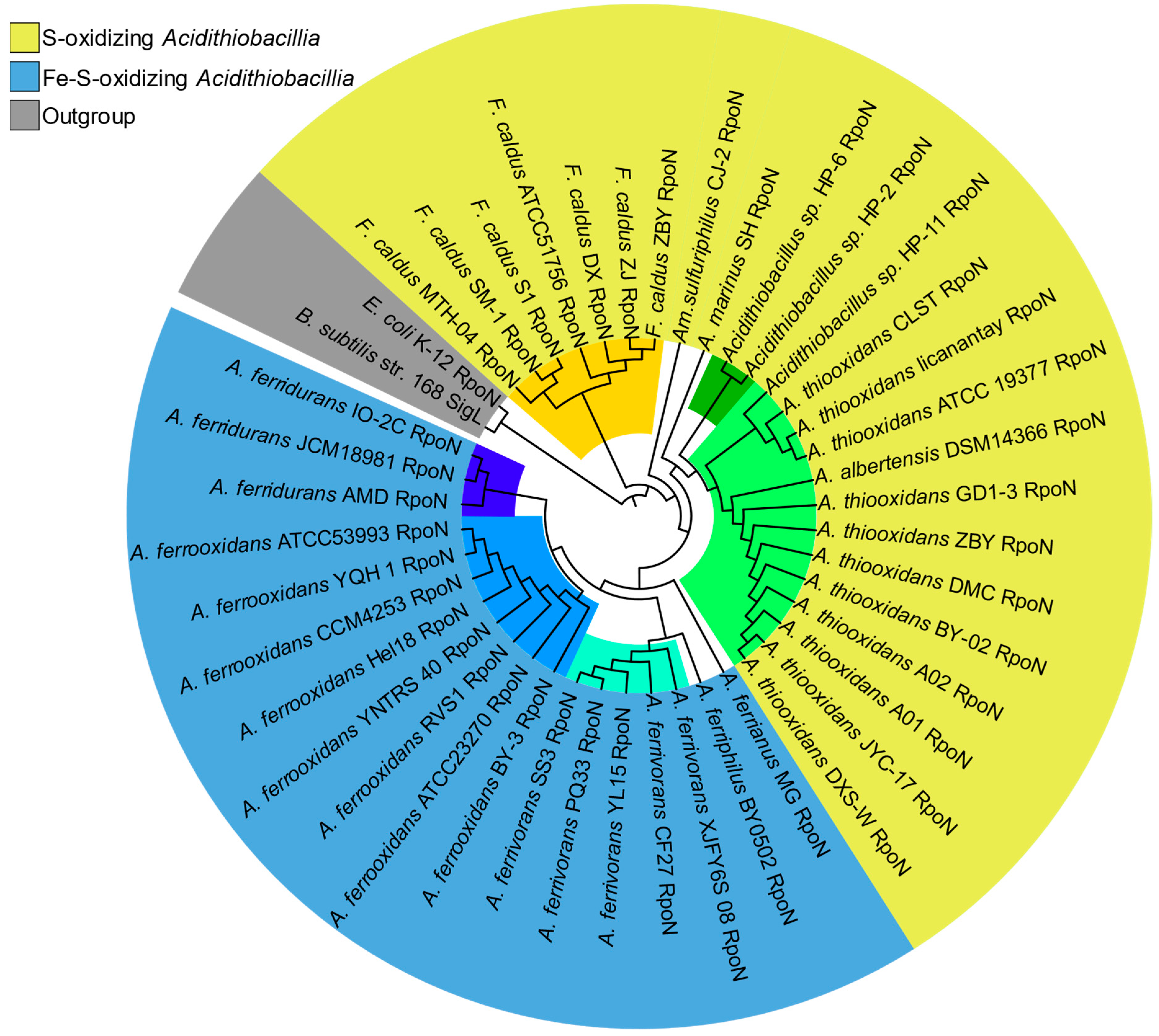
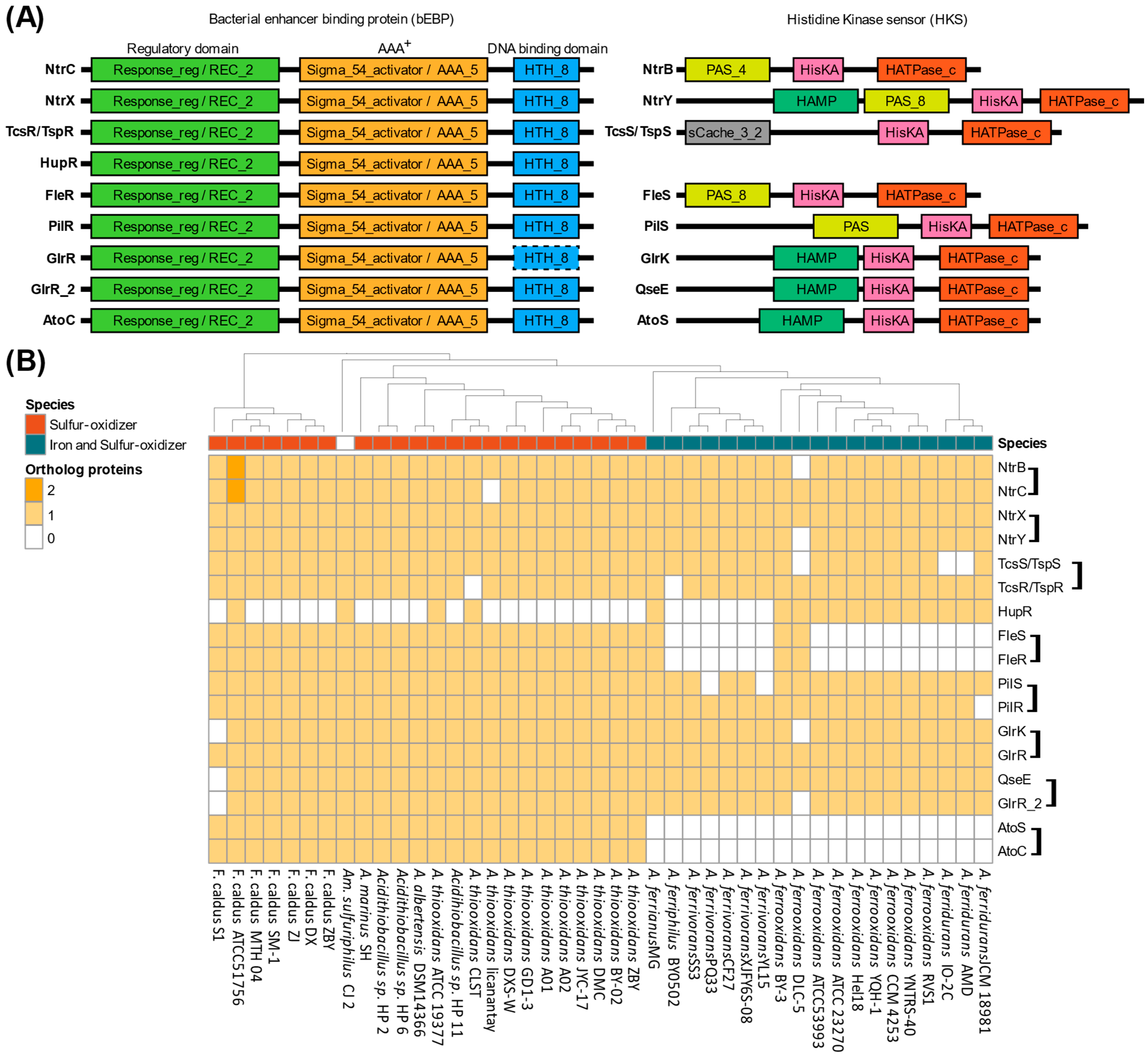
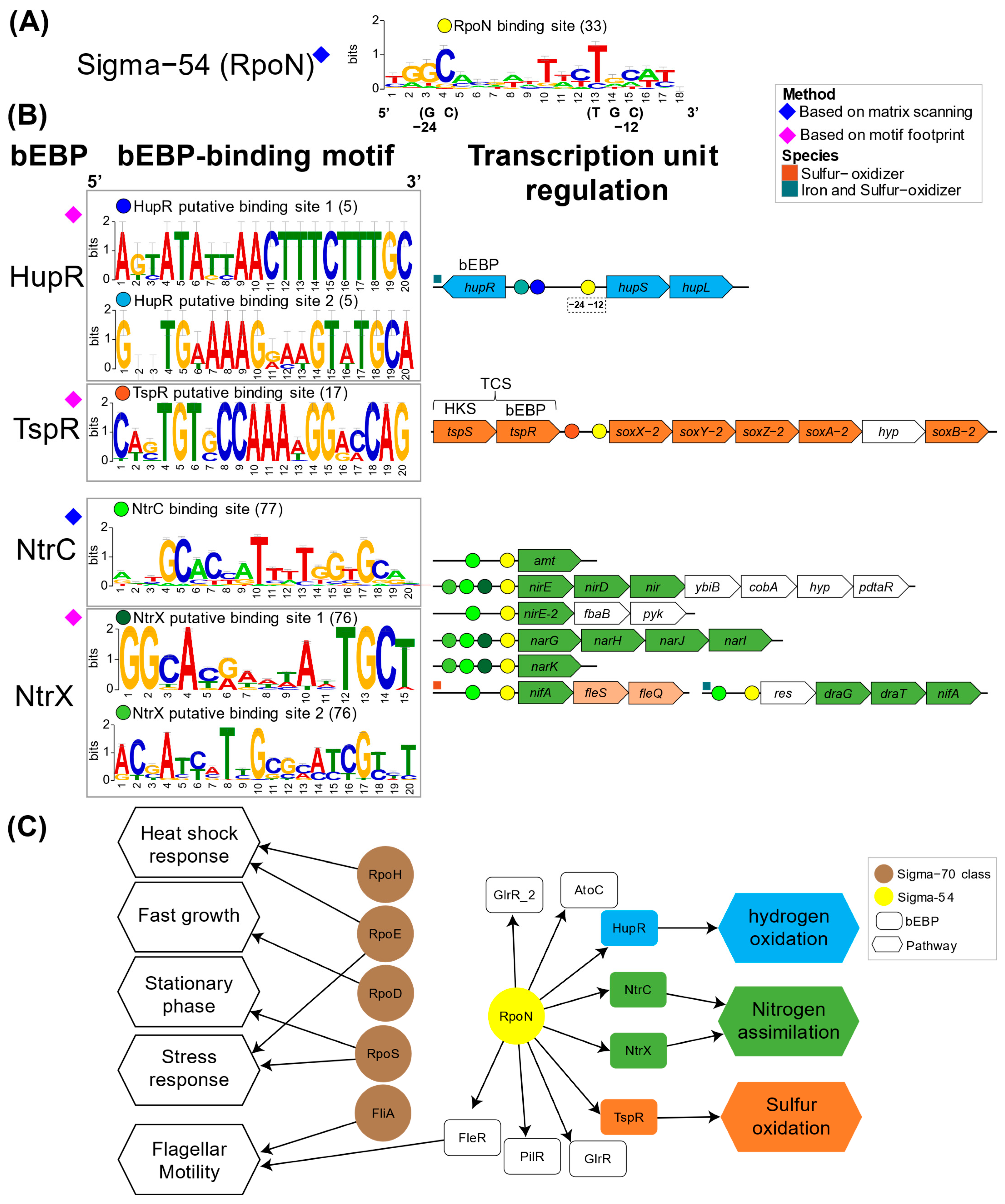
3.3. Sigma54: A Master Regulator for Energy and Nutrient Assimilation in Acidithiobacillia?
4. Discussion
4.1. An Acidophilic Lifestyle Demands Robust Gene Regulation Circuitry
4.2. Sigma Factor Master Regulators Tend to Mirror Acidithiobacillia Evolution
4.3. Sigma54-Dependent Two-Component Systems in Acidithiobacillia Were Potentially Linked to Key Metabolic Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TF | transcription factor |
| TFBS | transcription factor binding site |
| TCS | two-component system |
| SF | sigma factor |
| bEBP | bacterial enhancer binding proteins |
| HKS | histidine kinase sensor |
| DBD | DNA-binding domain |
| OG | orthologous group |
| TU | transcriptional unit |
| PWM | Position Weight Matrix |
References
- Goltsman, D.S.A.; Comolli, L.R.; Thomas, B.C.; Banfield, J.F. Community Transcriptomics Reveals Unexpected High Microbial Diversity in Acidophilic Biofilm Communities. ISME J. 2015, 9, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Quatrini, R.; Johnson, D.B. Microbiomes in Extremely Acidic Environments: Functionalities and Interactions That Allow Survival and Growth of Prokaryotes at Low pH. Curr. Opin. Microbiol. 2018, 43, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, X.T.; Jiang, Z.; Liang, Z.L.; Wang, P.; Liu, Z.H.; Li, L.Z.; Yin, H.Q.; Jia, Y.; Huang, Z.S.; et al. Key Factors Governing Microbial Community in Extremely Acidic Mine Drainage (pH < 3). Front. Microbiol. 2021, 12, 761579. [Google Scholar] [CrossRef]
- Méndez-García, C.; Peláez, A.I.; Mesa, V.; Sánchez, J.; Golyshina, O.V.; Ferrer, M. Microbial Diversity and Metabolic Networks in Acid Mine Drainage Habitats. Front. Microbiol. 2015, 6, 475. [Google Scholar] [CrossRef]
- Jerez, C.A. Biomining of Metals: How to Access and Exploit Natural Resource Sustainably. Microb. Biotechnol. 2017, 10, 1191–1193. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Won, S.; Ha, M.G.; Nguyen, D.D.; Kang, H.Y. Bioleaching for Environmental Remediation of Toxic Metals and Metalloids: A Review on Soils, Sediments, and Mine Tailings. Chemosphere 2021, 282, 131108. [Google Scholar] [CrossRef]
- Nuñez, H.; Moya-Beltrán, A.; Covarrubias, P.C.; Issotta, F.; Cárdenas, J.P.; González, M.; Atavales, J.; Acuña, L.G.; Johnson, D.B.; Quatrini, R. Molecular Systematics of the Genus Acidithiobacillus: Insights into the Phylogenetic Structure and Diversification of the Taxon. Front. Microbiol. 2017, 8, 30. [Google Scholar] [CrossRef]
- Williams, K.P.; Kelly, D.P. Proposal for a New Class within the Phylum Proteobacteria, Acidithiobacillia Classis Nov., with the Type Order Acidithiobacillales, and Emended Description of the Class Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013, 63, 2901–2906. [Google Scholar] [CrossRef] [PubMed]
- Moya-Beltrán, A.; Beard, S.; Rojas-Villalobos, C.; Issotta, F.; Gallardo, Y.; Ulloa, R.; Giaveno, A.; Degli Esposti, M.; Johnson, D.B.; Quatrini, R. Genomic Evolution of the Class Acidithiobacillia: Deep-Branching Proteobacteria Living in Extreme Acidic Conditions. ISME J. 2021, 15, 3221–3238. [Google Scholar] [CrossRef]
- González-Rosales, C.; Vergara, E.; Dopson, M.; Valdés, J.H.; Holmes, D.S. Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle. Front. Microbiol. 2022, 12, 822229. [Google Scholar] [CrossRef]
- Sepúlveda-Rebolledo, P.; González-Rosales, C.; Dopson, M.; Pérez-Rueda, E.; Holmes, D.S.; Valdés, J.H. Comparative Genomics Sheds Light on Transcription Factor-Mediated Regulation in the Extreme Acidophilic Acidithiobacillia Representatives. Res. Microbiol. 2024, 175, 104135. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Johnson, D.B. Biodiversity, Metabolism and Applications of Acidophilic Sulfur-Metabolizing Microorganisms. Environ. Microbiol. 2012, 14, 2620–2631. [Google Scholar] [CrossRef]
- Esparza, M.; Jedlicki, E.; Dopson, M.; Holmes, D.S. Expression and Activity of the Calvin-Benson-Bassham Cycle Transcriptional Regulator CbbR from Acidithiobacillus ferrooxidans in Ralstonia eutropha. FEMS Microbiol. Lett. 2015, 362, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Kupka, D.; Liljeqvist, M.; Nurmi, P.; Puhakka, J.A.; Tuovinen, O.H.; Dopson, M. Oxidation of Elemental Sulfur, Tetrathionate and Ferrous Iron by the Psychrotolerant Acidithiobacillus strain SS3. Res. Microbiol. 2009, 160, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, S.; Johnson, D.B. Acidithiobacillus ferridurans Sp. Nov., an Acidophilic Iron-, Sulfur- and Hydrogen-Metabolizing Chemolithotrophic Gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 4018–4025. [Google Scholar] [CrossRef]
- Leyn, S.A.; Novichkov, P.S.; Stepanova, V.V.; Ravcheev, D.A.; Kazakov, A.E.; Suvorova, I.A.; Rodionov, D.A. Comparative Genomics and Evolution of Transcriptional Regulons in Proteobacteria. Microb. Genom. 2016, 2, e000061. [Google Scholar] [CrossRef]
- Paget, M.S. Bacterial Sigma Factors and Anti-Sigma Factors: Structure, Function and Distribution. Biomolecules 2015, 5, 1245–1265. [Google Scholar] [CrossRef]
- Zhang, N.; Buck, M. A Perspective on the Enhancer Dependent Bacterial RNA Polymerase. Biomolecules 2015, 5, 1012–1019. [Google Scholar] [CrossRef]
- Doucleff, M.; Malak, L.T.; Pelton, J.G.; Wemmer, D.E. The C-Terminal RpoN Domain of Sigma54 Forms an Unpredicted Helix-Turn-Helix Motif Similar to Domains of sigma70. J. Biol. Chem. 2005, 280, 41530–41536. [Google Scholar] [CrossRef]
- Paget, M.S.B.; Helmann, J.D. The Σ70 Family of Sigma Factors. Genome Biol. 2003, 4, 203. [Google Scholar] [CrossRef]
- Otani, H.; Udwary, D.W.; Mouncey, N.J. Comparative and Pangenomic Analysis of the Genus Streptomyces. Sci. Rep. 2022, 12, 18909. [Google Scholar] [CrossRef]
- Bush, M.; Dixon, R. The Role of Bacterial Enhancer Binding Proteins as Specialized Activators of σ 54-Dependent Transcription. Microbiol. Mol. Biol. Rev. 2012, 76, 497–529. [Google Scholar] [CrossRef] [PubMed]
- Shingler, V. Signal Sensing by Sigma-54-Dependent Regulators Derepression as a Control Mechanism. Mol. Microbiol. 1996, 19, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, A.; Severinova, E.; Darst, S.A. Crystal Structure of a Σ70 Subunit Fragment from E. coli RNA Polymerase. Cell 1996, 87, 127–136. [Google Scholar] [CrossRef]
- Haldenwang, W.G. The Sigma Factors of Bacillus subtilis. Microbiol. Mol. Biol. Rev. 1995, 59, 1–30. [Google Scholar] [CrossRef]
- Sauer, U.; Santangelo, J.D.; Treuner, A.; Buchholz, M.; Dürre, P. Sigma Factor and Sporulation Genes in Clostridium. FEMS Microbiol. Rev. 1995, 17, 331–340. [Google Scholar] [CrossRef]
- Feklístov, A.; Sharon, B.D.; Darst, S.A.; Gross, C.A. Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective. Annu. Rev. Microbiol. 2014, 68, 357–376. [Google Scholar] [CrossRef]
- Clark, A.E.; Adamson, C.C.; Carothers, K.E.; Roxas, B.A.P.; Viswanathan, V.K.; Vedantam, G. The Alternative Sigma Factor SigL Influences Clostridioides difficile Toxin Production, Sporulation, and Cell Surface Properties. Front. Microbiol. 2022, 13, 871152. [Google Scholar] [CrossRef]
- Kazakov, A.E.; Rajeev, L.; Chen, A.; Luning, E.G.; Dubchak, I.; Mukhopadhyay, A.; Novichkov, P.S. Σ54-Dependent Regulome in Desulfovibrio vulgaris Hildenborough. BMC Genom. 2015, 16, 919. [Google Scholar] [CrossRef]
- Mangold, S.; Rao Jonna, V.; Dopson, M. Response of Acidithiobacillus caldus toward Suboptimal pH Conditions. Extremophiles 2013, 17, 689–696. [Google Scholar] [CrossRef]
- Yang, C.L.; Chen, X.K.; Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Lin, J.Q.; Chen, L.X. Essential Role of σ Factor Rpof in Flagellar Biosynthesis and Flagella-Mediated Motility of Acidithiobacillus caldus. Front. Microbiol. 2019, 10, 1130. [Google Scholar] [CrossRef]
- Gao, R.; Stock, A.M. Biological Insights from Structures of Two-Component Proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Fu, L.J.; Lin, J.Q.; Pang, X.; Liu, X.M.; Wang, R.; Wang, Z.B.; Lin, J.Q.; Chen, L.X. The Σ54-Dependent Two-Component System Regulating Sulfur Oxidization (Sox) System in Acidithiobacillus caldus and Some Chemolithotrophic Bacteria. Appl. Microbiol. Biotechnol. 2017, 101, 2079–2092. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.; Volkman, B.F.; Luginbuhl, P.; Nohaile, M.J.; Kustu, S.; Wemmer, D.E. Structure of a Transiently Phosphorylated Switch in Bacterial Signal Transduction. Nature 1999, 402, 894–897. [Google Scholar] [CrossRef]
- Sallai, L.; Tucker, P.A. Crystal Structure of the Central and C-Terminal Domain of the Σ54-Activator ZraR. J. Struct. Biol. 2005, 151, 160–170. [Google Scholar] [CrossRef]
- Park, S.; Meyer, M.; Jones, A.D.; Yennawar, H.P.; Yennawar, N.H.; Nixon, B.T. Two-Component Signaling in the AAA+ ATPase DctD: Binding Mg2+ and BeF3− Selects between Alternate Dimeric States of the Receiver Domain. FASEB J. 2002, 16, 1964–1966. [Google Scholar] [CrossRef]
- Kilkenny, C.A.; Berger, D.K.; Rawlings, D.E. Isolation of the Thiobacillus ferrooxidans NtrBC Genes Using a T. ferrooxidans NifH-LacZ Fusion. Microbiology 1994, 140, 2543–2553. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Parks, D.H.; Tyson, G.W.; Hugenholtz, P. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of Nearly 8000 Metagenome-Assembled Genomes Substantially Expands the Tree of Life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; Von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by EggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- González, C.; Lazcano, M.; Valdés, J.; Holmes, D.S. Bioinformatic Analyses of Unique (Orphan) Core Genes of the Genus Acidithiobacillus: Functional Inferences and Use as Molecular Probes for Genomic and Metagenomic/Transcriptomic Interrogation. Front. Microbiol. 2016, 7, 2035. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Valdés, J.; Veloso, F.; Jedlicki, E.; Holmes, D. Metabolic Reconstruction of Sulfur Assimilation in the Extremophile Acidithiobacillus ferrooxidans Based on Genome Analysis. BMC Genom. 2003, 4, 51. [Google Scholar] [CrossRef]
- Quatrini, R.; Valdés, J.; Jedlicki, E.; Holmes, D.S. The Use Of Bioinformatics And Genome Biology To Advance Our Understanding Of Bioleaching Microorganisms. In Microbial Processing of Metal Sulfides; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–314. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Dodson, R.J.; Tettelin, H.; Blake, R.; Eisen, J.A.; Holmes, D.S. Acidithiobacillus ferrooxidans Metabolism: From Genome Sequence to Industrial Applications. BMC Genom. 2008, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.E.; Field, B. Operons. Cell. Mol. Life Sci. 2009, 66, 3755–3775. [Google Scholar] [CrossRef] [PubMed]
- Moinier, D.; Byrne, D.; Amouric, A.; Bonnefoy, V. The Global Redox Responding RegB/RegA Signal Transduction System Regulates the Genes Involved in Ferrous Iron and Inorganic Sulfur Compound Oxidation of the Acidophilic Acidithiobacillus ferrooxidans. Front. Microbiol. 2017, 8, 1277. [Google Scholar] [CrossRef]
- Neira, G.; Cortez, D.; Jil, J.; Holmes, D.S. AciDB 1.0: A Database of Acidophilic Organisms, Their Genomic Information and Associated Metadata. Bioinformatics 2020, 36, 4970–4971. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef]
- Crooks, G.; Hon, G.; Chandonia, J.; Brenner, S. WebLogo: A Sequence Logo Generator Gavin. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.T.; Contreras-moreira, B.; Castro-mondragon, J.A.; Santana-garcia, W.; Ossio, R.; Robles-espinoza, C.D.; Bahin, M.; Collombet, S.; Vincens, P.; Thieffry, D.; et al. RSAT 2018: Regulatory Sequence Analysis Tools 20th Anniversary. Nucleic Acids Res. 2018, 46, 209–214. [Google Scholar] [CrossRef]
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That Are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. J. 2016, 12, e1004862. [Google Scholar] [CrossRef]
- Davis, M.C.; Kesthely, C.A.; Franklin, E.A.; MacLellan, S.R. The Essential Activities of the Bacterial Sigma Factor. Can. J. Microbiol. 2017, 63, 89–99. [Google Scholar] [CrossRef]
- Liu, X.; Matsumura, P. An Alternative Sigma Factor Controls Transcription of Flagellar Class-III Operons in Escherichia coli: Gene Sequence, Overproduction, Purification and Characterization. Gene 1995, 164, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.L.; Peng, A.A.; He, H.; Yang, Y.; Liu, X.D.; Qiu, G.Z. A New Strain Acidithiobacillus albertensis BY-05 for Bioleaching of Metal Sulfides Ores. Trans. Nonferrous Met. Soc. China 2007, 17, 168–175. (In English) [Google Scholar] [CrossRef]
- Valdés, J.H.; Pedroso, I.; Quatrini, R.; Hallberg, K.B.; Valenzuela, P.D.T.; Holmes, D.S. Insights into the Metabolism and Ecophysiology of Three Acidithiobacilli by Comparative Genome Analysis. Adv. Mater. Res. 2007, 20–21, 439–442. [Google Scholar] [CrossRef]
- Valdés, J.; Pedroso, I.; Quatrini, R.; Holmes, D.S. Comparative Genome Analysis of Acidithiobacillus ferrooxidans, A. Thiooxidans and A. caldus: Insights into Their Metabolism and Ecophysiology. Hydrometallurgy 2008, 94, 180–184. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, L.; Xing, W.; Chen, P.; Zhang, Y.; Wang, W. Acidithiobacillus ferrooxidans and Its Potential Application. Extremophiles 2018, 22, 563–579. [Google Scholar] [CrossRef]
- Rowe, O.F.; Sánchez-España, J.; Hallberg, K.B.; Johnson, D.B. Microbial Communities and Geochemical Dynamics in an Extremely Acidic, Metal-Rich Stream at an Abandoned Sulfide Mine (Huelva, Spain) Underpinned by Two Functional Primary Production Systems. Environ. Microbioly 2007, 9, 1761–1771. [Google Scholar] [CrossRef]
- Wang, R.; Lin, J.Q.; Liu, X.M.; Pang, X.; Zhang, C.J.; Yang, C.L.; Gao, X.Y.; Lin, C.M.; Li, Y.Q.; Li, Y.; et al. Sulfur Oxidation in the Acidophilic Autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 10, 3290. [Google Scholar] [CrossRef]
- Wang, Z.B.; Li, Y.Q.; Lin, J.Q.; Pang, X.; Liu, X.M.; Liu, B.Q.; Wang, R.; Zhang, C.J.; Wu, Y.; Lin, J.Q.; et al. The Two-Component System RsrS-RsrR Regulates the Tetrathionate Intermediate Pathway for Thiosulfate Oxidation in Acidithiobacillus caldus. Front. Microbiol. 2016, 7, 1755. [Google Scholar] [CrossRef] [PubMed]
- Vignais, P.M.; Billoud, B. Occurrence, Classification, and Biological Function of Hydrogenases: An Overview. Chem. Rev. 2007, 107, 4206–4272. [Google Scholar] [CrossRef]
- Vignais, P.M.; Elsen, S.; Colbeau, A. Transcriptional Regulation of the Uptake [NiFe]Hydrogenase Genes in Rhodobacter capsulatus. Biochem. Soc. Trans. 2005, 33, 28–32. [Google Scholar] [CrossRef]
- Mackintosh, M.E. Nitrogen Fixation by Thiobacillus ferrooxidans. J. Gen. Microbiol. 1978, 105, 215–218. [Google Scholar] [CrossRef]
- Bonato, P.; Alves, L.R.; Osaki, J.H.; Rigo, L.U.; Pedrosa, F.O.; Souza, E.M.; Zhang, N.; Schumacher, J.; Buck, M.; Wassem, R.; et al. The NtrY–NtrX Two-Component System Is Involved in Controlling Nitrate Assimilation in Herbaspirillum seropedicae Strain SmR1. FEBS J. 2016, 283, 3919–3930. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen Regulation in Bacteria and Archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef]
- Drepper, T.; Groß, S.; Yakunin, A.F.; Hallenbeck, P.C.; Masepohl, B.; Klipp, W. Role of GlnB and GlnK in Ammonium Control of Both Nitrogenase Systems in the Phototrophic Bacterium Rhodobacter capsulatus. Microbiology 2003, 149, 2203–2212. [Google Scholar] [CrossRef]
- Levicán, G.; Ugalde, J.A.; Ehrenfeld, N.; Maass, A.; Parada, P. Comparative Genomic Analysis of Carbon and Nitrogen Assimilation Mechanisms in Three Indigenous Bioleaching Bacteria: Predictions and Validations. BMC Genom. 2008, 9, 581. [Google Scholar] [CrossRef]
- Ishida, M.L.; Assumpção, M.C.; Machado, H.B.; Benelli, E.M.; Souza, E.M.; Pedrosa, F.O. Identification and Characterization of the Two-Component NtrY/NtrX Regulatory System in Azospirillum brasilense. Braz. J. Med. Biol. Res. 2002, 35, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Minamino, T. Flagella-Driven Motility of Bacteria. Biomolecules 2019, 9, 279. [Google Scholar] [CrossRef]
- Jyot, J.; Dasgupta, N.; Ramphal, R. FleQ, the Major Flagellar Gene Regulator in Pseudomonas aeruginosa, Binds to Enhancer Sites Located Either Upstream or Atypically Downstream of the RpoN Binding Site. J. Bacteriol. 2002, 184, 5251–5260. [Google Scholar] [CrossRef]
- Jacobi, S.; Schade, R.; Heuner, K. Characterization of the Alternative Sigma Factor Σ54 and the Transcriptional Regulator FleQ of Legionella pneumophila, Which Are Both Involved in the Regulation Cascade of Flagellar Gene Expression. J. Bacteriol. 2004, 186, 2540–2547. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sepúlveda-Rebolledo, P.; González-Rosales, C.; Dopson, M.; Pérez-Rueda, E.; Holmes, D.S.; Valdés, J.H. Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments. Microorganisms 2025, 13, 1199. https://doi.org/10.3390/microorganisms13061199
Sepúlveda-Rebolledo P, González-Rosales C, Dopson M, Pérez-Rueda E, Holmes DS, Valdés JH. Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments. Microorganisms. 2025; 13(6):1199. https://doi.org/10.3390/microorganisms13061199
Chicago/Turabian StyleSepúlveda-Rebolledo, Pedro, Carolina González-Rosales, Mark Dopson, Ernesto Pérez-Rueda, David S. Holmes, and Jorge H. Valdés. 2025. "Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments" Microorganisms 13, no. 6: 1199. https://doi.org/10.3390/microorganisms13061199
APA StyleSepúlveda-Rebolledo, P., González-Rosales, C., Dopson, M., Pérez-Rueda, E., Holmes, D. S., & Valdés, J. H. (2025). Comparative Genomics of Sigma Factors in Acidithiobacillia Sheds Light into the Transcriptional Regulatory Networks Involved in Biogeochemical Dynamics in Extreme Acidic Environments. Microorganisms, 13(6), 1199. https://doi.org/10.3390/microorganisms13061199







