Abstract
Understanding the genetic characteristics of Aeromonas jandaei in Brazilian aquaculture is crucial for developing effective control strategies against this fish pathogen. The present study conducted a genomic analysis of Brazilian A. jandaei strains with the objective of investigating their virulence potential and resistance profiles. Four Brazilian isolates were subjected to sequencing, and comparative genomic analyses were conducted in conjunction with 48 publicly available A. jandaei genomes. The methods employed included quality assessment, de novo assembly, annotation, and analyses of antimicrobial resistance and virulence factors. The results demonstrated the presence of fluoroquinolone resistance genes within the core genome. Notably, these antibiotics are not authorized for use in aquaculture in Brazil, suggesting that their resistance determinants may originate from other selective pressures or horizontal gene transfer unrelated to aquaculture practices. The analysis identified significant virulence mechanisms, including T2SS, T3SS, and notably T6SS (vgrG3 gene), which was more prevalent in Brazilian isolates. Additionally, genes associated with motility, adhesion, and heavy metal resistance were identified. These findings highlight the enhanced adaptability of Brazilian A. jandaei strains and raise concerns about antimicrobial resistance in aquaculture, emphasizing the need for improved regulatory oversight and control strategies.
1. Introduction
Aquaculture has witnessed a significant expansion in recent decades, becoming an increasingly crucial aspect of global food security and economic development. In 2022, aquaculture surpassed capture fisheries in terms of aquatic animal production for the first time, reaching 94.4 million tonnes and accounting for 51% of the global total [1]. However, intensive production faces significant challenges, particularly disease outbreaks caused by high-density and stressful conditions, which frequently result in substantial economic losses [2]. Obtaining accurate data on Brazilian fish production, disease outbreaks, and associated losses remains challenging. However, an estimate suggests that direct and indirect losses amount to approximately USD$ 84 million annually [3].
Among the bacterial pathogens affecting aquaculture, the genus Aeromonas, particularly Aeromonas jandaei, has emerged as a significant concern in freshwater environments, particularly affecting Nile tilapia (Oreochromis niloticus) [4]. This genus consists of Gram-negative, facultative anaerobic bacteria widely distributed in aquatic ecosystems. Phenotypic identification methods often fail to differentiate between species due to the complexity of the genus’s taxonomy [5]. In fish, Aeromonas infections, particularly Motile Aeromonad Septicaemia (MAS), are among the most frequent bacterial diseases, especially in freshwater species. Stress or immune suppression increases susceptibility to MAS, which presents with non-specific clinical signs such as fin rot, ulcers, hemorrhages, exophthalmia, and ascitis [6].
A. jandaei has also been reported in Brazilian native species such as pirarucu (Araipamas gigas) and tambaqui (Colossoma macropomum) [7,8,9], and also in walking catfish (Clarias sp.) [10], European eel [11], freshwater crocodiles [12], as well as soft-shell turtles [13]. Its virulence is driven by multiple factors, including haemolysin, temperature-sensitive protease, haemolysin-aerolysin, and nuclease, which facilitate tissue invasion and immune evasion [4]. If not promptly and adequately treated, Aeromonas infections can result in significant mortality among affected fish populations [14].
The emergence of antimicrobial resistance in aquaculture pathogens, including Aeromonas spp., has become a growing concern globally. This situation is aggravated by the limited number of approved antibiotics for aquaculture use [15]. Understanding the genetic basis of virulence and antimicrobial resistance in these pathogens has become crucial for developing effective control strategies. Recent years have witnessed a shift in understanding bacterial pathogens through genomic approaches. Bioinformatics and genomic analyses have become essential tools for characterizing virulence factors, antimicrobial resistance mechanisms, and evolutionary relationships among bacterial strains. In Brazil, where aquaculture represents a growing sector of the economy, understanding the genetic characteristics of local A. jandaei isolates becomes crucial for developing effective control strategies. This study employs comprehensive genomic analysis to investigate Brazilian A. jandaei strains, aiming to provide insights into their virulence potential and resistance profiles, contributing to the broader understanding of this important fish pathogen.
2. Materials and Methods
2.1. Isolation and Genome Sequencing
Four isolates were obtained from Nile tilapia (Oreochromis niloticus) exhibiting signs of disease from the São Francisco River in the northeast of Brazil. Two of the isolates (designated Aer_On4M and Aer_On5M) were obtained in 2009 in Sobradinho, in the state of Bahia. The remaining two isolates (GTBM29 and GT15) were derived from fish collected in Petrolina, Pernambuco, and were obtained in 2011. They were isolated from the kidneys of different fish.
Isolates were grown in Tryptic Soy Agar (TSA) at 28 °C for 24 h. All isolates were stored in brain-heart infusion (BHI) broth supplemented with glycerol at −80 °C until analysis. The DNA was extracted from the samples using a salting-out method [16]. Sequencing was conducted on the Illumina Hi-Seq 2500 platform (Illumina, San Diego, CA, USA) using a 2 × 150 bp paired-end library (500 bp insert) at the University of Göttingen, Germany.
2.2. Quality Control, Assembly, Completeness, and Annotation
The sequencing quality of the samples was evaluated using FastQC (v0.11.2) metrics [17]. The adapters were removed using Trimmomatic (v0.39) [18].
Subsequently, Unicycler (v0.4.7) [19] was employed for assembly using the default parameters for the de novo method. The quality of the assembled genomes was verified using QUAST (v5.0.2) [20] with the default parameters. To ascertain the completeness of all genomes, a BUSCO (Benchmarking Universal Single-Copy Orthologs) (v4.1.2) [21] analysis was conducted using the gammaproteobacteria_odb10 database and a minimum completeness threshold of 95%. Prokka (1.14.6) [22] was employed for both functional and structural annotation of all genomes to achieve better standardization.
2.3. Data Acquisition
All the 48 A. jandaei genomes available at the public genome repository of the National Center for Biotechnology (NCBI) [23] were acquired in .fna files to complete this study database on 5 August 2024. All genomes were classified by strain, size (mb), assembly, and host/isolation source (Table 1).

Table 1.
Genomes of A. jandaei downloaded from NCBI database.
2.4. Species Classification
Following this, an Average Nucleotide Identity (ANI) analysis was conducted on all genomes, including the four initial ones and the 48 obtained from NCBI, using the MUMmer alignment method with pyANI (0.2.12) [24]. A similarity threshold of 96% or greater [5] was employed to ascertain whether the genomes in question belonged to the same species.
The phylogenomic tree was constructed using Orthofinder (v2.5.5) [25]. An alignment was conducted with DIAMOND [26] (e-value 5 × 10−6), followed by a multiple sequence alignment (MSA). For the alignment of sequences after the identification of clusters of orthologous genes, MAFFT [27] was employed. The phylogenetic tree was constructed using FastTree, and the resulting tree was visualized and edited using iTOL (v7.1) [28].
2.5. Resistance and Virulence Genes
The identification of resistance and virulence genes was conducted using PanViTa (v1.1.8) [29]. Genes exhibiting over 70% identity compared to reference sequences were classified as present. The analysis involved comparisons with three databases: for resistance genes, the Comprehensive Antibiotic Resistance Database (CARD) [30] was utilized, while for virulence genes, the Virulence Factors of Pathogenic Bacteria (VFDB) [31], and finally for metal resistance, the Bacterial Metal Resistance Database (BacMet) [32] were employed.
3. Results
3.1. Genomic Metrics
The initial quality control assessment demonstrated that all genomes exhibited a PHRED quality score of 30 or higher. The BUSCO analysis established a minimum completeness threshold of 95%. Six strains (SEL_ont, SRR12456170_bin.18_metaWRAP_v1.3_MAG, 3299, 3348, Colony25, and Colony119) failed to reach this threshold and were consequently excluded from subsequent analyses. All study subjects achieved a completeness score of 99.7%. Additionally, adapter sequences were removed from all samples. The assembly of four genomes (GTBM29, GT15, On4M, and On5M) showed the results presented in Table 2.

Table 2.
Results (bp, contigs, N50, L50, and GC content) obtained on Quast from the four genomes (GTBM29, GT15, On4M, On5M).
These genomes are available on GenBank/NCBI, and their BioProject and BioSample are shown in Table 3.

Table 3.
BioProjects and BioSamples from GTBM29, GT15, On4M, and On5M.
3.2. Genome Nucleotide Similarity
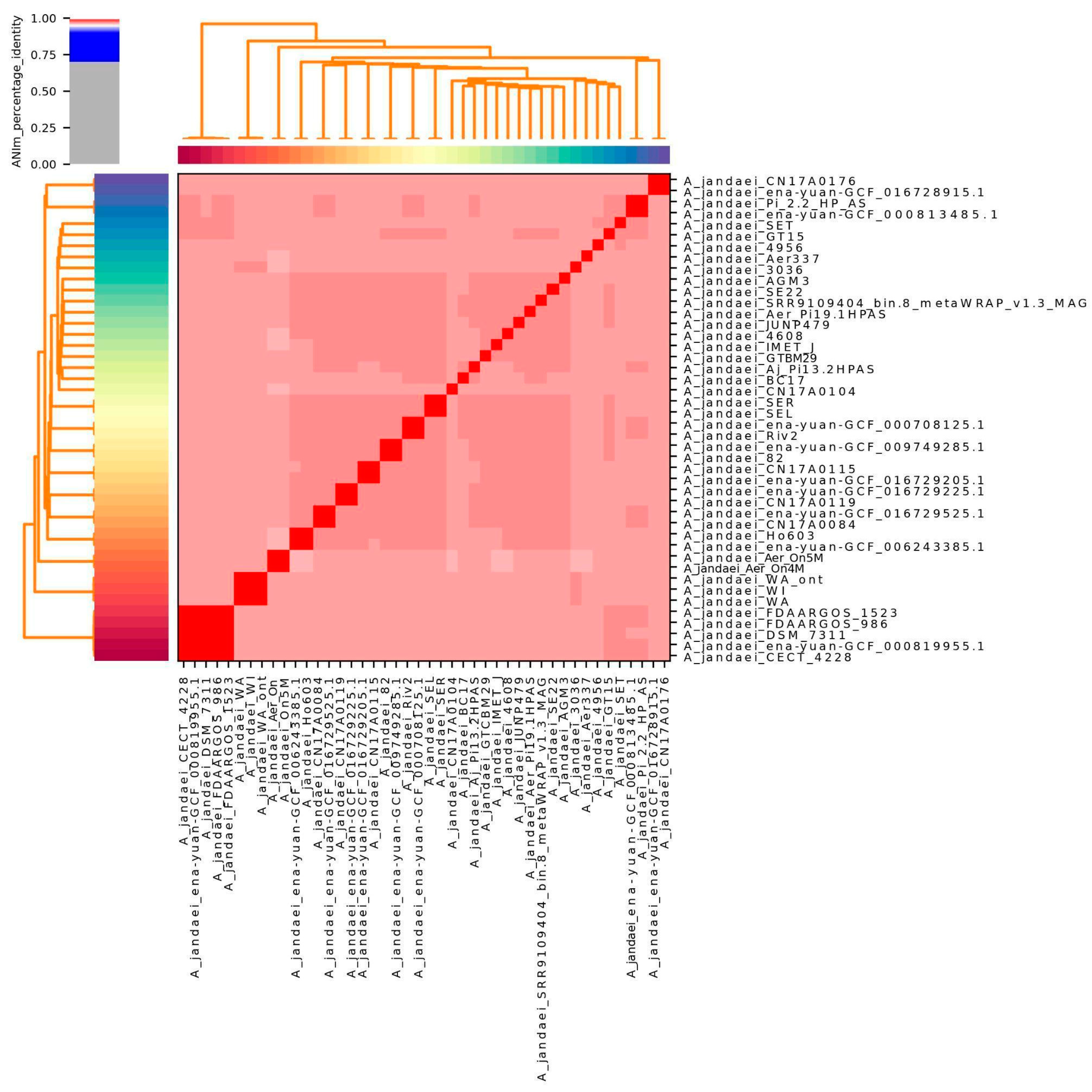
The average nucleotide identity analysis of 46 strains revealed that 45 isolates exhibited similarity values exceeding 96%, as shown in Figure 1. Two strains (L14h and ena-yuan-GCF_019348715.1) were omitted from the visualization due to lower similarity values. This analysis confirmed that our samples are indeed from Aeromonas jandaei. Supplementary Table S1 reveals strain similarities, demonstrating that the two Sobradinho strains (Aer_On4M and Aer_On5M) exhibit high genetic proximity, although they cannot be classified as clonal.
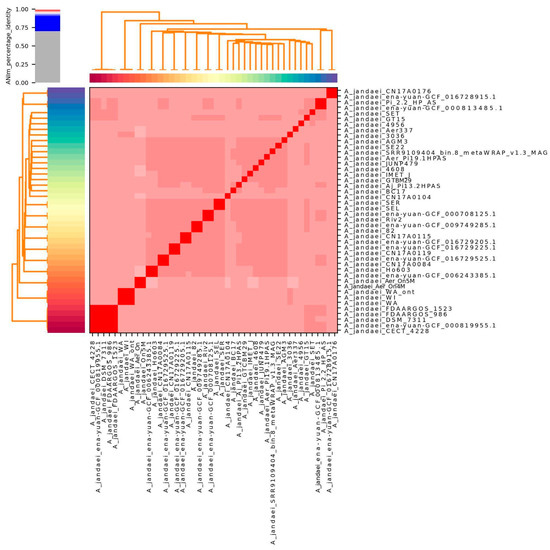
Figure 1.
Average Nucleotide Identity (ANI) analysis of 42 Aeromonas jandaei genomes from NCBI and 4 from this study. Heatmap shows high genomic similarity (>96%), indicating they belong to the same species.
3.3. Phylogenomic Analysis of Orthologous Genes
The phylogenomic tree of orthologous genes is shown in Figure 2. The tree demonstrates the genetic relationships among A. jandaei strains, including our four Brazilian isolates from this study: GT15 and GTBM29 (Petrolina isolates) form a distinct clade with high bootstrap support values, suggesting their close evolutionary relationship, which aligns with their temporal and geographical origin. However, On4M and On5M (Sobradinho isolates), despite being isolated from the same geographical region (northeastern Brazil) as shown in Supplementary Figure S1, cluster separately from the Sobradinho isolates, indicating significant genetic diversity among Brazilian A. jandaei strains even within the same region. This suggests that geographical proximity does not necessarily reflect phylogenetic relatedness in this species. The presence of A. veronii FDAARGOS_632 as an outgroup helps to root the tree and provides context for the evolutionary relationships within A. jandaei species. The tree topology suggests significant genetic diversity among A. jandaei strains, with several distinct lineages being observed.

Figure 2.
Phylogenomic tree of Aeromonas strains constructed using OrthoFinder. The tree includes 41 genomes from public databases and 4 genomes from the current study (shown in purple). The bootstrap values, which indicate the statistical support for each clade, are displayed next to the corresponding branches. The most prevalent resistance mechanisms for each isolate are shown alongside their respective branches, with color-coding indicating different resistance classes.
To explore potential genomic variation across isolates, we examined the accessory genome composition. To maintain clarity, we present the complete accessory genome matrix, showing presence/absence patterns for all accessory genes, as Supplementary Figure S2.
3.4. Resistance Genes
3.4.1. Antimicrobial Resistance Results
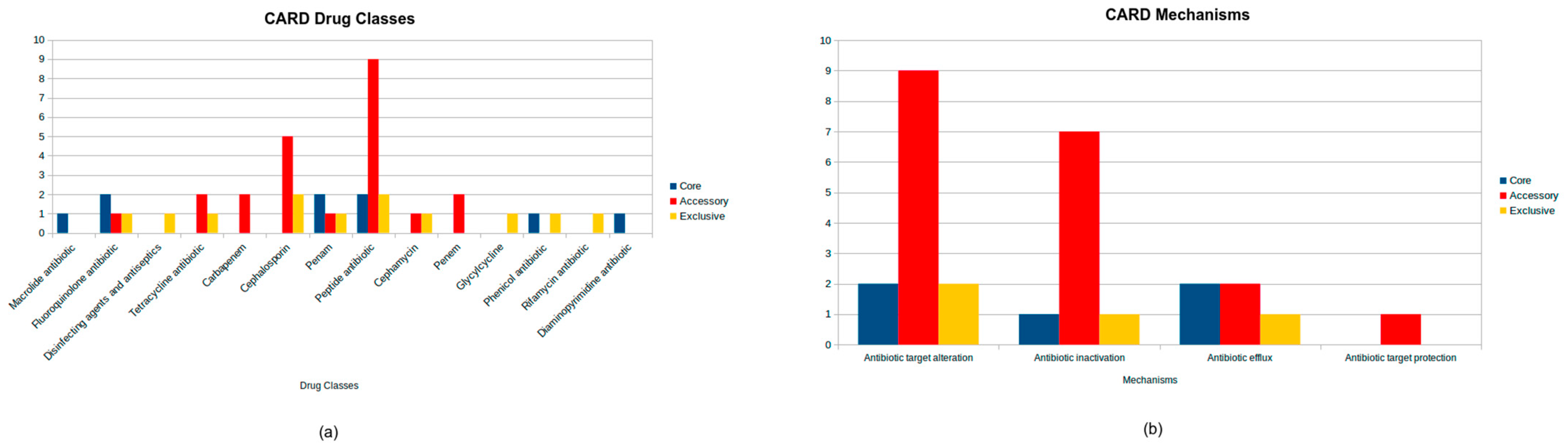
The CARD analysis revealed a total of 28 genes. Five genes were found on the core genome (bacA, OXA-12, MCR-7.1, CRP, and rsmA). Genes bacA and MCR-7.1 are involved in antibiotic target alteration and confer resistance to peptide antibiotics, as well as the OXA-12 gene responsible for antibiotic inactivation and providing resistance to penam antibiotics. Additionally, the strains with the CRP and rsmA genes are associated with antibiotic efflux and mediate resistance to a broader range of antibiotics, including penams, fluoroquinolones, macrolides, phenicols, and diaminopyrimidines.
The accessory genome contained 19 antibiotic resistance genes with diverse functions. This included 9 genes involved in antibiotic target alteration, such as the MCR-3 family (MCR-3.1, MCR-3.28, MCR-3.37, MCR-3.9, MCR-3.2, MCR-3.6, MCR-3.36, and MCR-3.33) and arnA conferring resistance to peptide antibiotics, as well as 7 genes responsible for antibiotic inactivation, including cphA3 and cphA7 targeting carbapenems, AQU-1, AQU-2, and AQU-3 active against cephalosporins, FOX-2 effective against cephamycins and cephalosporins, and TRU-1 providing resistance to penams and cephalosporins. Additionally, 2 genes associated with efflux mechanisms, tet (E) and tet (A), were identified, providing resistance to tetracycline antibiotics. The accessory genome also had 1 gene, QnrS6, linked to protection of the antibiotic target and conferring resistance to fluoroquinolones.
In addition, 4 exclusive genes were identified: FOX-15, a gene for antibiotic inactivation effective against cephamycins and cephalosporins; ugd, a gene for antibiotic target alteration conferring resistance to peptide antibiotics; acrB, a gene for antibiotic efflux providing resistance to tetracyclines, disinfecting agents and antiseptics, phenicols, rifamycins, penams, glycylcyclines, cephalosporins, and fluoroquinolones; and PmrF, a gene for antibiotic target alteration also associated with peptide antibiotic resistance.
The drug classes and mechanisms related to all strains can be seen in Figure 3.
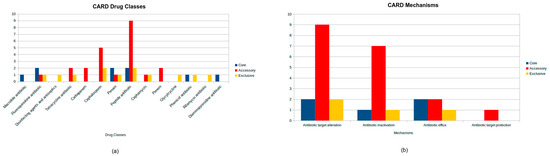
Figure 3.
CARD database antimicrobial resistance analysis in Aeromonas jandaei, showing the distribution of drug classes (a) and antimicrobial resistance mechanisms (b), highlighting the number of genes belonging to core, accessory, and exclusive genomes.
The PanViTa analysis (Supplementary Figure S3) shows random distribution of accessory AMR genes among the strains, with no shared resistance patterns. Most resistance genes were located in the core genome, while the few accessory AMR genes displayed strain-specific occurrences. When looking exclusively into the four Brazilian strains from this study, the gene distribution can be seen in Table 4.

Table 4.
Genes found on Aeromonas jandaei GT15, GTBM29, Aer_On4M and Aer_On5M strains through CARD analysis.
3.4.2. Heavy Metal Resistance Results
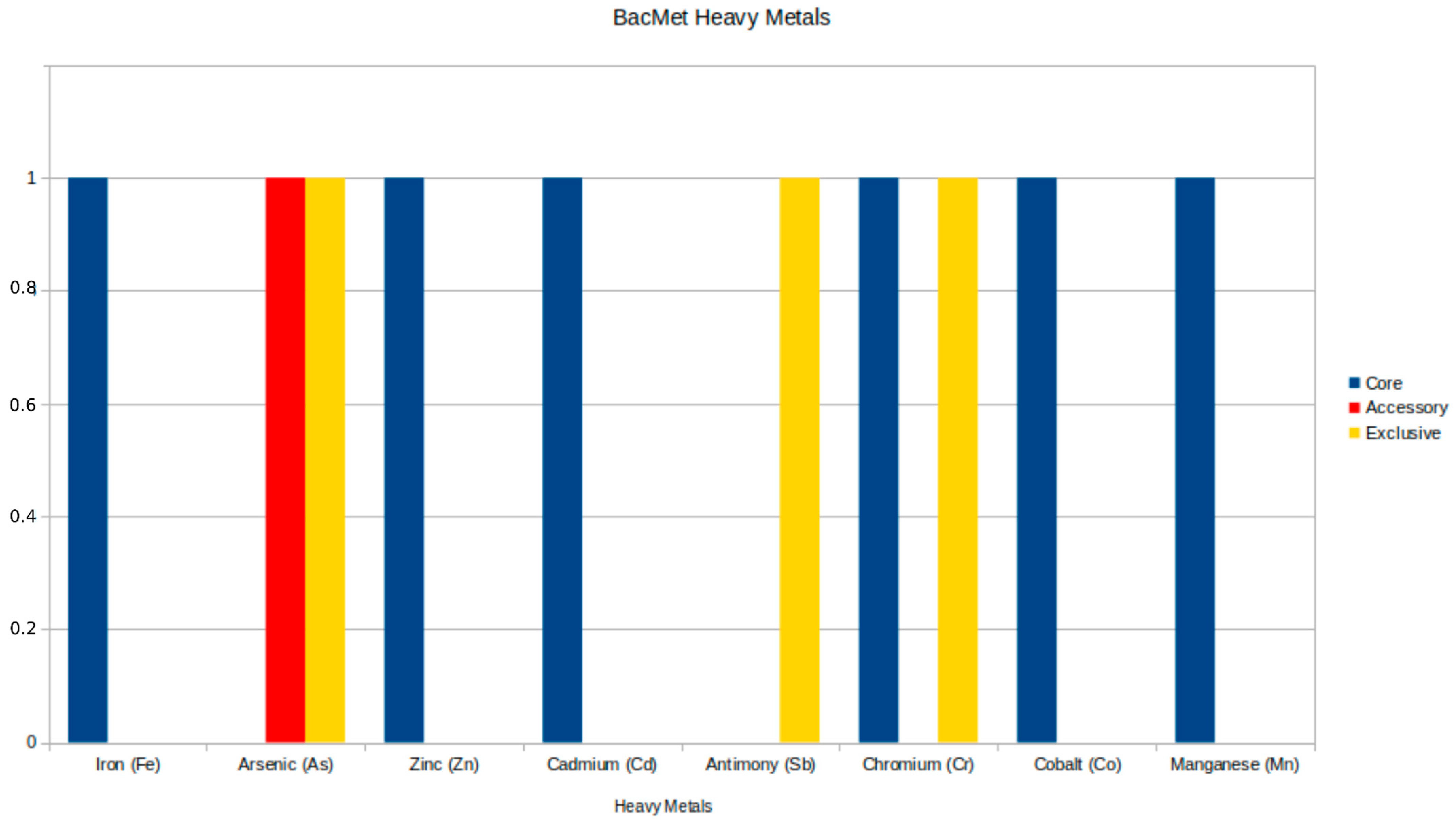
The analysis of resistance to heavy metals revealed five significant genes: two in the core genome and one in the accessory genome, as detailed in Table 5, along with two exclusive genes (arsA and recG) found in other genomes beyond the Brazilian samples. The core genome genes include the ruvB gene, which encodes an ATP-dependent DNA helicase from the Holliday junction and is associated with resistance to chromium (Cr), and the divalent cation transporter mntH/yfeP, which shows resistance to multiple metals, including manganese (Mn), iron (Fe), cadmium (Cd), cobalt (Co), and zinc (Zn). The arsC gene, an accessory gene, encodes the ArsC protein and is specifically related to resistance to arsenic (As). The distribution of heavy metal resistance genes is illustrated in Figure 4. Supplementary Figure S4 illustrates the distribution of genes across all strains, revealing that the majority of lineages, including the strains from this work, do not have accessory genes.

Table 5.
Three genes represent all heavy metal resistance genes identified in the four Brazilian genomes analyzed in this study.
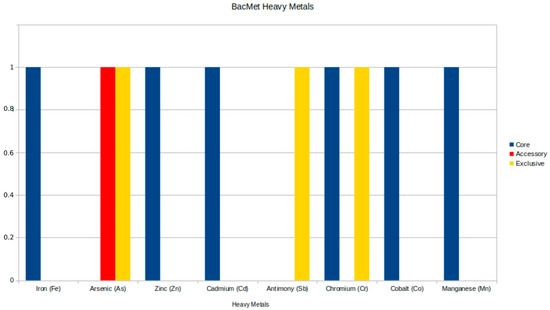
Figure 4.
Genomic distribution of heavy metal resistance genes from the BacMet database in Aeromonas jandaei, illustrating the prevalence and diversity of resistance mechanisms across different genome categories.
3.5. Virulence Genes
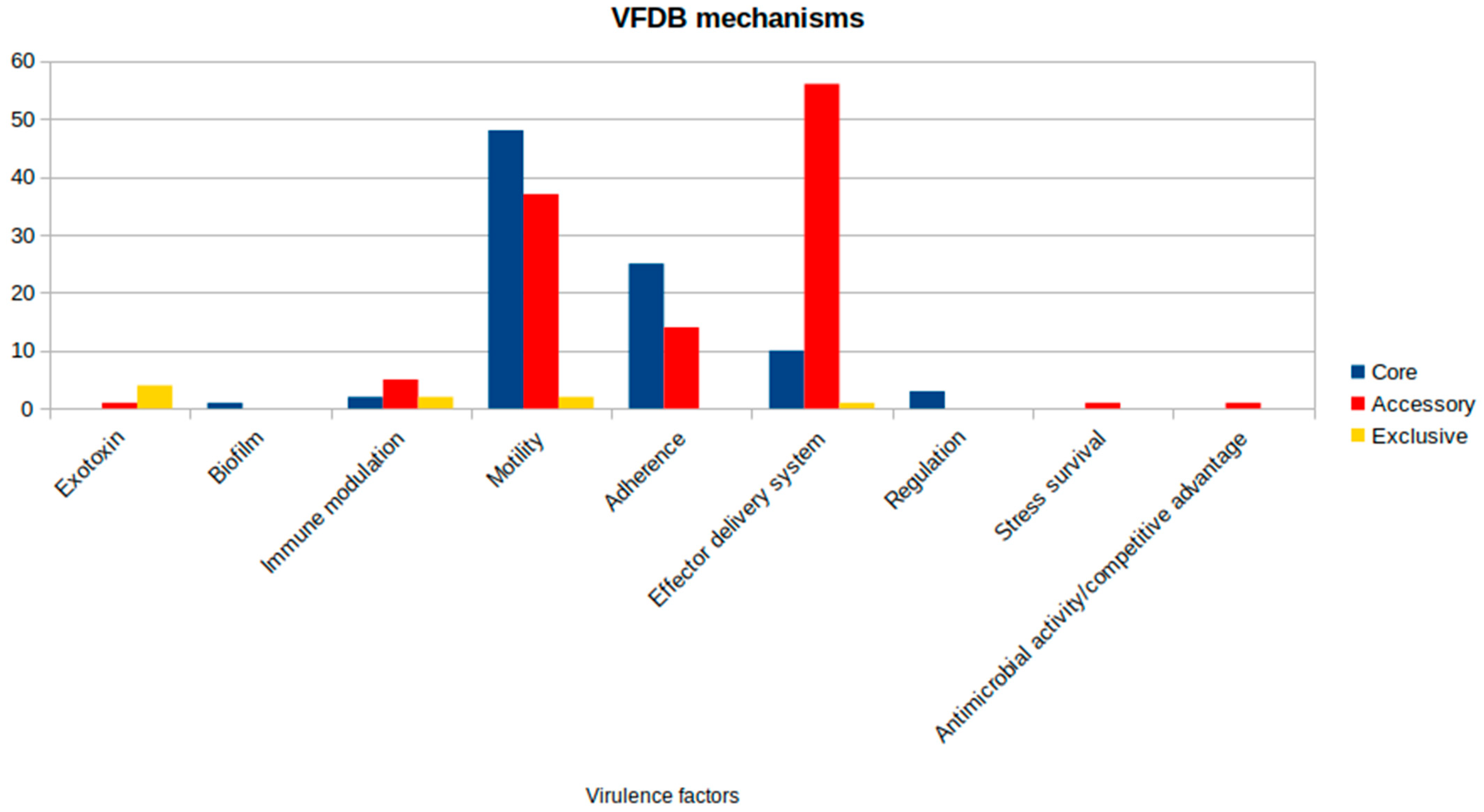
The virulence factors analysis revealed an amount of 213 genes present within the genetic profile of the subjected strains. Amongst this set, 89 genes are shared by all analyzed strains. Most of the core gene sets are related to motility (48) and adhesion (25) mechanisms. A less obvious portion is linked to the well-described effector delivery system (10). However, when looking at genes related to the accessory compartment, a greater prevalence of this class of virulence factors is highlighted, as shown in Figure 5. Considering this result, there are some attention-grabbing outcomes that could lead to a new hypothesis regarding the pathogenicity of the species. All identified genes and their associated virulence mechanisms are detailed in Supplementary Table S2. Their distribution across strains (Supplementary Figure S5) reveals a consistent pattern distinguishing the On4M and On5M isolate groups, with shared virulence profiles within each cluster that align with their phylogenetic relationships.
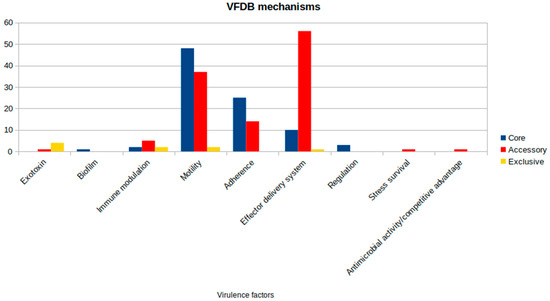
Figure 5.
Virulence factors distribution by class from the Virulence Factor Database (VFDB) in Aeromonas jandaei.
Regarding the highlighted Brazilian strains, a set composed of a few genes is intriguing. Five genes (rfbF, tppE, tppB, tapY1, and tppA) are shared by two out of four genomes that were isolated from the same geographical localization. In this context, four genes (tppE, tppB, tapY1, and tppA) are linked to adhesion processes through pilus formation, and one gene is related to immunomodulation (rfbF). On the other hand, the vgrG3 gene is present in all subjects and is more prevalent in Brazilian strains, considering isolates other than that directly related to this study.
4. Discussion
4.1. Resistome Data
It is essential to start pointing to the core genes that were found, as seen in Figure 3. There are three different classes showing a major presence in the core genomes: the fluoroquinolones, penams, and peptide antibiotics. The fluoroquinolones are interesting because of the fact that a previous study identified this class as very relevant in in vitro essays, exhibiting its high susceptibility when used against Aeromonas spp. isolated from fish [33]. However, a previous study [34] says otherwise: when a variety of Aeromonas were submitted to an antimicrobial test, they expressed a higher resistance than the previous study (>60%) to nalidixic acid, a fluoroquinolone. Changing to the genotypic characteristics, a work has done the resistome identification of five species of the Aeromonas genus, showing that the resistance genes for fluoroquinolones were very equally distributed among all of the five species [35]. In this way, it is possible to understand that it is not very surprising that these genes for fluoroquinolones are in the core genomes of the Aeromonas jandaei of this study, but it is still worrying for the fact that these kinds of antimicrobials are more recent and already have so many resistance genes distributed among the Aeromonas species that should be more rare [36].
Another concerning aspect regarding fluoroquinolones in Brazil is their current unregulated status in aquaculture. Fluoroquinolones are already approved for aquaculture in many other major producing countries, including the European Union, China, Japan, and Thailand. While some fluoroquinolones are regulated in Brazil for use in other livestock species, their use in aquaculture remains unregulated [37]. This regulatory gap is particularly worrying given that residual enrofloxacin has been detected in O. niloticus (tilapia) fillets [38], which indicates its illegal use [39,40] since only oxytetracycline [41] and florfenicol [42] are currently authorized. Furthermore, the bacterial isolates from Brazilian aquaculture in this study have demonstrated a possible resistance to fluoroquinolones, even though these antimicrobials are not officially approved for aquaculture use.
Resistance to fluoroquinolones is often linked to specific mutations in the gyrA and parC genes. These mutations have been well-documented in many bacterial species and are known to play a major role in fluoroquinolone resistance [43,44]. To check if our four A. jandaei isolates carried similar mutations, we analyzed the gyrA and parC genes using MEGAres, a database designed to detect antimicrobial resistance genes [45]. Our analysis showed no signs of the typical fluoroquinolone-resistance mutations found in other bacteria. This finding matches our earlier observation that fluoroquinolone resistance genes were missing from the accessory genomes of these isolates. The lack of these mutations suggests that changes in gyrA and parC are unlikely to be responsible for fluoroquinolone resistance in our A. jandaei isolates. Further research is needed to explore other potential mechanisms behind this resistance.
For penam resistance, a Brazilian study has used nine Aeromonas veronii from animal (fish), human, and environmental sources and some NCBI public genomes for the identification of resistance genes from different classes of antimicrobials. As a result, they have found that the resistance genes from penam were not so abundant; the majority of the isolates and the NCBI ones had only two genes linked to the penam, except one from NCBI that had 15 [46]. Other genotypic analyses, the previous work [47] that used 26 isolates of various species, 13 of them Aeromonas, that were isolated from different fishes and water sources, exhibit in PCR that genes for penam resistance were the second most present among the isolates, that gene being a representative from the bla family. Furthermore, a study on A.veronii isolated from a fish in comparison with other complete genomes from the Aeromonas genus has shown that the gene OXA-12, a gene from penam resistance, was one of the most present in the results, which is quite normal in Aeromonas spp. and has also been present in A. jandaei [48,49].
The last major core resistance class was peptide antibiotics, which in this case are defined for the genes from the MCR family, MCR-7.1 and MCR-3.1. Both of them are linked to the resistance to colistin; they are also found in the most diverse types of bacterial species and can originate or serve as a reservoir of these types of resistance in the environment or other ambients they are in [50].
4.2. Heavy Metal Resistance
Heavy metal resistance in Aeromonas has been documented in the literature, revealing variations among different studies. In our research, we identified resistance to several heavy metals, including chromium (Cr), arsenic (As), manganese (Mn), iron (Fe), cadmium (Cd), cobalt (Co), and zinc (Zn), alongside the identification of three resistance genes: ruvB, arsC, and mntH/yfeP.
Comparing these findings with those from other studies, it can be observed that cadmium often exhibits high resistance rates, ranging from 9.1% to 61% in isolates [51,52], while resistance to chromium is also significant, with up to 95.3% of isolates showing resistance [51]. Although zinc showed high resistance in some isolates (67.3% to 100%) [52,53], our data complement this information by indicating resistance to other metals, such as arsenic, which were not highlighted in previous studies. In addition, the genes identified in our study, such as arsC and mntH/yfeP, expand the understanding of the molecular mechanisms underlying heavy metal resistance, which in the literature also includes genes such as copA and czcA [52]. The differences in resistance levels and the presence of resistance genes reflect the complexity of the adaptation of Aeromonas to polluted habitats and suggest that further research is needed to understand the relationship between heavy metal resistance and environmental contamination [54].
4.3. Virulence Factors
The genetic profile analyzed offers a perspective on the virulence factors of Aeromonas jandaei. Focusing on specific factors, there is a significant relevance of elements related to motility and adhesion, such as the genes fimA, fimC, fimD, fimE, and fimF, which are associated with pili formation and host adhesion [55]. These genes are essential for free-living pathogens such as Aeromonas species [56,57], since host adhesion is crucial for the start of tissue colonization and enables bacterial cells to be protected from physical and chemical challenges through the formation of biofilm structures [57,58].
The isolates analyzed also exhibited a gene complex composed of flg and flr genes, which are linked to flagellar biosynthesis [59,60]. The importance of flagella for motility in the environment is mainly relevant for free-living pathogens, allowing them to migrate to a more suitable location and enhance surface contact, making this adaptation a significant evolutionary acquisition [59].
Another critical point is the presence of elements related to secretion systems. Genes associated with type II (T2SS) and type III (T3SS) secretion systems are fundamental to bacterial pathogenicity [61,62]. These systems allow the bacteria to release enzymes and toxins into the extracellular environment or directly into the cytoplasm of the host cells, promoting invasion and causing significant tissue damage [63].
An interesting finding is the higher prevalence of the vgrG3 gene in Brazilian isolates. It is a part of the type VI secretion system (T6SS), and this system is known for its aggressive capability, acting as a molecular harpoon that injects toxins and effectors directly into the cytoplasm of the target cells [64,65]. Beyond its role in pathogenicity, the T6SS is an important factor in bacterial competition [66], which in theory would enable Aeromonas jandaei to eliminate other competing bacteria, particularly in aquatic environments.
The presence of these genomic mechanisms suggests an increased competence of Aeromonas jandaei strains to colonize and persist in different environments and conditions, particularly in host interactions.
5. Conclusions
Genome analysis of Aeromonas jandaei isolates from Brazilian aquaculture environments has offered significant insights into the antimicrobial resistance potential, heavy metal resistance, and virulence mechanisms. The investigation of Aeromonas jandaei from Brazilian aquaculture environments has revealed the presence of genes related to multiple resistance mechanisms and virulence factors, which may pose a risk to both aquaculture production and public health. The identification of these genetic elements through bioinformatics approaches provides crucial information for the development of more effective disease prevention and control strategies in aquaculture settings.
The findings underscore the necessity for the establishment of comprehensive genomic surveillance programs in Brazilian aquaculture. The regular monitoring of bacterial populations through genomic tools can facilitate the early detection of emerging resistance patterns and potential pathogens, thereby enabling the implementation of effective control measures before they become significant problems. This approach is of particular importance given the economic significance of aquaculture in Brazil and the potential impact of bacterial diseases on production.
To mitigate these risks, it is essential to strengthen sanitary regulations and preventive measures. This includes implementing improved biosecurity protocols, enhancing water quality management, and developing more effective vaccination strategies. Furthermore, the establishment of clear guidelines for antimicrobial use in aquaculture, in conjunction with regular monitoring of bacterial populations, could help prevent the emergence and spread of resistant strains.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13051094/s1, Figure S1: Geographic location of bacterial isolation sites in the São Francisco River region; Figure S2: Presence/absence matrix of accessory genes among Aeromonas jandaei isolates, alongside a phylogenetic tree; Figure S3: Pangenome analysis revealing gene distribution and identity across bacterial strains; Figure S4: Clustering of heavy metal resistance genes across Aeromonas strains; Figure S5: Clustermap of virulence gene presence and identity across Aeromonas jandaei strains. Supplementary Table S1: Pairwise genomic similarity analysis of Aeromonas jandaei strains. The two Sobradinho isolates (Aer_On4M and Aer_On5M) show high genetic proximity (>99% ANI) yet distinct genomic features that preclude clonal classification, suggesting closely related but independently evolving strains. Values represent average nucleotide identity (ANI) percentages. Supplementary Table S2: Virulence-associated genes and their functional mechanisms.
Author Contributions
Conceptualization, M.M.d.C. and F.F.A.; methodology, M.L.O.D., D.L.N.R., G.B.V.d.L., J.C.A., G.V.G., J.J.d.S.G. and F.F.A.; formal analysis, M.L.O.D., D.L.N.R., G.B.V.d.L. and J.C.A.; software, M.L.O.D.; resources, V.A., B.B., M.M.d.C., U.d.P.P. and F.F.A.; writing—original draft preparation, M.L.O.D. and F.F.A.; writing—review and editing, M.L.O.D., G.V.G., J.J.d.S.G., V.A., B.B., E.G., G.C.T., M.M.d.C., U.d.P.P. and F.F.A.; supervision, F.F.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundo FUNDEP (Fundação de Desenvolvimento da Pesquisa), CNPq/MCTI/CT-Saúde Nº 52/2022—Processo: 408898/2022-4, CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior)—Processo: 88887.134782//2025-00, FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais), PROEX-UFMG, CNPq CABBIO—Processo 445840/2023-4 and L’Oréal-UNESCO For Women in Science.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All genomes are available at NCBI—a public database.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO. The State of World Fisheries and Aquaculture 2024; FAO: Rome, Italy, 2024; ISBN 978-92-5-138763-4. [Google Scholar]
- FAO (Ed.) The State of World Fisheries and Aquaculture 2022: Towards Blue Transformation; FAO: Rome, Italy, 2022; ISBN 978-92-5-136364-5. [Google Scholar]
- Tavares-Dias, M.; Martins, M.L. An Overall Estimation of Losses Caused by Diseases in the Brazilian Fish Farms. J. Parasit. Dis. 2017, 41, 913–918. [Google Scholar] [CrossRef]
- Assane, I.M.; de Sousa, E.L.; Valladão, G.M.R.; Tamashiro, G.D.; Criscoulo-Urbinati, E.; Hashimoto, D.T.; Pilarski, F. Phenotypic and Genotypic Characterization of Aeromonas jandaei Involved in Mass Mortalities of Cultured Nile Tilapia, Oreochromis niloticus (L.) in Brazil. Aquaculture 2021, 541, 736848. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- Jagoda, S.; Wijewardana, T.; Arulkanthan, A.; Igarashi, Y.; Tan, E.; Kinoshita, S.; Watabe, S.; Asakawa, S. Characterization and Antimicrobial Susceptibility of Motile Aeromonads Isolated from Freshwater Ornamental Fish Showing Signs of Septicaemia. Dis. Aquat. Organ. 2014, 109, 127–137. [Google Scholar] [CrossRef]
- Proietti-Junior, A.A.; Lima, L.S.; Roges, E.M.; Rodrigues, Y.C.; Lima, K.V.B.; Rodrigues, D.P.; Tavares-Dias, M. Experimental Co-infection by Aeromonas hydrophila and Aeromonas jandaei in Pirarucu Arapaima gigas (Pisces: Arapaimidae). Aquac. Res. 2021, 52, 1688–1696. [Google Scholar] [CrossRef]
- Medina-Morillo, M.; Sotil, G.; Arteaga, C.; Cordero, G.; Martins, M.L.; Murrieta-Morey, G.; Yunis-Aguinaga, J. Pathogenic Aeromonas spp. in Amazonian Fish: Virulence Genes and Susceptibility in Piaractus Brachypomus, the Main Native Aquaculture Species in Peru. Aquac. Rep. 2023, 33, 101811. [Google Scholar] [CrossRef]
- Pellin, G.P.; Martins, R.A.; Queiroz, C.A.D.; Sousa, T.F.; Muniz, A.W.; Silva, G.F.D.; Majolo, C. Aeromonas from Farmed Tambaqui from North Brazil: Molecular Identification and Pathogenic Potential. Ciênc. Rural 2023, 53, e20220151. [Google Scholar] [CrossRef]
- Mulia, D.S.; Dwi, N.R.; Suwarsito, S.; Muslimin, B. Molecular Characterization of Pathogenic Aeromonas Jandaei Bacteria Isolated from Cultured Walking Catfish (Clarias sp.). Biodivers. J. Biol. Divers. 2024, 25, 3. [Google Scholar] [CrossRef]
- Laufer, A.S.; Siddall, M.E.; Graf, J. Characterization of the Digestive-Tract Microbiota of Hirudo Orientalis, a European Medicinal Leech. Appl. Environ. Microbiol. 2008, 74, 6151–6154. [Google Scholar] [CrossRef] [PubMed]
- Pu, W.; Guo, G.; Yang, N.; Li, Q.; Yin, F.; Wang, P.; Zheng, J.; Zeng, J. Three Species of Aeromonas (A. dhakensis, A. hydrophila and A. jandaei) Isolated from Freshwater Crocodiles (Crocodylus siamensis) with Pneumonia and Septicemia. Lett. Appl. Microbiol. 2019, 68, 212–218. [Google Scholar] [CrossRef]
- Chen, M.; Xue, M.; Chen, J.; Xiao, Z.; Hu, X.; Zhang, C.; Jiang, N.; Fan, Y.; Meng, Y.; Zhou, Y. Isolation, Identification and Characterization of Aeromonas jandaei from Diseased Chinese Soft-Shell Turtles. J. Fish Dis. 2024, 47, e13919. [Google Scholar] [CrossRef]
- Pereira, C.; Duarte, J.; Costa, P.; Braz, M.; Almeida, A. Bacteriophages in the Control of Aeromonas sp. in Aquaculture Systems: An Integrative View. Antibiotics 2022, 11, 163. [Google Scholar] [CrossRef]
- Gazal, L.E.d.S.; Brito, K.C.T.d.; Kobayashi, R.K.T.; Nakazato, G.; Cavalli, L.S.; Otutumi, L.K.; Brito, B.G.d. Antimicrobials and Resistant Bacteria in Global Fish Farming and the Possible Risk for Public Health. Arq. Inst. Biológico 2020, 87, e0362019. [Google Scholar] [CrossRef]
- Gouveia, J.J.d.S.; Regitano, L.C.d.A. Extração de DNA; Embrapa Pecuária Sudeste: São Carlos, Brazil, 2007; pp. 3–8. [Google Scholar]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 September 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequences (RefSeq): A Curated Non-Redundant Sequence Database of Genomes, Transcripts and Proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Rodrigues, D.L.N.; Ariute, J.C.; Rodrigues da Costa, F.M.; Benko-Iseppon, A.M.; Barh, D.; Azevedo, V.; Aburjaile, F. PanViTa: Pan Virulence and resisTance Analysis. Front. Bioinform. 2023, 3, 1070406. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G.J. BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef]
- Esteve, C.; Alcaide, E.; Giménez, M.J. Multidrug-Resistant (MDR) Aeromonas Recovered from the Metropolitan Area of Valencia (Spain): Diseases Spectrum and Prevalence in the Environment. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 137–145. [Google Scholar] [CrossRef]
- Dhanapala, P.M.; Kalupahana, R.S.; Kalupahana, A.W.; Wijesekera, D.P.H.; Kottawatta, S.A.; Jayasekera, N.K.; Silva-Fletcher, A.; Jagoda, S.S.S.d.S. Characterization and Antimicrobial Resistance of Environmental and Clinical Aeromonas Species Isolated from Fresh Water Ornamental Fish and Associated Farming Environment in Sri Lanka. Microorganisms 2021, 9, 2106. [Google Scholar] [CrossRef]
- Roh, H.; Kannimuthu, D. Comparative Resistome Analysis of Aeromonas Species in Aquaculture Reveals Antibiotic Resistance Patterns and Phylogeographic Distribution. Environ. Res. 2023, 239, 117273. [Google Scholar] [CrossRef]
- Davies, J. Microbes Have the Last Word: A Drastic Re-evaluation of Antimicrobial Treatment Is Needed to Overcome the Threat of Antibiotic-resistant Bacteria. EMBO Rep. 2007, 8, 616–621. [Google Scholar] [CrossRef]
- Quesada, S.P.; Paschoal, J.A.R.; Reyes, F.G.R. Considerations on the Aquaculture Development and on the Use of Veterinary Drugs: Special Issue for Fluoroquinolones—A Review. J. Food Sci. 2013, 78, R1321–R1333. [Google Scholar] [CrossRef]
- Guidi, L.R.; Santos, F.A.; Ribeiro, A.C.S.R.; Fernandes, C.; Silva, L.H.M.; Gloria, M.B.A. Quinolones and Tetracyclines in Aquaculture Fish by a Simple and Rapid LC-MS/MS Method. Food Chem. 2018, 245, 1232–1238. [Google Scholar] [CrossRef]
- Rosário, A.E.C.D.; Barbanti, A.C.C.; Matos, H.C.; Maia, C.R.M.D.S.; Trindade, J.M.; Nogueira, L.F.F.; Pilarski, F.; Gallani, S.U.; Leal, C.A.G.; Figueiredo, H.C.P.; et al. Antimicrobial Resistance in Lactococcus spp. Isolated from Native Brazilian Fish Species: A Growing Challenge for Aquaculture 2024. Microorganisms 2024, 12, 2327. [Google Scholar] [CrossRef]
- Lulijwa, R.; Rupia, E.J.; Alfaro, A.C. Antibiotic Use in Aquaculture, Policies and Regulation, Health and Environmental Risks: A Review of the Top 15 Major Producers. Rev. Aquac. 2020, 12, 640–663. [Google Scholar] [CrossRef]
- Orlando, E.A.; Costa Roque, A.G.; Losekann, M.E.; Colnaghi Simionato, A.V. UPLC–MS/MS Determination of Florfenicol and Florfenicol Amine Antimicrobial Residues in Tilapia Muscle. J. Chromatogr. B 2016, 1035, 8–15. [Google Scholar] [CrossRef]
- Monteiro, S.H.; Francisco, J.G.; Campion, T.F.; Pimpinato, R.F.; Moura Andrade, G.C.R.; Garcia, F.; Tornisielo, V.L. Multiresidue Antimicrobial Determination in Nile Tilapia (Oreochromis niloticus) Cage Farming by Liquid Chromatography Tandem Mass Spectrometry. Aquaculture 2015, 447, 37–43. [Google Scholar] [CrossRef]
- Hooper, D.C. Emerging Mechanisms of Fluoroquinolone Resistance. Emerg. Infect. Dis. 2001, 7, 337–341. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of Quinolone Action and Resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Lakin, S.M.; Dean, C.; Noyes, N.R.; Dettenwanger, A.; Ross, A.S.; Doster, E.; Rovira, P.; Abdo, Z.; Jones, K.L.; Ruiz, J.; et al. MEGARes: An Antimicrobial Resistance Database for High Throughput Sequencing. Nucleic Acids Res. 2017, 45, D574–D580. [Google Scholar] [CrossRef]
- Maia, J.C.d.S.; Silva, G.A.d.A.; Cunha, L.S.d.B.; Gouveia, G.V.; Góes-Neto, A.; Brenig, B.; Araújo, F.A.; Aburjaile, F.; Ramos, R.T.J.; Soares, S.C.; et al. Genomic Characterization of Aeromonas Veronii Provides Insights into Taxonomic Assignment and Reveals Widespread Virulence and Resistance Genes throughout the World. Antibiotics 2023, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- Bergum, J.A. Profiling Antimicrobial Resistance Genes in Bacteria Species Isolated from Fish Foods Sold in Retail Outlets. Master’s Thesis, Nord University, Bodø, Norway, 2024. [Google Scholar]
- Das, S.; Sreejith, S.; Babu, J.; Francis, C.; Midhun, J.S.; Aswani, R.; Sebastain, K.S.; Radhakrishnan, E.K.; Mathew, J. Genome Sequencing and Annotation of Multi-Virulent Aeromonas veronii XhG1.2 Isolated from Diseased Xiphophorus hellerii. Genomics 2021, 113, 991–998. [Google Scholar] [CrossRef]
- Alksne, L.E.; Rasmussen, B.A. Expression of the AsbA1, OXA-12, and AsbM1 Beta-Lactamases in Aeromonas Jandaei AER 14 Is Coordinated by a Two-Component Regulon. J. Bacteriol. 1997, 179, 2006–2013. [Google Scholar] [CrossRef]
- Liu, D.; Song, H.; Ke, Y.; Xia, J.; Shen, Y.; Ou, Y.; Hao, Y.; He, J.; Li, X.; Zhou, Y.; et al. Co-Existence of Two Novel Phosphoethanolamine Transferase Gene Variants in Aeromonas jandaei from Retail Fish. Int. J. Antimicrob. Agents 2020, 55, 105856. [Google Scholar] [CrossRef] [PubMed]
- Akinbowale, O.L.; Peng, H.; Grant, P.; Barton, M.D. Antibiotic and Heavy Metal Resistance in Motile Aeromonads and Pseudomonads from Rainbow Trout (Oncorhynchus mykiss) Farms in Australia. Int. J. Antimicrob. Agents 2007, 30, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Dahanayake, P.S.; Hossain, S.; Wickramanayake, M.V.K.S.; Heo, G.-J. Antibiotic and Heavy Metal Resistance Genes in Aeromonas spp. Isolated from Marketed Manila Clam (Ruditapes philippinarum) in Korea. J. Appl. Microbiol. 2019, 127, 941–952. [Google Scholar] [CrossRef]
- Lee, S.W.; Wendy, W. Antibiotic and Heavy Metal Resistance of Aeromonas hydrophila and Edwardsiella tarda Isolated from Red Hybrid Tilapia (Oreochromis spp.) Coinfected with Motile Aeromonas Septicemia and Edwardsiellosis. Vet. World 2017, 10, 803–807. [Google Scholar] [CrossRef]
- Miranda, C.D.; Castillo, G. Resistance to Antibiotic and Heavy Metals of Motile Aeromonads from Chilean Freshwater. Sci. Total Environ. 1998, 224, 167–176. [Google Scholar] [CrossRef]
- Abdul Raheem Hasan, S.; Sajid Al-Jubori, S.; Abdul Sattar Salman, J. Molecular Analysis of fimA Operon Genes among UPEC Local Isolates in Baghdad City. Arch. Razi Inst. 2021, 76, 829–840. [Google Scholar] [CrossRef]
- Boyd, J.M.; Dacanay, A.; Knickle, L.C.; Touhami, A.; Brown, L.L.; Jericho, M.H.; Johnson, S.C.; Reith, M. Contribution of Type IV Pili to the Virulence of Aeromonas salmonicida subsp. Salmonicida in Atlantic Salmon (Salmo salar L.). Infect. Immun. 2008, 76, 1445–1455. [Google Scholar] [CrossRef]
- Stones, D.H.; Krachler, A.M. Against the Tide: The Role of Bacterial Adhesion in Host Colonization. Biochem. Soc. Trans. 2016, 44, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three Faces of Biofilms: A Microbial Lifestyle, a Nascent Multicellular Organism, and an Incubator for Diversity. npj Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef]
- Canals, R.; Ramirez, S.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S. Polar Flagellum Biogenesis in Aeromonas hydrophila. J. Bacteriol. 2006, 188, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Kazuhiro, K.; Tsutomu, O.; Tatsuki, Y.; Tetsuo, I. Sequence Analysis of the flgA Gene and Its Adjacent Region in Salmonella Typhimurium, and Identification of Another Flagellar Gene, flgN. Gene 1994, 143, 49–54. [Google Scholar] [CrossRef]
- Cambronne, E.D.; Roy, C.R. Recognition and Delivery of Effector Proteins into Eukaryotic Cells by Bacterial Secretion Systems. Traffic 2006, 7, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Kendall, M.M. Extra! Extracellular Effector Delivery into Host Cells via the Type 3 Secretion System. mBio 2017, 8, e00594-17. [Google Scholar] [CrossRef]
- Macion, A.; Wyszyńska, A.; Godlewska, R. Delivery of Toxins and Effectors by Bacterial Membrane Vesicles. Toxins 2021, 13, 845. [Google Scholar] [CrossRef]
- Brooks, T.M.; Unterweger, D.; Bachmann, V.; Kostiuk, B.; Pukatzki, S. Lytic Activity of the Vibrio Cholerae Type VI Secretion Toxin VgrG-3 Is Inhibited by the Antitoxin TsaB. J. Biol. Chem. 2013, 288, 7618–7625. [Google Scholar] [CrossRef]
- Coulthurst, S. The Type VI Secretion System: A Versatile Bacterial Weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef]
- Carruthers, M.D.; Nicholson, P.A.; Tracy, E.N.; Munson, R.S. Acinetobacter Baumannii Utilizes a Type VI Secretion System for Bacterial Competition. PLoS ONE 2013, 8, e59388. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).