Abstract
Virulence attributes and putative antibiotic resistance genes from enterococcal isolates from wastewater treatment facilities for sustainable reuse and the areas where they discharge treated water were assessed using phenotypic and molecular methods. This analysis was performed on 269 Enterococci, of which 202 were vancomycin-resistant Enterococcus (VRE). VRE strains show markedly higher resistance across multiple antibiotics, especially glycopeptides and beta-lactams, compared to the more susceptible profile observed in vancomycin-susceptible Enterococcus (VSE) strains. vanC was found in every instance of E. gallinarum among VRE and enterococci susceptible to vancomycin (VSE) isolates but not in VR E. faecium/faecalis. Among VRE, 127 (62.9%) possessed at least one of the tetK, tetL, tetM, or tetO, while 22 (17.3%) had two of these genes. The multidrug efflux pump gene emeA was detected in 27 out of 202 (13.4%) VRE isolates and 8 out of 67 (11.9%) VSE isolates. Exactly 69 (78.4%) possessed at least one of the virulence determinants tested, with 10 (11.4%) and seven (8%) positive for haemolysis and gelatinase activity respectively. The gelatinase gene, gelE, was detected in 16 (18.1%) isolates, while more isolates (n = 23; 26.1%) were positive for gelatinase activity. Cytolytic (cyl) genes (1.1%), Angiotensin-converting-enzyme genes (ace) (13.6%), endocarditis-specific antigen A genes (efaA) (25%), hyaluronidase (hyl) genes (9.1%), enterococcal surface protein (esp) genes (4.5%), among others, were detected. Gelatinase activity and the amplified virulence genes were further validated by sequencing the gel-positive amplicons, which were almost identical (98.97%), and the gelE gene of Enterococcus sp. strain SQ07C was deposited under the GenBank accession number PQ381122. Overall, our results showed that the enterococcal isolates were considered as potential pathogens of notable threat to human health via exposure through reuse, and there is a need for more stringent treatment protocols.
Keywords:
antibiotic resistance genes; virulence genes; wastewater; tet genes; Enterococcus; emeA gene; gelE 1. Introduction
The importance of enterococci as potential pathogens, causing urinary tract infections (UTIs), bacteremia and sepsis, endocarditis, and other infections, includes both their virulence attributes and their resistance to antibiotic therapy [1]. Pathogenic enterococci have long become an emerging public health concern due to their increased resistance to antimicrobial therapy [1,2]. Enterococci, which were ordinarily commensals, became pathogenic due to their ability to acquire genes easily and hence are referred to as emerging pathogens [3] or opportunistic pathogens [4,5]. They are implicated in a variety of infections, and the morbidity and mortality caused by these infections are higher with resistant strains [6,7,8].
Multidrug-resistant enterococci have been detected and reported from wastewater treatment plants [9,10] and from surface waters [10,11]. The antibiotic resistance in enterococci occurs as a result of specific genes conferring resistance. These resistance genes may be intrinsic or acquired, as exemplified in glycopeptide antibiotics such as vancomycin. Some strains of enterococci inherently exhibit a built-in resistance at a fundamental level against beta-lactam antibiotics, such as penicillins, because of their limited binding affinity to penicillin-binding proteins (PBPs) [12]. Alternatively, they may produce the enzyme beta-lactamase and are thus not susceptible to beta-lactam antibiotics like penicillin [13]. Furthermore, it is widely known that E. gallinarium is intrinsically resistant to vancomycin. Enterococci can also resist antibiotics by active efflux. Multi-drug efflux pump genes and thirty-four efflux pump genes were reportedly detected in Enterococcus [14,15,16]. These genes enhance their resistance to cephalosporins and penicillinase-resistant penicillins, low concentrations of aminoglycosides, clindamycin, fluoroquinolones, trimethoprim-sulfamethoxazole [16]. Enterococci lack cytochrome enzymes, which are necessary for the high-energy production needed for the active uptake of substances such as antibiotics into the cell. This confers some resistance to aminoglycosides at low levels [12]. Even though the genes conferring resistance to ciprofloxacin, gyrA and parC [17] do exist in enterococci, their resistance to fluoroquinolones has long been shown to be due to active efflux [18].
Between 60 and 80% of enterococci are resistant to tetracycline [10,11]. The resistance is mediated through two different mechanisms related to different genes. The tetracycline resistance genes are tetL, tetM, tetN, and tetO, where tetL mediates active efflux of tetracycline from cells while tetM and tetN shield the ribosomes, preventing them from tetracycline [11]. Luna and Robert [19] reported the detection of both tetM and tetO among Streptococcus pneumoniae isolates and predicted the potential for transmission to Enterococcus faecalis. Some tet genes play essential roles as efflux pump genes, exemplified by tetA, tetC, tetE, tetG, tetK, tetL, etc. Some others exemplified as tetM, tetO, tetQ, tetS, tetT, tetW, etc., are acknowledged as ribosomal protection protein genes, while tetX is essentially identified as an enzymatic modification. These genes are detectable in various environmental matrices with attendant public health impact [20,21,22].
Vancomycin has shown activity against a wide range of Enterococcus species that are resistant to other antibiotics. The importance of species-specific variations in the carriage of the vancomycin resistance gene among enterococci has long been reported [23]. Further reports [24,25,26] have shown that surveillance of van gene carriage among both Enterococcus and Staphylococcus species is imperative. The expression of these genes leads to the emergence of vancomycin resistance in both genera, which have been categorized with high priority by WHO [27].
Virulence factors that have been previously described in enterococci include gelatinase production, enterococcal surface protein (esp), aggregation substance (as), cytolysin, hyaluronidase, and biofilm synthesis [28,29]. While the genes gelE, esp, asa1, cylA, and hyl are linked to gelatinase, enterococcal surface protein, aggregation substance, cytolysin, and hyaluronidase production, respectively, that of biofilm formation involves several genes that are complex and multifactorial [30]. The biofilms have been associated with multitudes of infections as they afford them the potential to withstand harsh environmental conditions and to circumvent the action of the immune system or antimicrobial agents [31]. Consequently, we assessed the biofilm formation among Enterococcus species as well as virulence genes in this study.
Additional elements linked to virulence in Enterococcus encompass endocarditis-specific antigen A (efaA), collagen-binding protein (ace), serine protease, capsule, cell wall polysaccharide, and superoxide [3,9]. These virulence factors contribute to infection by facilitating adhesion, colonization, and invasion into host tissues, manipulating host immunity, and producing extracellular enzymes and toxins, thereby amplifying the severity of infections [32]. Adhesins in this context comprise aggregation substance, enterococcal surface protein, and endocarditis-specific antigen A, and collagen-binding proteins help enterococci to adhere to their host’s tissue, while cytolysin, gelatinase, and hyaluronidase affect the host’s tissue, leading to degradation and invasion [32].
The intricate pathways through which virulence and antibiotic resistance genes propagate involve various sources, including agricultural runoff [33], effluents from wastewater treatment plants [34], and improper disposal of pharmaceuticals [35]. These conduits introduce genetic elements into aquatic ecosystems, where they are assimilated by bacteria, endure, and have the potential to transfer to humans and animals. It is imperative to comprehend the migration patterns of virulence and antibiotic resistance genes within recipient environments. Agricultural fields [36], urban zones, and wildlife habitats may act as amplification hotspots for these genes. The interchange of genetic material among environmental bacteria, pathogens, and commensals heightens the risk of disease emergence [37,38].
The One Health framework underscores collaborative endeavors across diverse disciplines. Essential to this approach is monitoring and surveillance programs that track the prevalence and movement of genetic elements [39]. Implementing interventions at the source, such as enhanced wastewater treatment [40] and the adoption of sustainable agricultural practices, assumes critical importance in diminishing the influx of virulence and antibiotic-resistance genes into water systems.
Despite notable progress, significant research gaps persist. This research gap includes a paucity of datasets on virulence factors and antibiotic resistance genes borne by Enterococcus species recovered from wastewater to be reused in the study area. This gap makes it impossible to fully analyze the potential public health consequences of environmental contamination by such pathogens, particularly in the area under investigation. There is a need for intermittent studies in molecular epidemiology to elucidate the mechanisms of gene transfer, persistence, and their repercussions on public health. Employing such methodologies can augment our comprehension of the diversity and dynamics of virulence and antibiotic resistance genes within water ecosystems. The purpose of this research was to assess the virulence characteristics and antibiotic resistance genes of enterococci extracted from both wastewater treatment plants and the surrounding environment into which they discharge.
2. Materials and Methods
2.1. Collecting and Processing of Samples
The gathering of samples from influent, effluent, biofilter/clarifiers, final effluent, and recipient surface water in Durban, South Africa, along with the subsequent isolation and purification process using membrane-Enterococcus Slanetz and Bartley (mSB) agar (Oxoid, UK), was conducted following established bacteriological protocols of Rosenberg-Goldstein and his colleagues as modified, outlined, and cited in Adegoke et al. [10]. The verification of purified isolates involved phenotypical/biochemical characterization, polymerase chain reaction, and Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. (MALDI-TOF-MS) [10]. The disk diffusion method with modifications as previously outlined [10] was used, while a strain-based antibiogram was depicted using a heatmap plotted with Microsoft Excel 2016.
2.2. Detection of Van A, B, C1, and C2/3 Genes Specific to Each Species
Detection of van A, B, C1, and C2/3 genes specific to each species was carried out using Multiplex PCR, as established by Iweriebor et al. [41], including PCR primers and conditions, which are described in the Supplementary file (Table S1).
2.3. Detection of Other Resistance Genes Tet, gyrA, parC, and emeA Genes
Multiplex PCR was utilized to detect 14 tetracycline resistance (tet) genes. The multiplex groups, primer concentrations, and amplification conditions were as adapted by Jia et al. [14]. In Group I, tet(B), the concentration of primers employed was 0.25 µM, while tet(C) and tet(D) each had primers at a concentration of 0.25 µM and 2.0 µM, respectively. In Group II, tet(A), tet(E), and tet(G) were each targeted with primers at a concentration of 1.0 µM. Group III included tet(K), tet(L), tet(M), tet(O), and tet(S).
To identify ciprofloxacin resistance genes, the primer sequences and conditions from Nowroozi et al. [17] were employed for the gyrA and parC genes. The detection of the emeA gene, associated with a multidrug efflux pump, followed the method outlined by Jia et al. [14]. The PCR primers and conditions are described in the Supplementary file (Table S2).
2.4. Detection of Virulence Genes
DNA samples obtained from the isolates were assessed for the existence of the asa1 (Aggregation substance), gelE (Gelatinase), cylA (Cytolysin), esp (Enterococcal surface protein), and hyl (Hyaluronidase) using a modified multiplex PCR method based on the approach of Vankerckhoven et al. [29]. This was modified by using extracted DNA from the isolates as the DNA template instead of a whole cell suspension, along with adjustments to the initial denaturation temperature, reaction volume, and gel conditions. Each 25 µL PCR mixture comprised 5 µL of DNA template, 0.1 µM concentration for each primer specific to asa1, gelE, and hyl, and 0.2 µM concentration for either primer specific to cylA and esp. Additionally, 12.5 µL of Hot-Star Taq master mixture (ThermoFisher, Oxford, United Kingdom) was included, consisting of 2.5 U of Hot-StarTaq DNA polymerase, 1.5 mM MgCl2, and 200 µM deoxynucleoside triphosphates, with an extra 1.0 mM MgCl2. The cycling parameters included an initial activation phase at 95 °C for 5 min, accompanied by 30 cycles of denaturation (94 °C for 60 s), annealing (56 °C for 60 s), and extension (72 °C for 60 s), concluding with a cycle of 10 min at 72 °C.
For the detection of the ace and efaA genes, monoplex PCR was employed using the primer sequences and cycling conditions from Iweriabor et al. [9]. Each 20 µL of cocktail volume contains 10 µL of master mix, as well as 1 µL of each primer, and 5 µL of DNA template. Cycling conditions for ace included an initial denaturation at 94 °C for 3 min, in which the next was 35 cycles of amplification (93 °C/1 min, 50 °C/1 min, 73 °C/1 min) and a final extension at 72 °C for 10 min. The same conditions were applied for efaA, then an annealing temperature of 56.5 °C for 1 min. The resolution of products was conducted as previously described. The primers utilized were described previously by Iweriabor et al. [9] and Vankerckhoven et al. [29].
2.5. Validation of Virulence Gene by Sequencing Gelatinase Gene, gelE Amplicon
Successfully amplified products were purified and sequenced using a PCR purification kit and Sanger sequencing. The sequenced data was analysed using BioEdit 7 software (Version 7.7.1) for sequence alignment and to determine the nucleotide sequences, identify genetic variations, compare them with known gelE sequences in public databases, and submitted to the National Center for Biotechnology Information (NCBI) Bankit. The phylogenetic reconstruction was performed employing the Neighbor-Joining algorithm, yielding an optimal phylogenetic tree with a cumulative branch length of 2.285. Genetic divergence between sequences was calculated through the Maximum Composite Likelihood approach, with evolutionary distances expressed as the number of nucleotide substitutions per site. Bootstrap support values, indicating the percentage of sites containing unambiguous bases for each clade, are displayed adjacent to internal nodes.
The investigation incorporated 18 protein-coding DNA sequences, analyzing first, second, and third codon positions along with non-coding regions. Ambiguous positions were handled through pairwise deletion during sequence alignment, producing a final dataset containing 1730 homologous sites. All evolutionary computations were executed using MEGA 12 (Version 12.0.11) software.
2.6. Statistical Examination
IBM SPSS version 23 was utilized to analyze presumed antibiotic resistance genes through descriptive statistics and correlation analysis. To compare variations across multiple groups, one-way analysis of variance (ANOVA) was utilized, and post-hoc analysis was conducted using the Duncan t-test. Statistical value was established with a significance level of p ≤ 0.05. Correlation analysis between the tested phenotypic antimicrobials, resistance genes, and virulence genes was performed, and a correlation matrix was plotted using Version 3.8 of Python.
3. Results
We analysed 269 enterococci isolates, consisting of 202 Enterococci resistant to vancomycin (VRE) and enterococci susceptible to vancomycin (VSE), which were isolated from sewage and recipient river samples from the two wastewater treatment plants. The 202 VRE consisted of 91 wastewater and 111 river isolates, while the 67 VSE consisted of 36 wastewater and 31 river isolates.
3.1. Antibiotic Resistance Across the Sampling Points and Species
The antimicrobial susceptibility testing used for fifteen antibiotics has been previously described [10]. There was no remarkable difference in the resistance pattern of the isolates, which was related neither to the sampling points nor to the species. However, eight isolates exhibited resistance to all fifteen antibiotics assessed. The hierarchical clustering heatmaps (Figure 1 and Figure 2) illustrate the antibiotic susceptibility profiles of Enterococcus species, differentiating between Vancomycin-sensitive (VSE) and Vancomycin-resistant (VRE) strains, respectively, across a panel of 15 antibiotics. While both VRE and VSE strains exhibit varying levels of resistance, VRE strains show markedly higher resistance across multiple antibiotics, especially glycopeptides and beta-lactams, compared to the more susceptible profile observed in VSE strains. Seven of these isolates were E. faecium and one E. faecalis. Five of the seven E. faecium super-resistant isolates were isolated from the chlorinated effluent and the clarifiers of Plant II on the same sampling date. The E. faecalis was isolated from Plant II influent, while the remaining two E. faecium isolates came from Plant I influent.
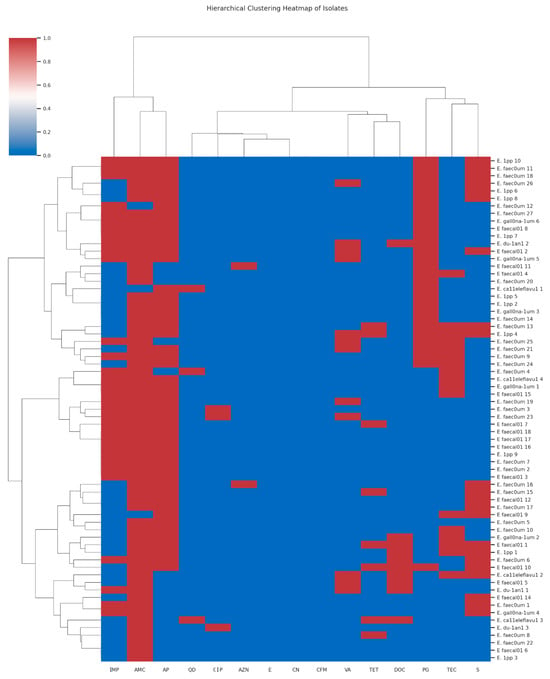
Figure 1.
The hierarchical clustering heatmap of the antimicrobial susceptibility patterns of Vancomycin-susceptible Enterococcus (VSE) isolates across multiple antibiotics [Red indicates resistance (R), corresponding to a value of −1; white represents intermediate resistance (I), denoted by a value of 0; and blue signifies sensitivity (S), corresponding to a value of 1; The antibiotics CN = Gentamycin; AZN = Aztreonam; CIP = Ciprofloxacin; TEC = Teicoplanin; AMC = Amoxicillin-Clavulanic Acid; VA = Vancomycin; TET = Tetracycline; CFM = Cefixime; S = Streptomycin; E = Erythromycin; QD = Quinupristin-Dalfopristin; IMP = Imipenem; DOC = Doxycycline; AP = Ampicillin and PG = Penicillin G].
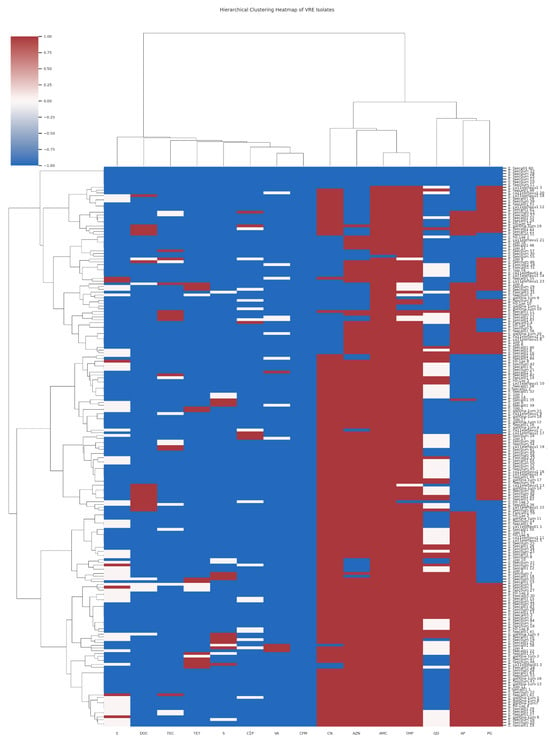
Figure 2.
The hierarchical clustering heatmap of the antimicrobial susceptibility patterns of Vancomycin Resistant Enterococcus (VRE) isolates across multiple antibiotics [Red indicates resistance (R), corresponding to a value of −1; white represents intermediate resistance (I), denoted by a value of 0; and blue signifies sensitivity (S), corresponding to a value of 1; The antibiotics CN = Gentamycin; AZN = Aztreonam; CIP = Ciprofloxacin; TEC = Teicoplanin; AMC = Amoxicillin-Clavulanic Acid; VA = Vancomycin; TET = Tetracycline; CFM = Cefixime; S = Streptomycin; E = Erythromycin; QD = Quinupristin-Dalfopristin; IMP = Imipenem; DOC = Doxycycline; AP = Ampicillin and PG = Penicillin G].
3.2. Vancomycin Resistance Genes Based on Species Diversity
Four different vancomycin resistance genes were detected within the group of VREs. All the VRE isolates except one were shown to possess a van gene, with a dominance of vanA (73.8%) (Table 1). This gene also occurred within the VSE group, but at a lower frequency. Van C2/3 occurred in both groups at similar frequencies. In the sensitive group (VSE), no resistance genes were found in most of the isolates (67%). Noteworthy that vanC (vanC1 and vanC2/3) was found in all E. gallinarum that were VRE and VSE isolates, but not in VRE E. faecium/faecalis. In addition, all vancomycin-susceptible E. gallinarum possessed at least a van gene, but vanA was not found in any of the E. gallinarum isolates. vanC was detected across all species of VSE. Not more than one van gene was identified in the isolates. Worth noting is the detection of VSE E. gallinarum from vancomycin-supplemented plates and confirmed disk diffusion test, due to their loss or lack of vanC1, known to give them intrinsic resistance.

Table 1.
Vancomycin resistance genes were detected among species of VRE and VSE.
3.3. Detection of Other Resistance Genes
Although there is remarkably high resistance to ciprofloxacin among the isolates, the detection of the two ciprofloxacin resistance genes, gyrA and parC, was very low. The gyrA gene was detected only in 14 VRE isolates, while parC was detected in five. Contrastingly, tetracycline resistance genes were detected in greater numbers among the isolates. Four out of the 14 tet genes tested were detected, which include tetK, tetL, tetM, and tetO. One hundred and twenty-seven (62.9%) of the 202 VRE possessed at least one of these tet genes, while 22 (which included the eight isolates that exhibited resistance to all the antibiotics) out of 127 had two tet genes (tetL and tetM). Among the 67 VSE, 21 (31.3%) had at least one tet gene, while two tet genes were detected in 4 out of 21 (three with tetL and tetM and one with tetK and tetL). Multidrug efflux pump gene, emeA, was also detected in an appreciable number of the isolates. Table 2 summarizes the detection of the resistance genes.

Table 2.
Tetracycline resistance and efflux pump responsible for multiple drug resistance genes detected in Enterococcal isolates.
3.4. Detection of gelE and Gelatinase Activity
Gelatinase gene, gelE, was detected in 16 (18.1%) isolates, while 23 (26.1%) were positive for gelatinase activity. Only eight (50%) of the 16 isolates that possessed gelE were positive for gelatinase activity, while the additional 15 (65%) isolates that hydrolysed gelatine had no detectable gelE gene. The eight isolates that were both positive for gelE and gelatinase activity consisted of six E. faecalis and two E. faecium. However, not all gelE-positive E. faecalis were positive for gelatinase activity. The selected sequenced amplicon Enterococcus sp. strain Isol_07c_gelatinase_(gelE)_gene with accession number PQ381122 validated the presence of this gene as seen in the gel electrophoresis. The dendrogram showing hierarchical relationships of Enterococcus sp. strain SQ07C with other referenced strains with gelatinase genes is depicted in Figure 3.

Figure 3.
Dendrogram showing hierarchical relationships of Enterococcus sp. strain SQ07C (indicated with red arrow) with other referenced strains with gelatinase genes.
3.5. Detection of cylA and Haemolytic Activity
The cyl gene was detected only in one isolate (E. casseliflavus), which did not show haemolytic activity. Rather, haemolysis was seen in 24 isolates in which the cyl gene was not detected. These isolates included E. faecalis and E. faecium and other species such as E. gallinarum, E. casseliflavus, and E. hirae.
3.6. Detection of Ace, efaA, asa1, hyl, Esp Genes and Biofilm Formation
Angiotensin-converting enzyme genes, ace, were detected in 13.6% (12) of the isolates, while endocarditis-specific antigen A genes, efaA, were the most prevalent virulence genes among the isolates (25%; n = 22). The species that harbored these genes were mostly E. faecalis, followed by E. faecium and E. hirae. The asa1 was detected in 10 (11.4%) of the isolates, with a majority of E faecalis. Hyaluronidase gene, hyl, and enterococcal surface protein genes, esp, were also detected in 9.1% (8) and 4.4% (4) of the isolates, respectively. The hyl was detected only in E. faecium isolates, and the esp only in E. faecalis. The isolates were all either weak or non-biofilm formers except for two isolates, one each of E. faecalis and E. faecium, which exhibited strong and moderate biofilm formation, respectively. Figure 4 shows the prevalence of virulence genes/factors detected among the isolates, while the virulence genes within different species are presented in Table 3.
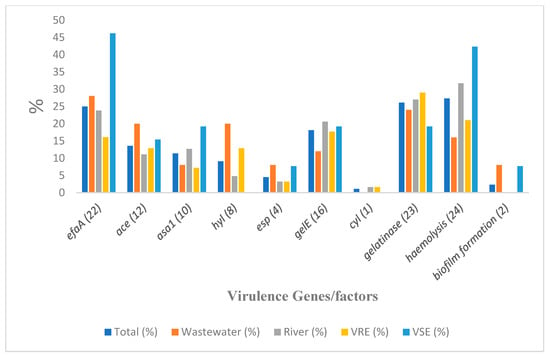
Figure 4.
Virulence determinants detected from specific sample sources and Enterococcus isolates’ types. Note: number of isolates from wastewater = 25; river = 63; VRE = 62; VSE = 26.

Table 3.
Prevalence of the virulence genes and the Enterococcus species involved.
Among the 88 isolates, 69 (78.4%) possessed at least one of the virulence determinants tested, with 10 (11.4%) and seven (8%) positive for haemolysis and gelatinase activity respectively, as the only strains with virulence factors and without harboring any of the virulence genes. None of these last two groups included any E. faecalis isolates. Furthermore, nine out of the 69 positive isolates were E. faecalis and found to be highly virulent, possessing between four and seven virulence genes/factors. There was no notable disparity in the prevalence of any of the six virulence genes (gelE, esp, asa1, hyl, efaA, and ace) between enterococcal isolates when wastewater and river samples were compared (p > 0.05), and between VRE and VSE (p > 0.05), but the prevalence of these virulence genes among E. faecalis and E. faecium was significantly different (p = 0.05). Figure 4 and Table 3 show the prevalence of the virulence gene among the general isolates and species, respectively. For ease of visualization of tested genes, Figure 4 depicts the virulence determinants detected from specific sample sources (wastewater and river) and their distribution into VRE and VSE isolates.
Key findings of the correlation analysis revealed co-resistance patterns. There was a strong correlation between gentamycin (CN) and aztreonam (AZN) resistance, which suggests possible cross-resistance or linked resistance mechanisms. The tet(M) gene shows the strongest association with phenotypic tetracycline resistance among the tet genes analyzed. In species-specific virulence, E. faecalis isolates are strongly associated with efaA and gelE, while E. faecium shows a specific association with the hyl gene. Vancomycin resistance associations were also obvious. VRE isolates showed higher prevalence of tet(L) (30.2% vs. 10.4% in VSE) and tet(O) (5.0% vs. 0% in VSE), suggesting possible genetic linkage or co-selection. Details are depicted in Figure 5, which contains the antibiotic resistance genes and virulence genes. The correlation matrix of the resistance and virulence factors is depicted in Figure 6.
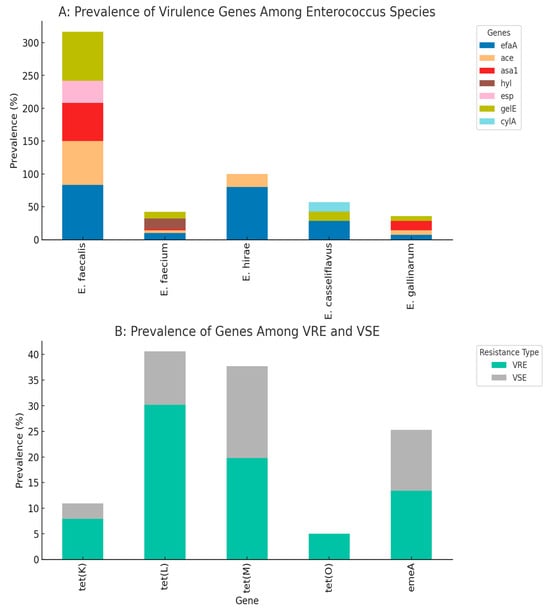
Figure 5.
Prevalence of virulence genes among different Enterococcus species (Plot (A)). Distribution of resistance genes among VRE (Vancomycin-Resistant Enterococci) and VSE (Vancomycin-Susceptible Enterococci) (Plot (B)).
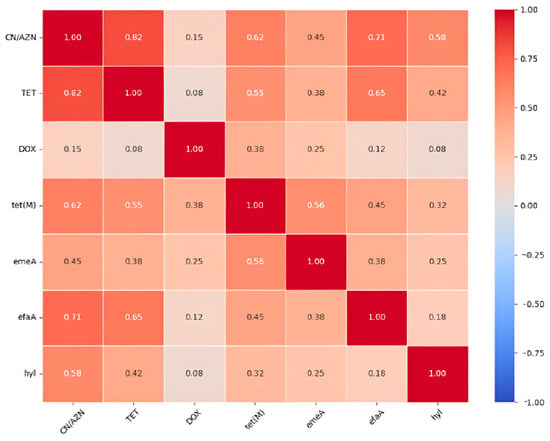
Figure 6.
Correlation matrix of the resistance and virulence factors.
4. Discussion
Surface water faces contamination with harmful substances originating from various sources, alongside discharges from wastewater such as runoff from farms, animal waste, and other human-related endeavors [42]. The enterococcal isolates from the river samples may have originated from these sources. The isolates from the two Wastewater treatment plants (WWTPs) and their connected surface water recipients in this study were multidrug resistant. About 50% of all the isolates were resistant to six antibiotics, besides vancomycin and teicoplanin, while 3.4% exhibited outright resistance characteristics to all the antibiotics.
An increasing resistance to antibiotics by enterococci due to the acquisition of resistance genes was noted in publications elsewhere [9,43,44]. Although the intrinsic resistance to quinupristin-dalfopritin by E. faecalis [14,45,46] contributed to some extent to the high resistance against this antibiotic, its resistance by E. faecium is of concern, as this has been an alternative drug in the treatment of vancomycin-resistant E. faecium infections [46]. However, the total resistance against the fifteen antibiotics by eight isolates, consisting of seven E. faecium and one E. faecalis, is a serious concern [14]. Each of these isolates had vanA and two tet genes, confirming their increased antibiotic resistance. E. faecium and E. faecalis are prolific species among others, and their importance and severity in human infection and their antibiotic resistance are well known. E. faecium is known for its high resistance to antibiotics, while E. faecalis is more virulent [7].
It is known that some strains of enterococci possess an intrinsic resistance to beta-lactam antibiotics such as penicillins due to their low affinity to penicillin binding proteins (PBPs) or by producing the enzyme beta-lactamase against the beta-lactam agents [12,47].
The antibiotic resistance patterns observed in Enterococcus isolates, particularly E. faecalis and E. faecium, underscore the importance of a One Health approach to comprehensively address the implications for human and animal health. According to Martin et al. [47], while most E. faecalis isolates exhibit susceptibility to penicillin or ampicillin within concentrations of 1 to 8 µg/mL, E. faecium isolates demand higher concentrations, typically ranging from 16 to 64 µg/mL for effective growth inhibition. However, certain isolates within these species display heightened resistance, emphasizing the evolving nature of antibiotic resistance.
The study at hand reinforces these findings, revealing resistance rates of 25% to 55% to penicillins among both vancomycin-resistant (VRE) and vancomycin-sensitive (VSE) isolates, primarily dominated by E. faecalis and E. faecium, with a minor presence in E. gallinarum. This highlights the need for collaborative efforts under the One Health paradigm, recognizing the interconnectedness of human and animal health.
The intriguing observation of higher sensitivity to co-amoxiclav, with only 4.5/4.7% resistance in VRE/VSE isolates. Comparing the increased resistance to ampicillin and penicillin G, the findings suggest that clavulanic acid may be effective against penicillinase activity in the same Enterococcus isolates. This finding reinforces the importance of a multidisciplinary approach, as it not only sheds light on the mechanisms behind resistance but also suggests avenues for developing interventions that could be beneficial for both human and animal populations [47].
Although most of the bacteria exhibited resistance to ciprofloxacin, the frequency of detection of gyrA and parC genes was low. Thus, the resistance to ciprofloxacin by the enterococci isolates could not be associated with the possession of the corresponding resistance gene. The reason could be either that the genes were present but were not detected or that enterococci resist ciprofloxacin by another mechanism, including active efflux, as shown previously by Mahapatra et al. [18]. This could be partly affirmed by the detection of a number of efflux genes, emeA and tetL, among the isolates, and has also been shown by other authors [11,14]. Additional mechanisms may also contribute to the high resistance to ciprofloxacin since the number of isolates that possess either gyrA, parC, or emeA genes was lower than those that exhibited resistance to ciprofloxacin.
Enterococci isolates, which did not grow upon sub-culturing on vancomycin-supplemented medium, were considered to be VSE and facilitated the observation of the possession of resistance genetic determinants. In a study by Figueira et al. [48], on characterization of quinolone resistance observed in Aeromonas strains originating from aquatic environments, the gene aacA6-ib-cr was found in isolates selected with ciprofloxacin. This was similar to the detection of the vanA gene in our study. The van genes, particularly vanA, were detected mostly among VRE, which were isolated on Slanetz and Bartley agar supplemented with vancomycin. However, in the case of the bacteria isolated with antibiotic-supplemented medium, their true resistance profiles to other antibiotics seem to be affected. The antibiotic in the primary isolation medium seemed to play a pre-synergic effect when exposed to other antibiotics during AST, which explains the difference in the resistance pattern to some antibiotics between the VRE and VSE isolates in this study. The isolates from vancomycin-supplemented media were more susceptible to gentamicin, azithromycin, quinupristin-dalfopristin, and imipenem than those from non-vancomycin media. This also could be due to the fact that the organism must have been weakened by the antibiotics in the primary medium and then succumbed more easily to some other antibiotics during AST. Furthermore, the synergic effect of vancomycin with an aminoglycoside [49] and quinopristin-dalfopristin with a cell wall-active agent [45] against enterococci has been shown.
The detection of the vanA gene on some VSE isolates may seem unexpected. The presence of these genes in organisms without subsequently being expressed is well documented [50]. However, resistance marked by these isolates upon AST could be due to the fact that the putative vanA gene was not actively expressed. This slow expression could lead to misinterpretation. Enterococci isolates, which did not grow upon sub-culturing on vancomycin-supplemented medium, were taken to be VSE. Therefore, those isolates, which carry van genes that were poorly expressed, may not grow on vancomycin-supplemented medium and thus be classified as VSE. Hence, upon AST, following the standard reference guideline, the measured zones of inhibition were not large enough to be interpreted as susceptible and thus were regarded as resistant. Therefore, for proper determination of antibiotic resistance/susceptibility of an organism, both AST and detection of resistance genes, when possible, should be employed.
Enterococcus species is the third-most etiological agent of hospital-based infections globally [51] due to their virulence attributes coded for by virulence genes.
It is noteworthy to state that the isolates revealed the presence of seven virulence genes, namely ace, asa1, cylA, efaA, esp, gelE, and hyl, with 74% of the isolates positive for at least one virulence gene/factor. The efaA gene was the most prevalent (25%) while the cylA gene was the least (1.1%) virulence gene detected. In as much as the cylA gene was the least prevalent, hemolysin (27.3%) was the highest among the three virulence factors detected. In addition, the number of isolates positive for gelatinase activity was higher than the number of isolates in which the associated gene (gelE) was detected. Therefore, there may be the possibility of false negative results in the detection of the virulence genes. The detection of the gelE gene in enterococcal strains does not correlate with the ability to produce gelatinase, as shown in several investigations [30,50,52]. This was also the situation in this study, where only 50% of the isolates in which the gelE gene was detected actually expressed gelatinase. This could be a result of “silent genes”. However, the E. faecalis fsr quorum-sensing system was shown to control the production of gelatinase [50]. Disabling the gelE gene, which is regulated by fsr and encodes the zinc-metalloprotease known as gelatinase [53], led to gelE not being expressed. gelE is shown to be associated with E. faecalis [29], but Lopes et al. [54] mention that gelE was observed in species different from either E. faecalis or E. faecium, which was also the case in this study.
The cytolysin operon contains five genes: cylL1, cylL2, cylM, cylB, which are responsible for the physical effect of the L component, and cylA for expressing the activator protein A [29]. This activator (A) is a serine protease, which is responsible for further cleavage and activation of cyl1 and cyl2 subunit extracellularly for subsequent full expression [55,56]. In this study, detection of the cylA gene was chosen (because of the role of the activator protein A in cytolysin expression) rather than the whole cytolysin gene operon. Cytolysin is associated with haemolytic activity [55,56,57]. Thus, β-haemolytic isolates indicate possession of cylA. However, β-haemolysis in 24 isolates and the detection of the cylA gene only in one isolate could not be linked. Either the isolates have this gene, but it was not detected in the PCR used, or the haemolytic activity of these isolates is a result of factors other than cytolysin.
Although Comerlato et al. [5] reported that there was no association between the connection of the origin of isolation and the existence as well as the functionality of any virulence factor, some virulence attributes may be more common in enterococci isolated from a particular kind of infection [58]. Certain adhesins, particularly agg/asa1 and efaA, showed notably higher occurrence within non-invasive Enterococcus spp. Isolates, such as in the reported genital tract infection (GTI) isolates, wound isolates, and isolates from urinary tract infections (UTIs), in contrast to isolates causing invasive bacteremia [32]. The ace/acm exhibited a notably increased prevalence by enhancing the potential to invade their host, as opposed to those from non-invasive genitourinary and wound sources. However, for environmental isolates, the type and frequency of virulence genes might not differ among isolates from different sites. This study did not show any difference in the type and the occurrence of virulence genes among the isolates from wastewater treatment plants and those of the river isolates. Previous reports have shown no differences in the presence of virulence genes in both VRE and VSE, in accordance with this study [5]. However, since Biswas et al. [30] showed a higher prevalence of hyl gene among VRE than VSE, the detection of hyl genes only among the E. faecium isolates of VRE may be considered relevant. These pathogens and their genes may constitute serious human health hazards and the development of superbugs [59].
The detection of vancomycin-susceptible E. gallinarum is notable, despite the expected intrinsic resistance, and is in line with observations made at different times by other researchers [60,61]. The vanC1 gene, located on its chromosome, mediates a mechanism of low resistance; its clinical detection and interpretation are challenging [62]. The vanC1 gene leads to the production of a D-Ala-D-Ser peptidoglycan precursor that is altered and of lowered affinity for vancomycin, yielding minimum inhibitory concentrations (MICs) typically 4–32 µg/mL [63]. Despite this natural resistance, existing evidence supports phenotypic heterogeneity with decreased MIC isolates that may be vancomycin susceptible on regular testing and effective therapeutic applications [61]. This necessitates the genotypic confirmation while identifying van genes in vancomycin resistance studies in E. gallinarum, as was performed in this study.
Exposure pathways of Enterococcus in wastewater may most likely be occupational [64], though residents of close settlements to the WWTPs may inhale aerosols containing virulence/antibiotic gene harboring Enterococcus from the WWTPs in a form of domestic exposure [2,65]. Such aerosols may settle in exposed food or, rarely, possibly on open wounds of the residents [66]. Using their virulence genes like those reported in our studies among others, both E. faecalis and E. faecium are recognized for 9 out of 10 clinical cases [6,67], because the virulence genes, when expressed, enhance their degree of pathogenicity. On the global level, these two species of Enterococcus are number 3 and 4 in terms of their pathogenicity [68]. The Cytolytic (cyl) and endocarditis-specific antigen A genes detected in our study make the bacteria a public health concern for invasion of the blood and organs. For example, Enterococcus species have been placed as the third most prevalent aetiological agent of bacteremia (morbidity: approx. 11–13%) in Europe and North America [69,70]. Gelatinase gene, gelE, which was detected in some of our isolated Enterococcus species, might play an active role in colonizing a wound when such a wound is exposed to contaminated water. gelE codes for gelatinase enzymes, which play vital roles in biofilm formation and wound colonization [71]. Enterococci are highly prevalent in wound infections [72,73]. So, there is a high risk associated with the reuse of wastewater effluent containing such bacteria for recreation, where individuals with wounds may be exposed.
The Cytolytic (cyl), Gelatinase (gelE), and Endocarditis-Specific Antigen A genes play pivotal roles in bacterial pathogenesis. The Cyl gene, associated with hemolysin production, contributes to the virulence of various bacteria, impacting both human and animal health. Gelatinase, encoded by gelE, facilitates tissue invasion and immune evasion, influencing the severity of infections. These virulence factors are particularly significant in Enterococcus species, where they contribute to the pathogenesis of infectious diseases in humans and animals.
The Endocarditis-Specific Antigen A (cylA) gene serves as a marker for endocarditis, a condition affecting both humans and animals, highlighting its relevance in the One Health perspective that emphasizes the interconnectedness of human, animal, and environmental health. Research by Sava et al. [74] demonstrates the importance of these genes in the context of Enterococcus infections, showcasing their impact on both human and veterinary medicine. By unraveling the role of Cyl, gelE, and endocarditis-specific antigen A genes (cylA), this knowledge can inform strategies for disease surveillance, prevention, and treatment, aligning with the holistic One Health perspective that emphasizes the interconnectedness of human, animal, and environmental health.
The previously identified and reported Enterococcus species were further analysed for virulence genes and antibiotic resistance genes. Various species of Enterococci possess both genes and phenotypic virulence attributes in various patterns. Since organisms possessing virulence genes or expressing virulence factors are potentially virulent.
5. Conclusions
It can be concluded that most of the enterococcal isolates from wastewater and river samples from the study depicted phenotypic and genotypic virulence attributes, and thus, they are considered a potential risk to the health of the community that reuses the water. With the majority of the isolates coming from river samples, the reuse of this water for irrigation or other activities may contribute significantly to the spread of these pathogenic and resistant enterococci. It may also lead to infection through direct or indirect exposure. Therefore, frequent surveillance is essential for appropriate mitigation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13051045/s1. Table S1: List of primers for detection of resistance genes; Table S2: Primers for the detection of virulence genes.
Author Contributions
Conceptualization was performed by A.A.A.; C.E.M.; P.R. and T.A.S.; Research was performed by A.A.A. and C.E.M.; supervised by A.A.A.; P.R. and T.A.S.; Draft written and analysis performed by A.A.A. and C.E.M. with support from P.R.; T.A.S.; O.K.F. and A.I.O. A.I.O. coordinated final compliance with South African standards. All authors edited the final draft. The research was supervised by A.A.A.; P.R. and T.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information. The list of primers for the detection of resistance genes is provided in Supplementary Table S1, while Primers for the detection of virulence genes are presented in Table S2, along with the original reference used in this study. The data presented in this study are openly available in [GenBank] at NCBI, accession number PQ381122.
Acknowledgments
Acknowledgements are extended to the personnel at the two sewage treatment facilities for their support in sample gathering. Gratitude is expressed to the SARChI for funding this investigation and to the DUT for providing a conducive research setting. The authors also acknowledge the Nigerian National Research Fund (NRF) of the TETFUND, Project Code TETF/DR&DCE/NRF/2020/SETI/99/VOL.1, for the availability of funds that enabled the sequencing of the selected isolate. The results of this study will provide useful information for the related ongoing funded research in Nigeria.
Conflicts of Interest
All authors declare that there are no financial and non-financial competing interests.
References
- Tendulkar, S.R.; Baghdayan, A.S.; Shankar, N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J. Bacteriol. 2006, 185, 2063–2072. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Stenstrom, T.A. Septic Systems. In Global Water Pathogen Project; Rose, J.B., Jiménez-Cisneros, B., Mihelcic, J.R., Verbyla, M.E., Eds.; Michigan State University: East Lansing, MI, USA; UNESCO: London, UK, 2019. [Google Scholar] [CrossRef]
- Giridhara Upadhyaya, P.M.; Ravikumar, K.L.; Umapathy, B.L. Review of virulence factors of Enterococcus: An emerging nosocomial pathogen. Indian J. Med. Microbiol. 2009, 27, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahdal, M.N.; Abozaid, S.M.; Al-Shammary, H.F.; Bohol, M.F.; Al-Thawadi, S.I.; Al-Jaberi, A.A.; Senok, A.C.; Shibl, A.M.; Al-Qahtani, A.A. Characterization of Enterococcus faecium isolates and first report of vanB phenotype–vanA genotype incongruence in the Middle East. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3223–3229. [Google Scholar] [CrossRef]
- Comerlato, C.B.; de Resende, M.C.C.; Caierão, J.; Alves d’Azevedo, P. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem. Inst. Oswaldo Cruz 2013, 108, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Billington, E.O.; Phang, S.H.; Gregson, D.B.; Pitout, J.D.D.; Ross, T.; Church, D.L.; Laupland, K.B.; Parkins, M.D. Incidence, risk factors, and outcomes for Enterococcus spp. bloodstream infections: A population-based study. Int. J. Infect. Dis. 2014, 26, 76–82. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C. The ecology, epidemiology, and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef]
- Diazgranados, C.A.; Zimmer, S.M.; Klein, M.; Jernigan, J.A. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: A meta-analysis. Clin. Infect. Dis. 2005, 41, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Virulence and antimicrobial resistance factors of Enterococcus spp. isolated from faecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol. 2015, 15, 136. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Madu, C.E.; Reddy, P.; Stenström, T.A.; Okoh, A.I. Prevalence of vancomycin-resistant Enterococcus in wastewater treatment plants and their recipients for reuse using PCR and MALDI-ToF MS. Front. Environ. Sci. 2022, 9, 797992. [Google Scholar] [CrossRef]
- Molale, L.G.; Bezuidenhout, C.C. Antibiotic resistance, efflux pump genes and virulence determinants in Enterococcus spp. from surface water systems. Environ. Sci. Pollut. Res. Int. 2016, 23, 21501–21510. [Google Scholar] [CrossRef]
- Klare, I.; Konstabel, C.; Badstübner, D.; Werner, G.; Witte, W. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 2003, 88, 269–290. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Li, G.; Wang, W. Prevalence and antimicrobial resistance of Enterococcus species: A hospital-based study in China. Int. J. Environ. Res. Public Health 2014, 11, 3424–3442. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R.; McAlpine, J.B.; Pazole, C.J.; Talbot, M.K.; Alder, E.A.; White, C.; Jonas, B.M.; Murray, B.E.; Weinstock, G.M.; Rogers, B. Enterococcus faecalis multi-drug resistance transporters: Applications for antibiotic discovery. J. Microbiol. Biotechnol. 2001, 3, 179–184. [Google Scholar]
- Jonas, B.M.; Murray, B.E.; Weinstock, G.M. Characterization of emeA, a NorA homolog and multidrug resistance efflux pump, in Enterococcus faecalis. Antimicrob. Agents Chemother. 2001, 45, 3574–3579. [Google Scholar] [CrossRef]
- Nowroozi, J.; Akhavan Sepahi, A.; Sabokbar, A. Comparison of gyrA and parC mutations in ciprofloxacin-resistant and -susceptible Enterococcus faecalis isolates. J. Med. Microbiol. 2014, 63, 68–74. [Google Scholar]
- Mahapatra, A.; Raj Kumar Patro, A.; Khajuria, A.; Dhal, S.; Praharaj, A.K. Ciprofloxacin-resistant Gram-negative isolates from a tertiary care hospital in Eastern India with novel gyrA and parC gene mutations. Med. J. Armed Forces India 2022, 78, 24–31. [Google Scholar] [CrossRef]
- Luna, V.A.; Roberts, M.C. The presence of the tet(O) gene in both tetracycline-resistant and -susceptible strains of Streptococcus pneumoniae. J. Antimicrob. Chemother. 1998, 42, 613–619. [Google Scholar] [CrossRef]
- Koike, S.; Krapac, I.G.; Oliver, H.D.; Yannarell, A.C.; Chee-Sanford, J.C.; Aminov, R.I.; Mackie, R.I. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 2007, 73, 4813–4823. [Google Scholar] [CrossRef]
- Macauley, J.J.; Qiang, Z.; Adams, C.D.; Surampalli, R.; Mormile, M.R. Disinfection of swine wastewater using chlorine, ultraviolet light and ozone. Water Res. 2007, 41, 855–863. [Google Scholar] [CrossRef]
- Stenström, T.A.; Okoh, A.I.; Adegoke, A.A. Antibiogram of environmental isolates of Acinetobacter calcoaceticus from Nkonkobe Municipality, South Africa. Fresenius Environ. Bull. 2016, 25, 3059–3065. [Google Scholar] [CrossRef]
- Mahmoudpour, A.; Rahimi, S.; Sina, M.; Soroush, M.H.; Shahisa, S.; Asl-Aminabadi, N. Isolation and identification of Enterococcus faecalis from necrotic root canals using multiplex PCR. J. Oral Sci. 2007, 49, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Mundy, L.M.; Sahm, D.F.; Gilmore, M. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 2000, 13, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Okoh, A.I. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol. 2014, 59, 133–140. [Google Scholar] [CrossRef]
- Depardieu, F.; Perichon, B.; Courvalin, P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 2004, 42, 5857–5860. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 13 February 2025).
- Chuang, O.N.; Schlievert, P.M.; Wells, C.L.; Manias, D.A.; Tripp, T.J. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect. Immun. 2009, 77, 539–548. [Google Scholar] [CrossRef]
- Lee, M.G.; Kang, M.J.; Kim, S.; Jeong, H.; Kang, D.K.; Paik, H.D.; Park, Y.S. Safety Assessment of Levilactobacillus brevis KU15006: A Comprehensive Analysis of its Phenotypic and Genotypic Properties. Prob. Antimicrob. Prot. 2024, 1–15. [Google Scholar] [CrossRef]
- Biswas, P.P.; Dey, S.; Sen, A.; Adhikan, L. Molecular characterization of virulence genes in vancomycin-resistant and vancomycin-sensitive enterococci. J. Glob. Infect. Dis. 2016, 8, 16–24. [Google Scholar] [CrossRef]
- Kim, S.J.; Shin, S.Y.; Kang, S.J.; Kim, T.H. Biofilm formation and virulence factors in clinical Enterococcus faecalis isolates from patients with urinary tract infections in Korea. J. Med. Microbiol. 2016, 65, 1165–1173. [Google Scholar] [CrossRef]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016, 20, 127–133. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J. Pollution from drug manufacturing: Review and perspectives. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130571. [Google Scholar] [CrossRef]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Krone, S.M.; Larsson, D.G.; Li, X.Z.; Robinson, J.A.; Simonet, P.; Tiedje, J.M. Influence of humans on evolution and mobilization of environmental antibiotic resistome. Emerg. Infect. Dis. 2011, 17, 1205. [Google Scholar] [CrossRef]
- Finley, R.L.; Collignon, P.; Larsson, D.G.; McEwen, S.A.; Li, X.Z. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef]
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.F.; Yannarell, A.C.; Fate, G.D. Fate and transport of antibiotic residues and antibiotic resistance genes following land application of manure waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef]
- Muziasari, W.I.; Pitkänen, L.K.; Sorum, H.; Stedtfeld, R.D.; Tiedje, J.M.; Virta, M. The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below marine fish farms. Front. Microbiol. 2014, 5, 465. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Macrolide, glycopeptide resistance and virulence genes in Enterococcus species isolates from dairy cattle. J. Med. Microbiol. 2016, 65, 641–648. [Google Scholar] [CrossRef]
- Pignata, C.; Fea, E.; Rovere, R.; Degan, R.; Lorenzi, E.; de Ceglia, M.; Schilirò, T.; Gilli, G. Chlorination in a wastewater treatment plant: Acute toxicity effects of the effluent and of the recipient water body. Environ. Monit. Assess. 2012, 184, 2091–2103. [Google Scholar] [CrossRef]
- Osman, K.; Alvarez-Ordóñez, A.; Ruiz, L.; Badr, J.; ElHofy, F.; Al-Maary, K.S.; Moussa, I.M.; Hessain, A.M.; Orabi, A.; Saad, A.; et al. Antimicrobial resistance and virulence characterization of Staphylococcus aureus and coagulase-negative staphylococci from imported beef meat. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 35. [Google Scholar] [CrossRef] [PubMed]
- Kotzamanidis, C.; Zdragas, A.; Kourelis, A.; Moraitou, E.; Papa, A.; Yiantzi, V.; Pantelidou, C.; Yiangou, M. Characterization of vanA-type Enterococcus faecium isolates from urban and hospital wastewater and pigs. J. Appl. Microbiol. 2009, 107, 997–1005. [Google Scholar] [CrossRef]
- Li, S.; Zhou, Y.; He, F.; Raheem, A.; Yang, H.; Pan, Y.; Pan, Z. Highly efficient capture of antibiotic resistance genes in wastewater using novel biochar-based hybrid adsorbents. Sci. Total Environ. 2023, 863, 160738. [Google Scholar] [CrossRef]
- Johnson, A.P.; Warner, M.; Hallas, G.; Livermore, D.M. Susceptibility to quinupristin/dalfopristin and other antibiotics of van-comycin-resistant enterococci from the UK, 1997 to mid-1999. J. Antimicrob. Chemother. 2000, 46, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.F.; Alvarez-Alvarez, R.; Liras, P. Penicillin-binding proteins, β-lactamases, and β-lactamase inhibitors in β-lactam-producing actinobacteria: Self-resistance mechanisms. Int. J. Mol. Sci. 2022, 23, 5662. [Google Scholar] [CrossRef]
- Figueira, V.; Vaz-Moreira, I.; Silva, M.; Manaia, C.M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and wastewater treatment plants. Water Res. 2011, 45, 5599–5611. [Google Scholar] [CrossRef]
- Said, L.B.; Klibi, N.; Lozano, C.; Dziri, R.; Slama, K.B.; Boudabous, A.; Torres, C. Diversity of enterococcal species and characterization of high-level aminoglycoside resistant enterococci of samples of wastewater and surface water in Tunisia. Sci. Total Environ. 2015, 530, 11–17. [Google Scholar] [CrossRef]
- Hashem, Y.A.; Abdelrahman, K.A.; Aziz, R.K. Phenotype–genotype correlations and distribution of key virulence factors in Enterococcus faecalis isolated from patients with urinary tract infections. Infect. Drug Res. 2021, 14, 1713–1723. [Google Scholar] [CrossRef]
- Kajihara, T.; Nakamura, S.; Iwanaga, N.; Oshima, K.; Takazono, T.; Miyazaki, T.; Izumikawa, K.; Yanagihara, K.; Kohno, N.; Kohno, S. Clinical characteristics and risk factors of enterococcal infections in Nagasaki, Japan: A retrospective study. BMC Infect. Dis. 2015, 15, 426. [Google Scholar] [CrossRef]
- Revathy, S.; Sridharan, K.S.; Elumalai, A.S.; Umasekar, U. Phenotypic detection of high-level aminoglycoside resistance (HLAR) in Enterococcus species in a tertiary care centre. J. Clin. Diagn. Res. 2009, 3, 1785–1790. [Google Scholar]
- La-Rosa, S.L.; Montealegre, M.C.; Singh, K.V.; Murray, B.E. Enterococcus faecalis Ebp pili are important for cell-cell aggregation and intraspecies gene transfer. Microbiology 2016, 162, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.; Gibbs, P.A.; Teixeira, P. Virulence factors among enterococci isolated from traditional fermented meat products produced in the North of Portugal. Food Control 2010, 21, 651–656. [Google Scholar] [CrossRef]
- Rahman, M.M.; Hasan, M.; Ahmed, A. Potential detrimental role of soluble ACE2 in severe COVID-19 comorbid patients. Rev. Med. Virol. 2021, 31, 1–12. [Google Scholar] [CrossRef]
- Maheshwari, M.; Ahmad, I.; Althubiani, A.S. Isolation and molecular characterization of multidrug-resistant Enterococcus faecalis from clinical samples. Trop. J. Pharm. Res. 2016, 15, 1207–1213. [Google Scholar]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence factors of Enterococcus spp. presented in food. LWT 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Gonzalez, B.; Pham, P.; Top, J.; Willems, R.J.L.; van Schaik, W.; van Passel, M.W.J.; Smidt, H. Characterization of Enterococcus isolates colonizing the intestinal tract of intensive care unit patients receiving selective digestive decontamination. Front. Microbiol. 2017, 8, 1596. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Faleye, A.C.; Stenstrom, T.A. Residual antibiotics, antibiotic-resistant superbugs, and antibiotic resistance genes in surface water catchments: Public health impact. Phys. Chem. Earth 2018, 105, 177–183. [Google Scholar] [CrossRef]
- Vincent, S.; Minkler, P.; Bincziewski, B.; Etter, L.; Shlaes, D.M. Vancomycin resistance in Enterococcus gallinarum. Antimicrob. Agents Chemother. 1992, 36, 1392–1399. [Google Scholar] [CrossRef]
- Hao, L.; Wang, H. Successful treatment of Enterococcus gallinarum infection in a neonate with vancomycin: A case report. BMC Pediatr. 2024, 24, 527. [Google Scholar] [CrossRef]
- Tharvornvee, W.; Pruksakorn, C.; Lekcharoensuk, P. Inducible vancomycin resistance is common in porcine Enterococcus gallinarum and E. casseliflavus isolates. Thai J. Vet. Med. 2016, 46, 627–635. [Google Scholar] [CrossRef]
- García-Solache, M.; Rice, L.B. The Enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef]
- Lioy, P.J. Exposure science: A view of the past and milestones for the future. Environ. Health Perspect. 2010, 118, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Keraita, B.; Amoah, P. Wastewater use in urban and peri-urban vegetable farming. In Wastewater Irrigation and Health; Drechsel, P., Scott, C.A., Raschid-Sally, L., Eds.; IWMI: Colombo, Sri Lanka, 2011; pp. 11–25. [Google Scholar]
- Bonetta, S.; Pignata, C.; Gasparro, E.; Richiardi, L.; Bonetta, S.; Carraro, E. Impact of wastewater treatment plants on microbiological contamination for evaluating the risks of wastewater reuse. Environ. Sci. Eur. 2022, 34, 20. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial resistance in Enterococcus spp. of animal origin. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Wiley: Hoboken, NJ, USA, 2018; pp. 185–227. [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC). European Centre for Disease Prevention and Control Publishes Annual Epidemiological Report 2011. Eurosurveillance 2011, 16, 20012. [Google Scholar]
- Ammerlaan, H.S.; Harbarth, S.; Buiting, A.G.; Crook, D.W.; Fitzpatrick, F.; Hanberger, H.; Herwaldt, L.A.; Van Keulen, P.H.; Kluytmans, J.A.; Kola, A.; et al. Secular trends in nosocomial bloodstream infections: Antibiotic-resistant bacteria increase the total burden of infection. Clin. Infect. Dis. 2013, 56, 798–805. [Google Scholar] [CrossRef] [PubMed]
- De Kraker, M.E.; Jarlier, V.; Monen, J.C.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Didem, K.A.R.T.; Kuştimur, A.S. Investigation of gelatinase gene expression and growth of Enterococcus faecalis clinical isolates in biofilm models. Turk. J. Pharm. Sci. 2019, 16, 356. [Google Scholar]
- Holá, V.; Ruzicka, F.; Horka, M. Microbial diversity in biofilm infections of the urinary tract with the use of sonication techniques. FEMS Immunol. Med. Microbiol. 2010, 59, 525–528. [Google Scholar] [CrossRef]
- Dworniczek, E.; Piwowarczyk, J.; Bania, J.; Kowalska-Krochmal, B.; Wałecka, E.; Seniuk, A.; Dolna, I.; Gościniak, G. Enterococcus in wound infections: Virulence and antimicrobial resistance. Acta Microbiol. Immunol. Hung. 2012, 59, 263–269. [Google Scholar] [CrossRef]
- Sava, I.; Heikens, E.; Toma, I.; Kropec, A.; Willems, R.; Hübner, J. Enterococcal surface protein is a virulence factor in bacteremia but is not a target of opsonic antibodies in E. faecium infection. In Proceedings of the American Society for Microbiology 109th General Meeting, Philadelphia, PA, USA, 17–21 May 2009. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).