Priority Colonization of Endophytic Fungal Strains Drives Litter Decomposition and Saprotroph Assembly via Functional Trait Selection in Karst Oak Forests
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Samples Collection
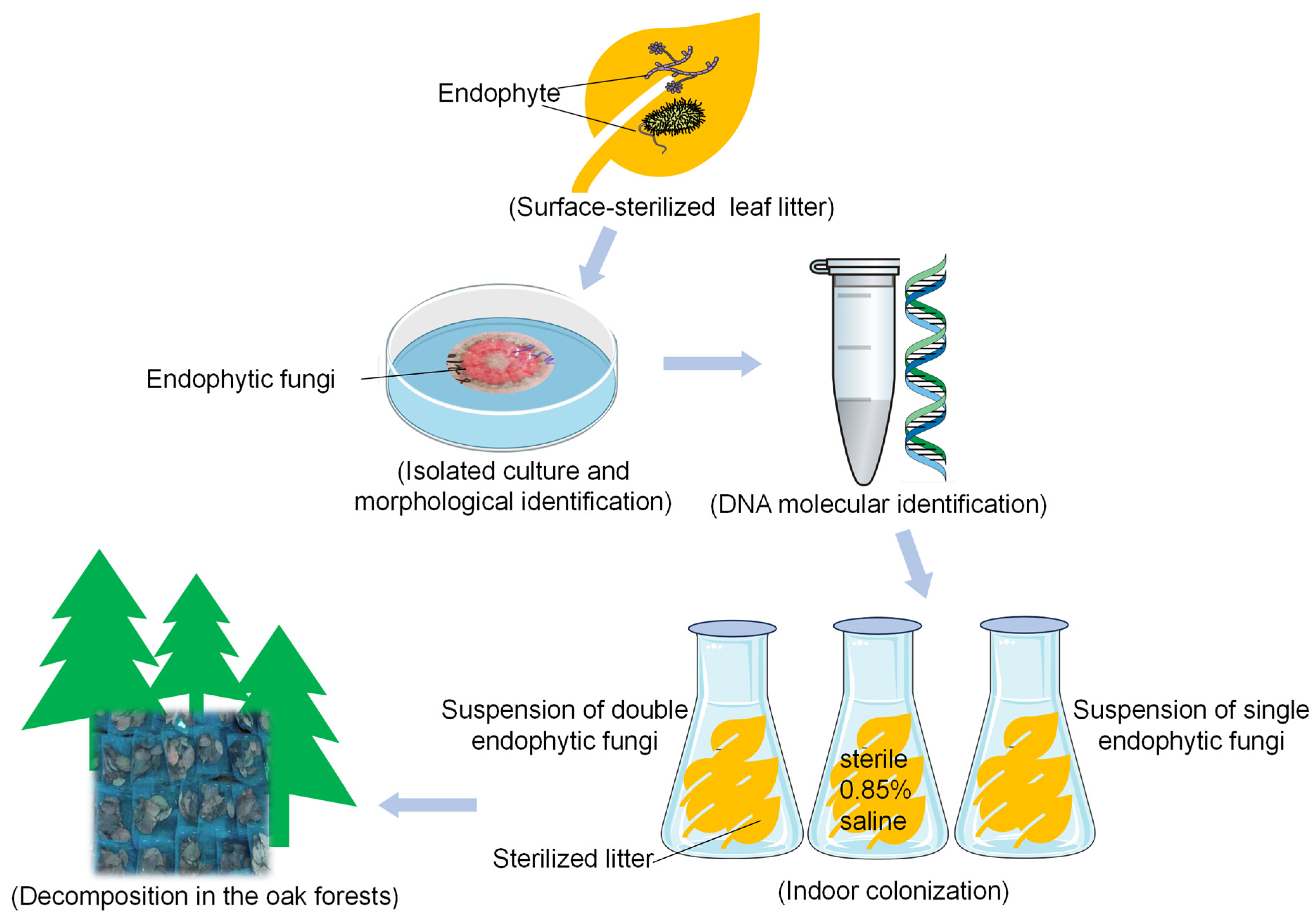
2.2. Isolation of Endophytic Fungi
2.3. Field Experiment Design for Litter Decomposition
2.4. Extracellular Enzyme Activity, Carbon Dioxide Release, and Mass Loss
2.5. DNA Extraction, PCR Amplification, and NovaSeq Sequencing
2.6. Leaf Litter Extracellular Enzyme Stoichiometry (EES)
2.7. Leaf Litter Organic Matter Quality
2.8. Statistical Analysis
2.8.1. Microbial Diversity
2.8.2. Microbial Community Assembly
2.8.3. Network Analysis
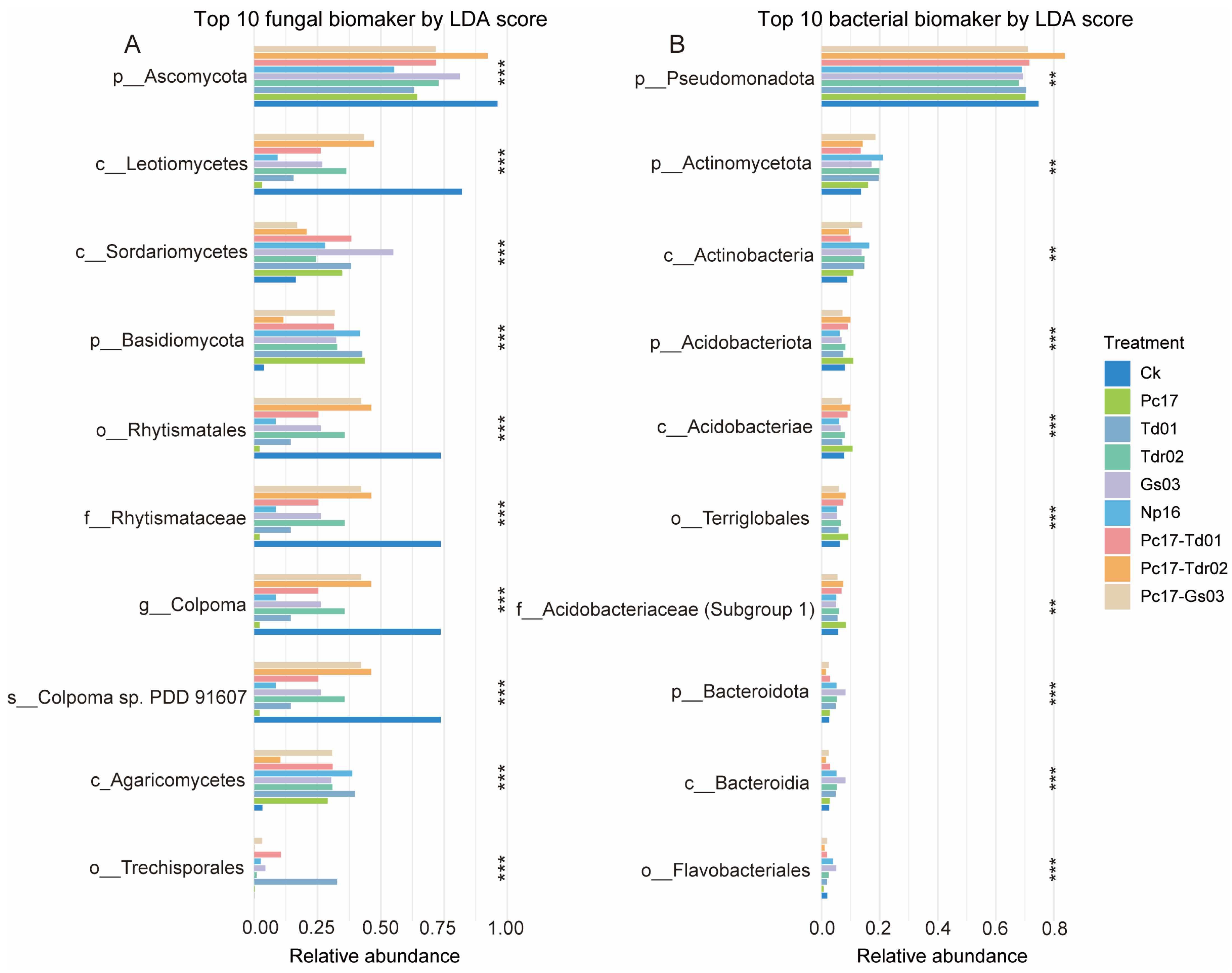
2.8.4. Differential Taxa Identification via LEfSe
2.8.5. Correlation Analyses of Endophytic Fungal Colonization to Microbial Functions
3. Results
3.1. CO2 Release, Mass Loss and Organic Matter Quality
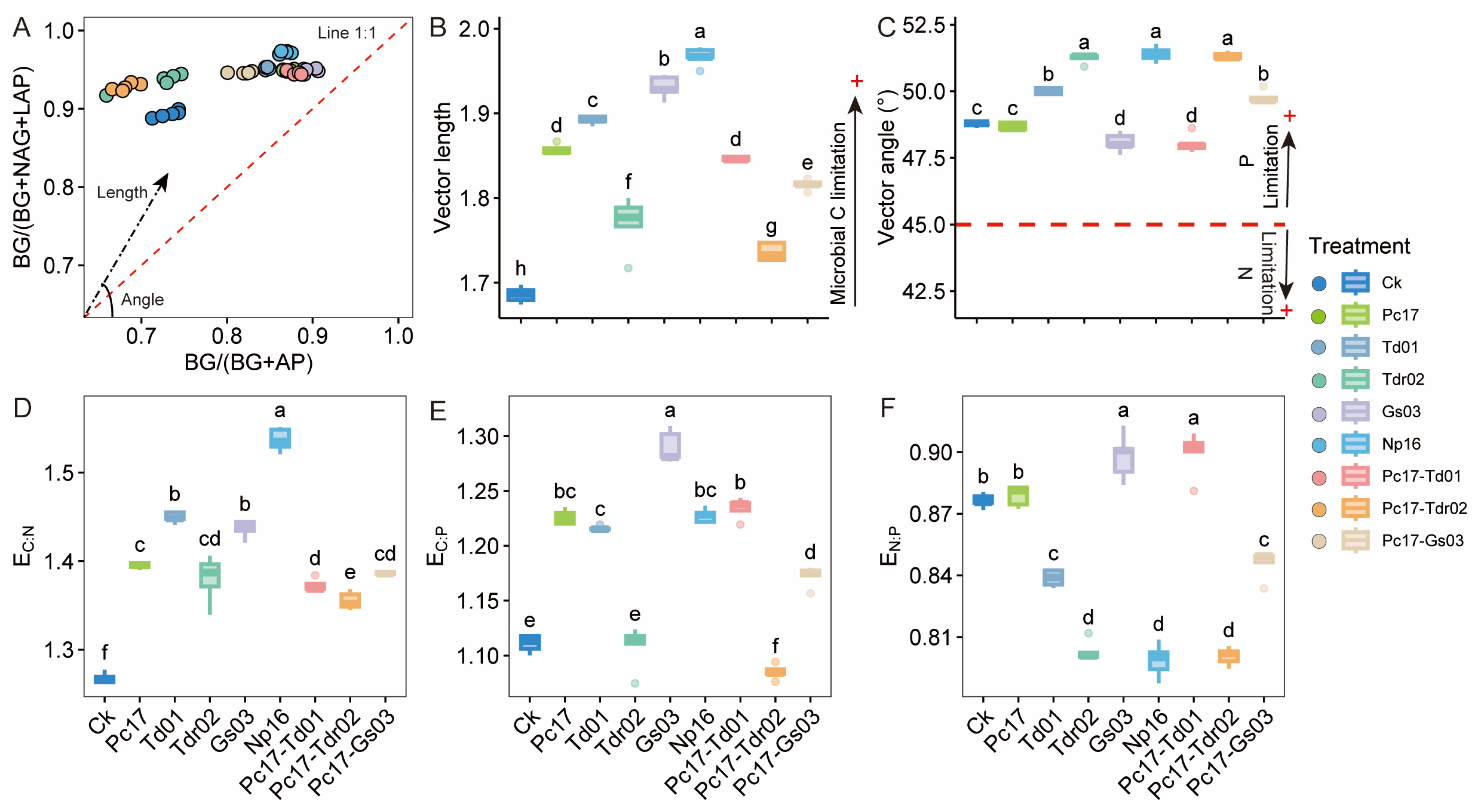
3.2. Extracellular Enzyme Activity and Stoichiometry
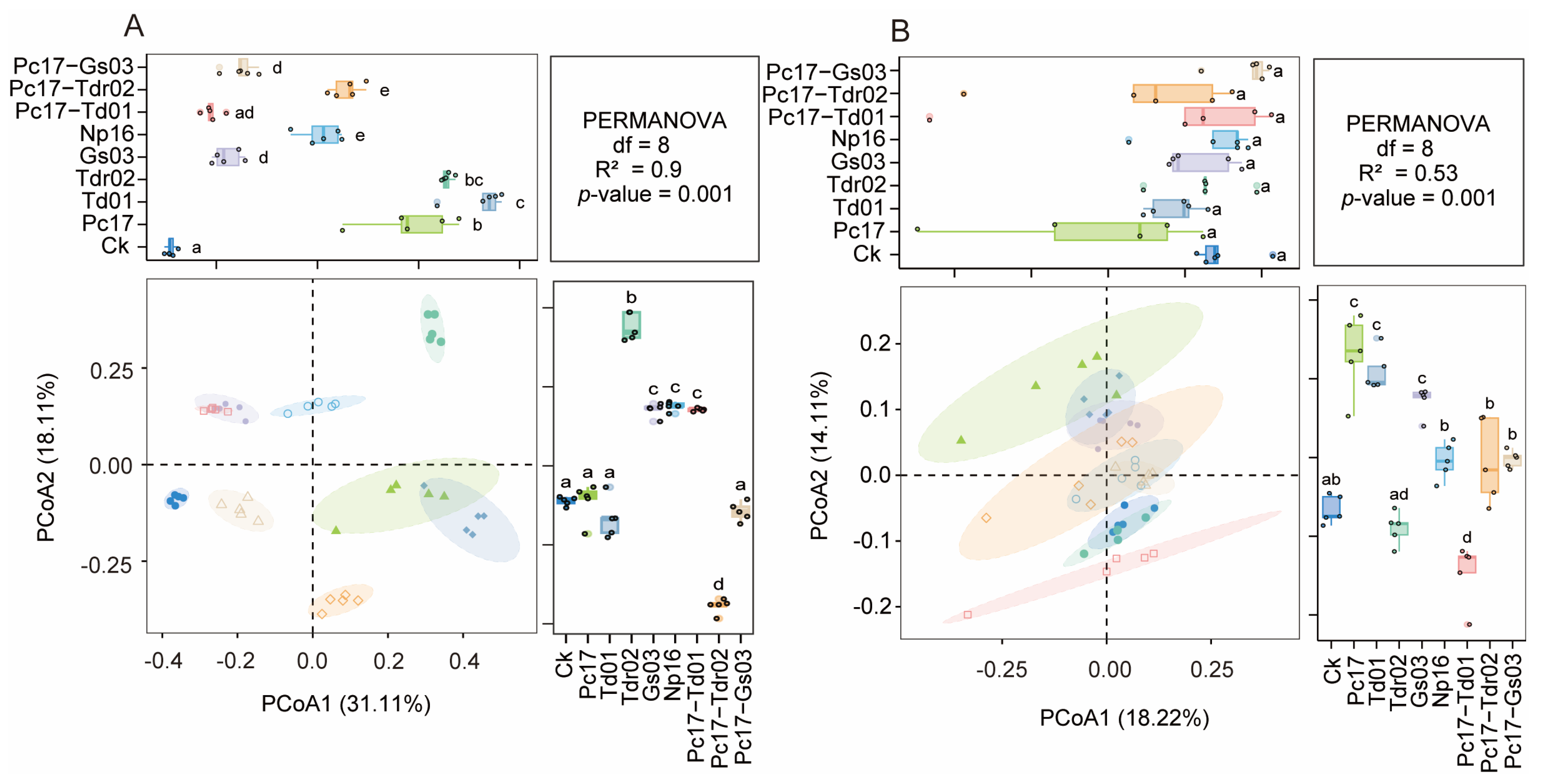
3.3. Microbial Community Structure and Diversity
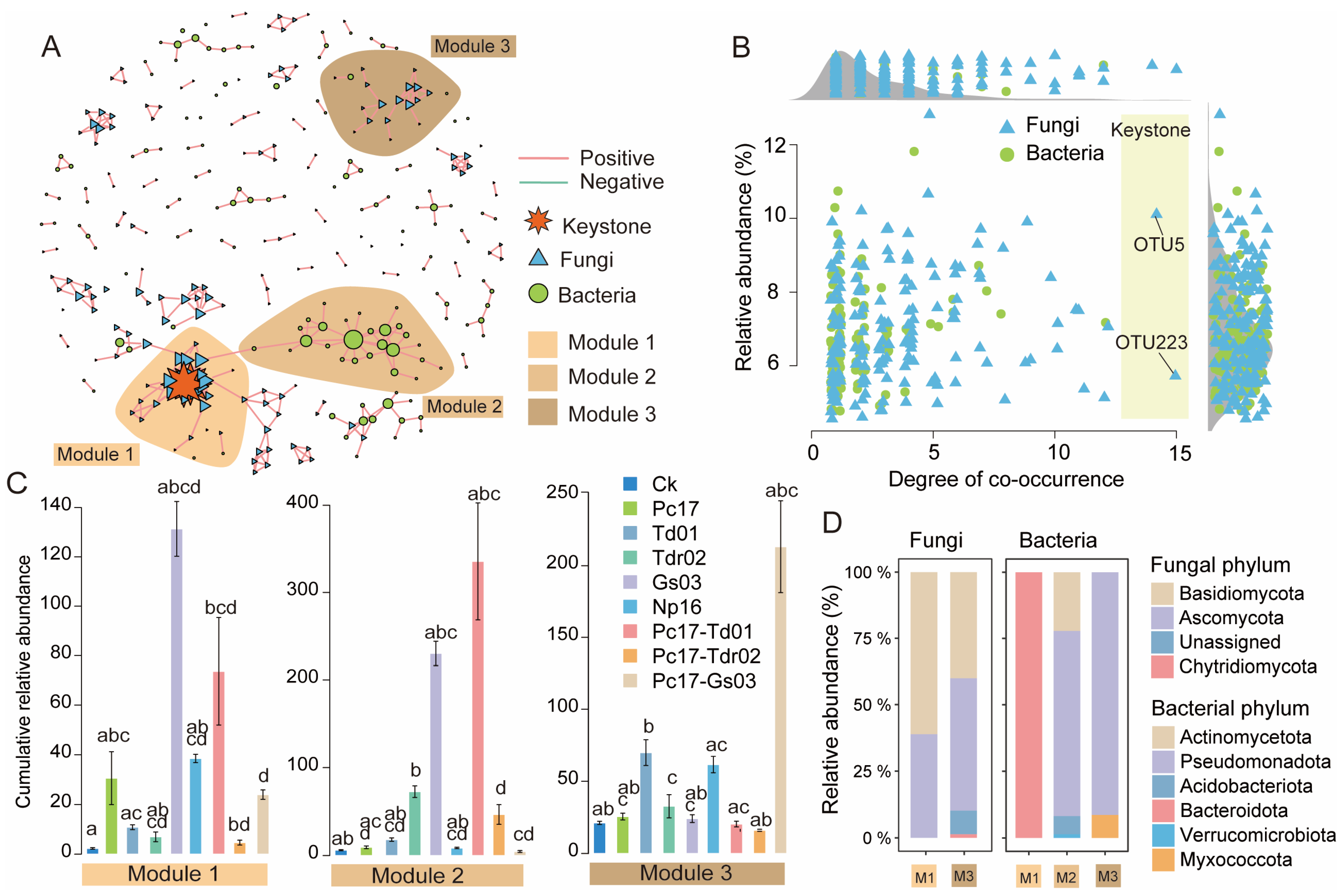
3.4. Microbial Co-Occurrence Network Analysis
3.5. Community Assembly
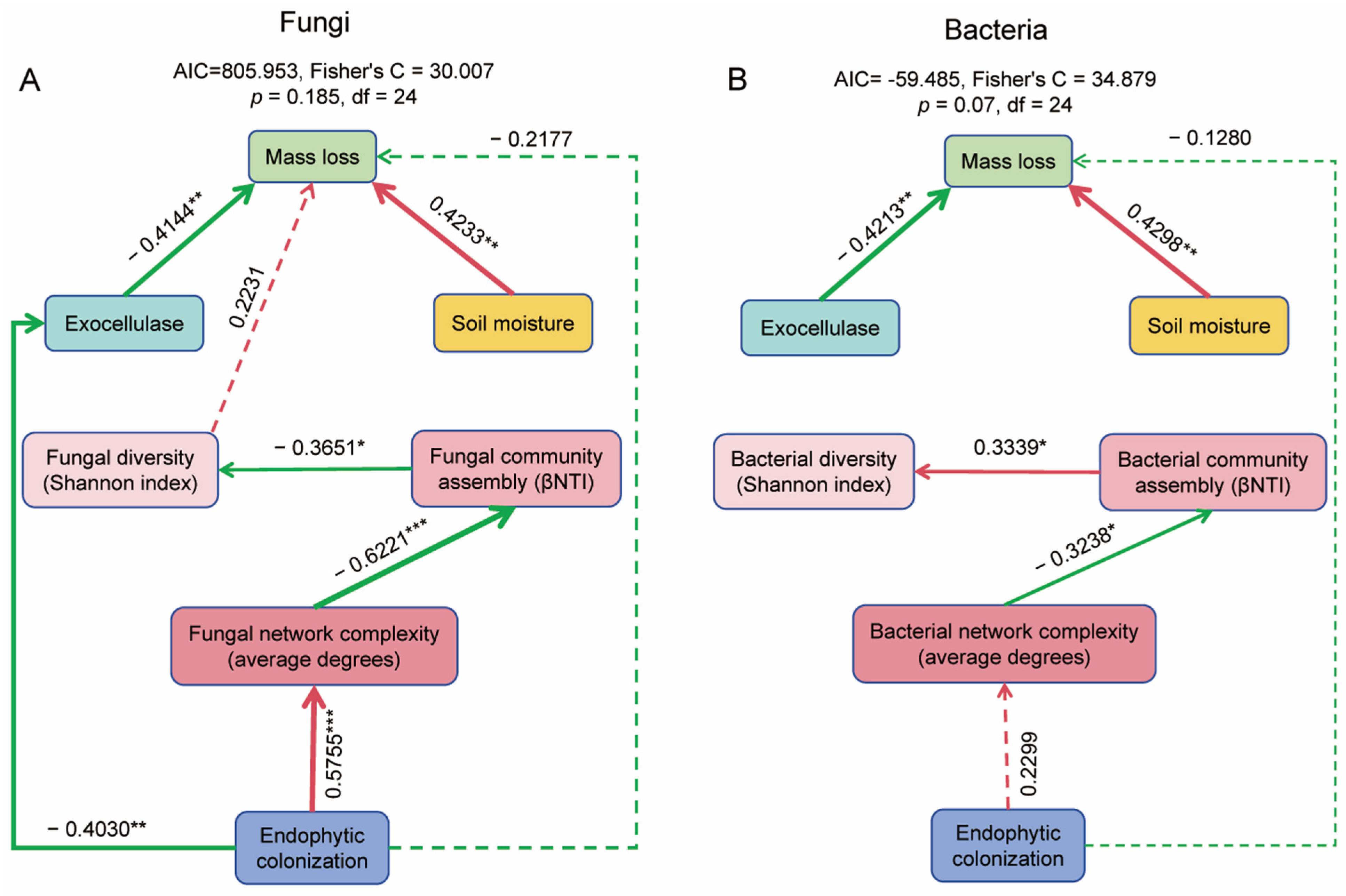
3.6. Relationship of Endophytic Colonization to Microbial Function
4. Discussion
4.1. Effect of Endophytic Colonization on Litter Decomposition
4.2. Effect of Endophytic Colonization on Microbial Composition and Diversity
4.3. Effect of Endophytic Colonization on Microbial Co-Occurrence Network and Community Assembly
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Swift, M.J.; Heal, O.W.; Anderson, J.M. Decomposition in terrestrial ecosystems. In Studies in Ecology; Blackwell Scientific Publications: Oxford, UK, 1979; Volume 5. [Google Scholar]
- Berg, B.; Laskowski, R. Decomposers: Soil Microorganisms and Animals. In Advances in Ecological Research; Elsevier: Amsterdam, The Netherlands, 2005; Volume 38, pp. 73–100. [Google Scholar] [CrossRef]
- Korkama-Rajala, T.; Müller, M.M.; Pennanen, T. Decomposition and fungi of needle litter from slow- and fast-growing Norway spruce (Picea abies) clones. Microb. Ecol. 2008, 56, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Promputtha, I.; Hyde, K.D.; McKenzie, E.H.C.; Peberdy, J.F.; Lumyong, S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 2010, 41, 89–99. [Google Scholar] [CrossRef]
- He, X.; Lin, Y.; Han, G.; Guo, P.; Tian, X. Diversity and decomposition potential of endophytes in leaves of a Cinnamomum camphora plantation in China. Ecol. Res. 2012, 27, 273–284. [Google Scholar] [CrossRef]
- Xiao, J.; He, Z.; He, X.; Lin, Y.; Kong, X. Tracing microbial community across endophyte-to-saportroph continuum of Cinnamomum camphora (L.) Presl leaves considering priority effect of endophyte on litter decomposition. Front. Microbiol. 2025, 15, 1518569. [Google Scholar] [CrossRef] [PubMed]
- Sieber-Canavesi, F.; Petrini, O.; Sieber, T.N. Endophytic Leptostroma species on Picea abies, Abies alba, and Abies balsamea: A cultural, biochemical, and numerical study. Mycologia 1991, 83, 89–96. [Google Scholar] [CrossRef]
- Fryar, S.C.; Yuen, T.; Hyde, K.D.; Hodgkiss, I.J. The influence of competition between tropical fungi on wood colonization in streams. Microb. Ecol. 2001, 41, 245–251. [Google Scholar] [CrossRef]
- Purahong, W.; Hyde, K.D. Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal Divers. 2011, 47, 1–7. [Google Scholar] [CrossRef]
- Fukami, T.; Dickie, I.A.; Wilkie, J.P.; Paulus, B.C.; Par, D.; Roberts, A.; Buchanan, P.K.; Allen, R.B. Assembly history dictates ecosystem functioning: Evidence from wood decomposer communities. Ecol. Lett. 2010, 13, 675–684. [Google Scholar] [CrossRef]
- Dickie, I.A.; Fukami, T.; Wilkie, J.P.; Allen, R.B.; Buchanan, P.K. Do assembly history effects attenuate from species to ecosystem properties? A field test with wood-inhabiting fungi. Ecol. Lett. 2012, 15, 133–141. [Google Scholar] [CrossRef]
- Kominoski, J.S.; Marczak, L.B.; Richardson, J.S. Riparian forest composition affects stream litter decomposition despite similar microbial and invertebrate communities. Ecology 2011, 92, 151–159. [Google Scholar] [CrossRef]
- Lin, Y.; He, X.; Ma, T.; Han, G.; Xiang, C. Priority colonization of Cinnamomum camphora litter by endophytes affects decomposition rate, fungal community and microbial activities under field conditions. Pedobiologia 2015, 58, 177–185. [Google Scholar] [CrossRef]
- Osono, T.; Takeda, H. Effects of organic chemical quality and mineral nitrogen addition on lignin and holocellulose decomposition of beech leaf litter by Xylaria sp. Eur. J. Soil Biol. 2001, 37, 17–23. [Google Scholar] [CrossRef]
- Unterseher, M.; Peršoh, D.; Schnittler, M. Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Divers. 2013, 60, 43–54. [Google Scholar] [CrossRef]
- Avolio, M.L.; Forrestel, E.J.; Chang, C.C.; Pierre, K.J.L.; Burghardt, K.T.; Simith, M.D. Demystifying dominant species. New Phytol. 2019, 223, 1106–1126. [Google Scholar] [CrossRef] [PubMed]
- Vannette, R.L.; Fukami, T. Historical contingency in species interactions: Towards niche-based predictions. Ecol. Lett. 2014, 17, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Polley, H.W.; Wilsey, B.J.; Derner, J.D. Dominant species constrain effects of species diversity on temporal variability in biomass production of tallgrass prairie. Oikos 2007, 116, 2044–2052. [Google Scholar] [CrossRef]
- Song, M.; Zong, N.; Jiang, J.; Shi, P.; Zhang, X.; Gao, J.-Q.; Loreau, M. Nutrient-induced shifts of dominant species reduce ecosystem stability via increases in species synchrony and population variability. Sci. Total Environ. 2019, 692, 441–449. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y. Abundant fungi adapt to broader environmental gradients than rare fungi in agricultural fields. Glob. Change Biol. 2020, 26, 4506–4520. [Google Scholar] [CrossRef]
- Fay, P.A.; Newingham, B.A.; Polley, H.W.; Morgan, J.A.; LeCain, D.R.; Nowak, R.S.; Smith, S.D. Dominant plant taxa predict plant productivity responses to CO2 enrichment across precipitation and soil gradients. AoB Plants 2015, 7, plv027. [Google Scholar] [CrossRef]
- Jiao, S.; Wang, J.; Wei, G.; Chen, W.; Lu, Y. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere 2019, 235, 248–259. [Google Scholar] [CrossRef]
- Liu, L.; Yang, J.; Yu, Z.; Wilkinson, D.M. The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J 2015, 9, 2068–2077. [Google Scholar] [CrossRef]
- Egidi, E.; Delgadobaquerizo, M.; Plett, J.M.; Wang, J.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-González, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef]
- Grime, J.P. Benefits of plant diversity to ecosystems: Immediate, filter and founder effects. J. Ecol. 1998, 86, 902–910. [Google Scholar] [CrossRef]
- Hoover, D.L.; Knapp, A.K.; Smith, M.D. Resistance and resilience of a grassland ecosystem to climate extremes. Ecology 2014, 95, 2646–2656. [Google Scholar] [CrossRef]
- Smith, M.D.; Koerner, S.E.; Knapp, A.K.; Avolio, M.L.; Chaves, F.A.; Denton, E.M.; Dietrich, J.; Gibson, D.J.; Gray, J.; Hoffman, A.M.; et al. Mass ratio effects underlie ecosystem responses to environmental change. J. Ecol. 2020, 108, 855–864. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Liang, Y.; Yu, X.; Pan, F. Plants Drive Microbial Biomass and Composition but Not Diversity to Promote Ecosystem Multifunctionality in Karst Vegetation Restoration. Microorganisms 2025, 13, 590. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Lin, Y.; He, Z.; He, X.; Kong, X. Priority effect of endophyte community in newly fallen Leaves of Quercus acutissima Carruth. on litter decomposition and saprotrophic microbial community. Forests 2025, 16, 249. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes: Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Agnihotri, S.; Dutt, D.; Tyagi, C.H.; Kumar, A.; Upadhyaya, J.S. Production and biochemical characterization of a novel cellulose-poor alkali-thermo-tolerant xylanase from Coprinellus disseminatus SW-1 NTCC 1165. World J. Microbiol. Biotechnol. 2010, 26, 1349–1359. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Antibus, R.K.; Linkins, A.E.; McClaugherty, C.A.; Rayburn, L.; Repert, D.; Weiland, T. Wood decomposition: Nitrogen and phosphorus dynamics in relation to extracellular enzyme activity. Ecology 1993, 74, 1586–1593. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Wang, H.; Tang, Y. Isolation, purification, and partial characterization of acid phosphatase from sweet potato leaves. Food Sci. 2015, 36, 152–157. (In Chinese) [Google Scholar] [CrossRef]
- Zhang, S.; Cai, X.; Luo, X.; Wang, S.; Guo, A.; Hou, J.; Wu, R. Molecular cloning and characterization of leucine aminopeptidase gene from Taenia pisiformis. Exp. Parasitol. 2018, 186, 1–9. [Google Scholar] [CrossRef]
- Papa, S.; Pellegrino, A.; Fioretto, A. Microbial activity and quality changes during decomposition of Quercus ilex leaf litter in three Mediterranean woods. Appl. Soil Ecol. 2008, 40, 401–410. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, H.; Sun, Y.; Shao, K.; Wang, X.; Ma, X.; Hu, A.; Zhang, H.; Fan, J. How habitat heterogeneity shapes bacterial and protistan communities in temperate coastal areas near estuaries. Environ. Microbiol. 2022, 24, 1775–1789. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Shah, J.J.F. Ecoenzymatic stoichiometry and ecological theory. Annu. Rev. Ecol. Evol. S 2012, 43, 313–343. [Google Scholar] [CrossRef]
- Moorhead, D.L.; Sinsabaugh, R.L.; Hill, B.H.; Weintraub, M.N. Vector analysis of ecoenzyme activities reveal constraints on coupled C, N and P dynamics. Soil Biol. Biochem. 2016, 93, 1–7. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, Y.; Duan, C.; Wang, X.; Zhang, X.; Ju, W.; Chen, H.; Yue, S.; Wang, Y.; Li, S.; et al. Ecoenzymatic stoichiometry reveals microbial phosphorus limitation decreases the nitrogen cycling potential of soils in semi-arid agricultural ecosystems. Soil Till. Res. 2020, 197, 104463. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, Z.; Zong, Y.; Li, W.; Chen, F.; Wang, G.G.; Li, J.; Fang, X. Mixing with coniferous tree species alleviates rhizosphere soil phosphorus limitation of broad-leaved trees in subtropical plantations. Soil Biol. Biochem. 2022, 175, 108853. [Google Scholar] [CrossRef]
- Abay, P.; Gong, L.; Luo, Y.; Zhu, H.; Ding, Z. Soil extracellular enzyme stoichiometry reveals the nutrient limitations in soil microbial metabolism under different carbon input manipulations. Sci. Total Environ. 2024, 913, 169793. [Google Scholar] [CrossRef]
- Veum, K.S.; Goyne, K.W.; Kremer, R.J.; Miles, R.J.; Sudduth, K.A. Biological indicators of soil quality and soil organic matter characteristics in an agricultural management continuum. Biogeochemistry 2014, 117, 81–99. [Google Scholar] [CrossRef]
- Margenot, A.J.; Calderón, F.J.; Bowles, T.M.; Parikh, S.J.; Jackson, L.E. Soil organic matter functional group composition in relation to organic carbon, nitrogen, and phosphorus fractions in organically managed tomato fields. Soil Sci. Soc. Am. J. 2015, 79, 772–782. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package (Version 2.5-7). 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 19 December 2024).
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef]
- Zheng, H.; Yang, T.; Bao, Y.; He, P.; Yang, K.; Mei, X.; Wei, Z.; Xu, Y.; Shen, Q.; Banerjee, S. Network analysis and subsequent culturing reveal keystone taxa involved in microbial litter decomposition dynamics. Soil Biol. Biochem. 2021, 157, 108230. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Clauset, A. Finding community structure in very large networks. Phys. Rev. E 2004, 70, 066111. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLOS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.-C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Li, C.; Li, H.; Yao, T.; Su, M.; Ran, F.; Han, B.; Li, J.; Lan, X.; Zhang, Y.; Yang, X.; et al. Microbial inoculation influences bacterial community succession and physicochemical characteristics during pig manure composting with corn straw. Bioresour. Technol. 2019, 289, 121653. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, T.; Li, C.; Song, F. Differences in microbial community and metabolites in litter layer of plantation and original Korean pine forests in north temperate zone. Microorganisms 2020, 8, 2023. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Delgado-Baquerizo, M.; Fanin, N.; Chen, X.; Zhou, Y.; Du, G.; Hu, F.; Jiang, L.; Hu, S.; Liu, M. Nutrient-induced acidification modulates soil biodiversity-function relationships. Nat. Commun. 2024, 15, 2858. [Google Scholar] [CrossRef]
- Zheng, Z.; Mamuti, M.; Liu, H.; Shu, Y.; Hu, S.; Wang, X.; Li, B.; Xu, L. Effects of nutrient additions on litter decomposition regulated by phosphorus-induced changes in litter chemistry in a subtropical forest, China. For. Ecol. Manag. 2017, 400, 123–128. [Google Scholar] [CrossRef]
- Hu, D.; Wang, M.; Zheng, Y.; Lv, M.; Zhu, G.; Zhong, Q.; Cheng, D. Leaf litter phosphorus regulates the soil meso- and micro- faunal contribution to home-field advantage effects on litter decomposition along elevation gradients. Catena 2021, 207, 105673. [Google Scholar] [CrossRef]
- Tie, L.; Hu, J.; Peñuelas, J.; Sardans, J.; Wei, S.; Liu, X.; Zhou, S.; Huang, C. The amounts and ratio of nitrogen and phosphorus addition drive the rate of litter decomposition in a subtropical forest. Sci. Total Environ. 2022, 833, 155163. [Google Scholar] [CrossRef] [PubMed]
- Angst, S.; Harantova, L.; Baldrian, P.; Angst, G.; Cajthaml, T.; Strakova, P.; Blahut, J.; Vesela, H.; Frouz, J. Tree species identity alters decomposition of understory litter and associated microbial communities: A case study. Biol. Fert. Soils 2019, 55, 525–538. [Google Scholar] [CrossRef]
- Barraclough, T.G. Species matter for predicting the functioning of evolving microbial communities—An eco-evolutionary model. PLoS ONE 2019, 14, e0218692. [Google Scholar] [CrossRef]
- Pacciani-Mori, L.; Giometto, A.; Suweis, S.; Maritan, A. Dynamic metabolic adaptation can promote species coexistence in competitive microbial communities. PLoS Comput. Biol. 2020, 16, e1007896. [Google Scholar] [CrossRef]
- Moroni, F.; Naya-Català, F.; Hafez, A.I.; Domingo-Bretón, R.; Soriano, B.; Llorens, C.; Pérez-Sánchez, J. Beyond microbial variability: Disclosing the functional redundancy of the core gut microbiota of farmed gilthead sea bream from a Bayesian network perspective. Microorganisms 2025, 13, 198. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, J.; Dai, S.; Zhang, W.; Wang, X.; Zhang, X.; Huang, Z. Functional redundancy enables a simplified consortium to match the lignocellulose degradation capacity of the orginal consortium. Environ. Res. 2025, 264, 120373. [Google Scholar] [CrossRef] [PubMed]
- Goberna, M.; Verdú, M. Phylogenetic-scale disparities in the soil microbial diversity-ecosystem functioning relationship. ISME J. 2018, 12, 2152–2162. [Google Scholar] [CrossRef] [PubMed]
- Žifčáková, L.; Větrovský, T.; Howe, A.; Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 2016, 18, 288–301. [Google Scholar] [CrossRef]
- Niwa, R.; Koyama, T.; Sato, T.; Adachi, K.; Tawaraya, K.; Sato, S.; Hirakawa, H.; Yoshida, S.; Ezawa, T. Dissection of niche competition between introduced and indigenous arbuscular mycorrhizal fungi with respect to soybean yield responses. Sci. Rep. 2018, 8, 7419. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.R.; Steidinger, B.S.; Bruns, T.D.; Peay, K.G. Competition-colonization tradeoffs structure fungal diversity. ISME J. 2018, 12, 1758–1767. [Google Scholar] [CrossRef]
- Wu, C.; Kong, X.; He, X.; Song, F.; Lin, Y.; Jia, Y.; Kurakov, A.V.; He, Z. The biotic and abiotic factors of regulation of arbuscular mycorrhizal fungi activity in litter decomposition: Review. Euras. Soil Sci. 2022, 55, 1446–1459. [Google Scholar] [CrossRef]
- Wang, W.G.; Li, A.; Yan, B.C.; Niu, S.B.; Tang, J.W.; Li, X.N.; Du, X.; Challis, G.L.; Che, Y.; Sun, H.D.; et al. LC-MS-Guided isolation of Penicilfuranone A: A new antifibrotic furancarboxylic acid form the plant endophytic fungus Penicillium sp. Sh18. J. Nat. Prod. 2016, 79, 149–155. [Google Scholar] [CrossRef]
- Mattoo, A.J.; Nonzom, S. Endophytic fungi: Understanding complex cross-talks. Symbiosis 2021, 83, 237–264. [Google Scholar] [CrossRef]
- Tao, J.; Gu, M.; Yu, S.; Shi, J.; Cheng, L.; Jin, J.; Lu, P.; Zhang, J.; Li, H.; Cao, P. The beneficial endophytic microbes enhanced tobacco defense system to resist bacterial wilt disease. Chem. Biol. Technol. Agric. 2024, 11, 21. [Google Scholar] [CrossRef]
- Manz, C.; Adamčík, S.; Looney, B.P.; Corrales, A.; Ovrebo, C.; Adamčíková, K.; Hofmann, T.A.; Hampe, F.; Piepenbring, M. Four new species of Russula subsection Roseinae from tropical montane forests in western Panama. PLoS ONE 2021, 16, e0257616. [Google Scholar] [CrossRef]
- Zlatković, M.; Sallmannshofer, M.; Schueler, S.; Cech, T.L.; Djilas, M.; Hoch, G.; Lapin, K.; Ogris, N.; Piškur, B.; Schwanda, K.; et al. Tubakia spp., Didymella macrostoma and Apiognomonia errabunda causing leaf spot and anthracnose of Quercus robur in the Mura-Drava-Danube Biosphere Reserve. Front. For. Glob. Change 2024, 7, 1363141. [Google Scholar] [CrossRef]



| Endophytic Fungal Species | Strain Number | Relative Abundance |
|---|---|---|
| Tubakia dryina | Td01 | 26.81% |
| Tubakia dryinoides | Tdr02 | 24.94% |
| Guignardia sp. | Gs03 | 19.89% |
| Aspergillus sydowii | As04 | 4.42% |
| Aspergillus sp. | As05 | 3.65% |
| Botryosphaeria dothidea | Bd06 | 3.56% |
| Phomopsis sp. | Ps07 | 3.17% |
| Colletotrichum boninense | Cb08 | 2.69% |
| Acrocalymma medicaginis | Am09 | 2.21% |
| Bjerkandera adusta | Ba10 | 2.16% |
| Schizophyllum sp. | Ss11 | 1.97% |
| Cytospora diatrypelloidea | Cd12 | 1.68% |
| Fungal sp. | Fs13 | 1.01% |
| Colletotrichum gigasporum | Cg14 | 0.77% |
| Nigrograna hydei | Nh15 | 0.48% |
| Neofusicoccum parvum | Np16 | 0.38% |
| Penicillium citrinum | Pc17 | 0.19% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; He, Z.; Lin, Y.; He, X.; Kong, X. Priority Colonization of Endophytic Fungal Strains Drives Litter Decomposition and Saprotroph Assembly via Functional Trait Selection in Karst Oak Forests. Microorganisms 2025, 13, 1066. https://doi.org/10.3390/microorganisms13051066
Yang D, He Z, Lin Y, He X, Kong X. Priority Colonization of Endophytic Fungal Strains Drives Litter Decomposition and Saprotroph Assembly via Functional Trait Selection in Karst Oak Forests. Microorganisms. 2025; 13(5):1066. https://doi.org/10.3390/microorganisms13051066
Chicago/Turabian StyleYang, Dongmei, Zaihua He, Yonghui Lin, Xingbing He, and Xiangshi Kong. 2025. "Priority Colonization of Endophytic Fungal Strains Drives Litter Decomposition and Saprotroph Assembly via Functional Trait Selection in Karst Oak Forests" Microorganisms 13, no. 5: 1066. https://doi.org/10.3390/microorganisms13051066
APA StyleYang, D., He, Z., Lin, Y., He, X., & Kong, X. (2025). Priority Colonization of Endophytic Fungal Strains Drives Litter Decomposition and Saprotroph Assembly via Functional Trait Selection in Karst Oak Forests. Microorganisms, 13(5), 1066. https://doi.org/10.3390/microorganisms13051066






