Aeromonas caviae subsp. aquatica subsp. nov., a New Multidrug-Resistant Subspecies Isolated from a Drinking Water Storage Tank
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Characterization and Identification of Isolates
2.3. Genome Sequencing and Bioinformatic Analysis
2.4. Genome Sequence Data Availability
3. Results and Discussion
3.1. Characterization of Isolates
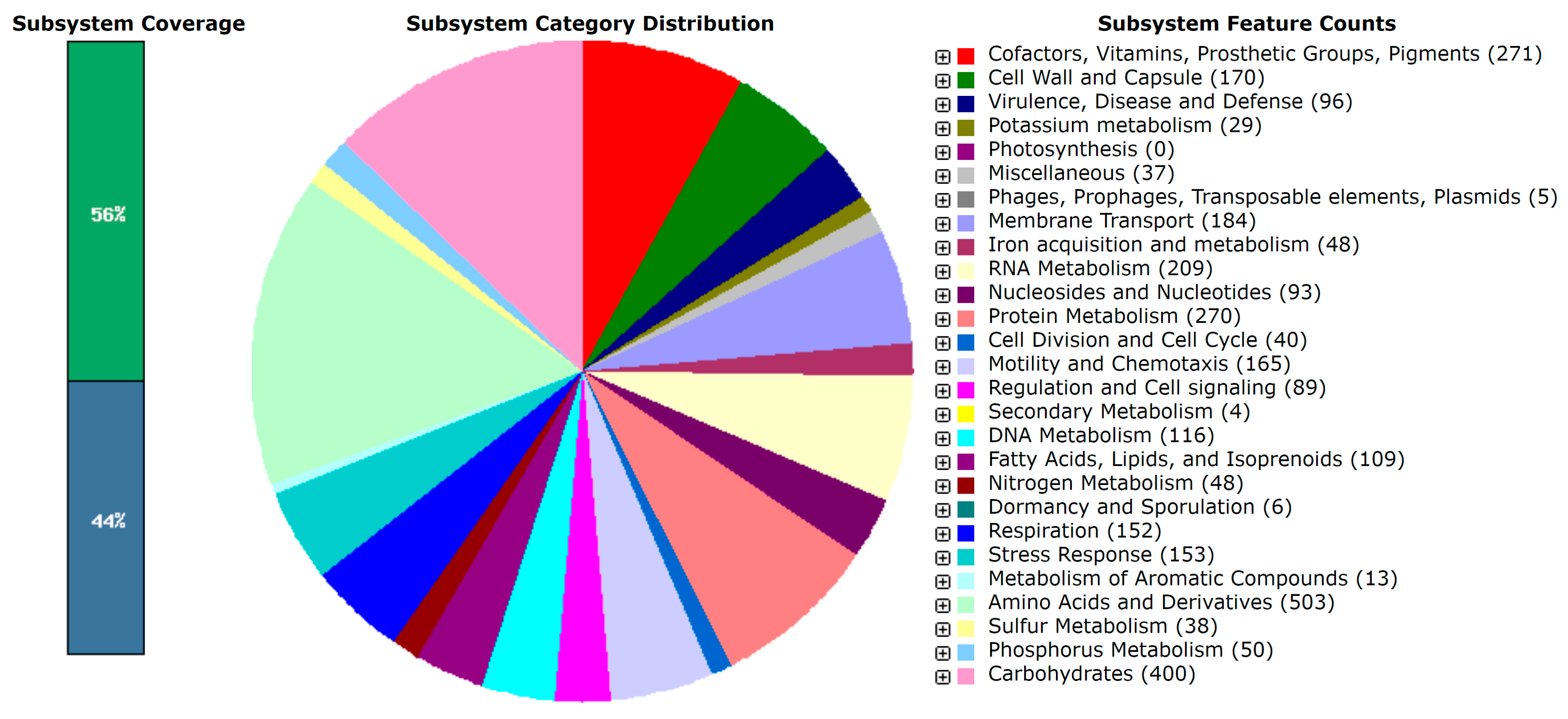
3.2. Genomic and Lipid Profiles
3.3. Antibiotic Resistance and Virulence Factors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Guidelines for Drinking-Water Quality, 4th ed.; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Egorov, A.I.; Best, J.M.; Frebis, C.P.; Karapondo, M.S. Occurrence of Aeromonas spp. in a random sample of drinking water distribution systems in the USA. J. Water Health 2011, 9, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, M.; Popowska, M. The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 2014, 64, 921–934. [Google Scholar] [CrossRef]
- Szewzyk, U.; Szewzyk, R.; Manz, W.; Schleifer, K.H. Microbiological safety of drinking water. Annu. Rev. Microbiol. 2000, 54, 81–127. [Google Scholar] [CrossRef]
- Nwachcuku, N.; Gerba, C.P. Emerging waterborne pathogens: Can we kill them all? Curr. Opin. Biotechnol. 2004, 15, 75–180. [Google Scholar] [CrossRef]
- Miyagi, K.; Sanob, K.; Hirala, I. Sanitary evaluation of domestic water supply facilities with storage tanks and detection of Aeromonas, enteric and related bacteria in domestic water facilities in Okinawa Prefecture of Japan. Water Res. 2017, 119, 171–177. [Google Scholar] [CrossRef]
- Wingender, J.; Flemming, H.C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health 2011, 214, 417–423. [Google Scholar] [CrossRef]
- Falkinham, J.O.; Pruden, A.; Edwards, M. Opportunistic Premise Plumbing Pathogens: Increasingly Important Pathogens in Drinking Water. Pathogens 2015, 4, 373–386. [Google Scholar] [CrossRef]
- Russo, S.C.S.; Shano, A.; Agapito, D.G.; da Silva, A.S.; Ferreira, J.V.R.; Cardoso, A.M. Isolation and identification of microbial strains with biotechnological potential for water quality control in aquaculture. Braz. J. Dev. 2021, 7, 117231–117241. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef]
- Hiransuthikul, N.; Tantisiriwat, W.; Lertutsahakul, K.; Vibhagool, A.; Boonma, P. Skin and Soft-Tissue Infections among Tsunami Survivors in Southern Thailand. Clin. Infect. Dis. 2005, 41, e93–e96. [Google Scholar] [CrossRef]
- Sinclair, H.A.; Edwards, F.; Harris, P.N.A.; Heney, C.; Laupland, K.B. Epidemiology of Aeromonas Species Bloodstream Infection in Queensland, Australia: Association with Regional and Climate Zones. Microorganisms 2022, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Grilo, M.L.; Pereira, A.; Sousa-Santos, C.; Robalo, J.I.; Oliveira, M. Climatic Alterations Influence Bacterial Growth, Biofilm Production and Antimicrobial Resistance Profiles in Aeromonas spp. Antibiotics 2021, 10, 1008. [Google Scholar] [CrossRef]
- Li, W.; Liu, C.; Ho, H.C.; Shi, L.; Zeng, Y.; Yang, X.; Huang, Q.; Pei, Y.; Huang, C.; Yang, L. Association between antibiotic resistance and increasing ambient temperature in China: An ecological study with nationwide panel data. Lancet Reg. Health West. Pac. 2022, 30, 100628. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, H.; Alizadeh, A.; Vafaie, M.; Maleki, M.R.; Khoei, S.G. An estimation of global Aeromonas infection prevalence in children with diarrhoea: A systematic review and meta-analysis. BMC Pediatr. 2023, 23, 254. [Google Scholar] [CrossRef] [PubMed]
- de Melo, B.S.T.; Mendes-Marques, C.L.; Campos, T.L.; Almeida, A.M.P.; Leal, N.C.; Xavier, D.E. High-resolution genome-wide analysis is essential for the identification of ambiguous Aeromonas strains. FEMS Microbiol Lett. 2019, 366, fnz245. [Google Scholar] [CrossRef]
- Hofer, E.; Reis, C.M.; Theophilo, G.N.; Cavalcanti, V.O.; Lima, N.V.; Henriques, M.F. Aeromonas associated with an acute diarrhea outbreak in São Bento do Una, Pernambuco. Rev. Soc. Bras. Med. Trop. 2006, 39, 217–220. [Google Scholar] [CrossRef]
- Kumar, M.R.; Venkatesh, V.N.; Sudhindra, K.S. Aeromonas species isolated from a case of meningitis. Indian J. Pathol. Microbiol. 2014, 57, 521–522. [Google Scholar] [CrossRef]
- Pessoa, R.B.G.; de Oliveira, W.F.; Correia, M.T.D.S.; Fontes, A.; Coelho, L.C.B.B. Aeromonas and Human Health Disorders: Clinical Approaches. Front. Microbiol. 2022, 13, 868890. [Google Scholar] [CrossRef]
- Schwartz, K.; Borowiak, M.; Strauch, E.; Deneke, C.; Richter, M.H. German Aeromonas Study Group. Emerging Aeromonas spp. infections in Europe: Characterization of human clinical isolates from German patients. Front. Microbiol. 2024, 15, 1498180. [Google Scholar] [CrossRef]
- Chao, C.M.; Lai, C.C.; Tang, H.J.; Ko, W.C.; Hsueh, P.R. Skin and soft-tissue infections caused by Aeromonas species. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.C.; LaMartina, E.L.; Lewis, J.R.; Dahl, A.J.; Nadig, N.; Szabo, A.; Newton, R.J.; Skwor, T.A. One Health and Global Health View of Antimicrobial Susceptibility through the “Eye” of Aeromonas: Systematic Review and Meta-Analysis. Int. J. Antimicrob. Agents 2023, 62, 106848. [Google Scholar] [CrossRef]
- Bojar, B.E.; Craig, A.T., 3rd; Leduc, A.; Blumenthal, M.; Mayo, B.; Ahmed, A.S.; Cahak, C.; Beattie, R.; Skwor, T. Similar antimicrobial resistance and virulence profiles among Aeromonas isolates from recreational beaches, post-chlorinated wastewater and clinical samples in Milwaukee, Wisconsin USA. Sci. Total Environ. 2025, 970, 179035. [Google Scholar] [CrossRef] [PubMed]
- Skwor, T.; Stringer, S.; Haggerty, J.; Johnson, J.; Duhr, S.; Johnson, M.; Seckinger, M.; Stemme, M. Prevalence of Potentially Pathogenic Antibiotic-Resistant Aeromonas spp. in Treated Urban Wastewater Effluents versus Recipient Riverine Populations: A 3-Year Comparative Study. Appl. Environ. Microbiol. 2020, 86, e02053-19. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, L.; Yan, C.; Lin, N.; Zhang, Y.; Sha, Y.; Zhao, J.; Lu, J.; Bao, Q.; Zhang, G. Whole-genome sequencing-based species classification, multilocus sequence typing, and antibiotic resistance mechanisms of the clinical Aeromonas complex. Front. Microbiol. 2025, 16, 1473150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef]
- Bianco, K.; de Farias, B.O.; Gonçalves-Brito, A.S.; Nascimento, A.P.A.; Magaldi, M.; Montenegro, K.; Flores, C.; Oliveira, S.; Monteiro, M.A.; Spisso, B.F.; et al. Mobile resistome of microbial communities and antimicrobial residues from drinking water supply systems in Rio de Janeiro, Brazil. Sci. Rep. 2022, 12, 19050. [Google Scholar] [CrossRef]
- Sobral, M.M.R.; Barreto, C.; Bianco, K.; Oliveira, S.S.; Clementino, M.M. Virulence determinants in genetically heterogeneous populations of Aeromonads recovered from an urban lagoon. J. Water Health 2019, 17, 380–392. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- VET04-A2; Methods for Broth Dilution Susceptibility Testing of Bacteria Isolated from Aquatic Animals; Approved Guideline, 2nd Edn. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014.
- Baron, S.; Granier, S.A.; Larvor, E.; Jouy, E.; Cineux, M.; Wilhelm, A.; Gassilloud, B.; Le Bouquin, S.; Kempf, I.; Chauvin, C. Aeromonas Diversity and Antimicrobial Susceptibility in Freshwater-An Attempt to Set Generic Epidemiological Cut-Off Values. Front. Microbiol. 2017, 8, 503. [Google Scholar] [CrossRef]
- Lee, C.; Cho, J.C.; Lee, S.H.; Lee, D.G.; Kim, S.J. Distribution of Aeromonas spp. as identified by 16S rDNA restriction fragment length polymorphism analysis in a trout farm. J. Appl. Microbiol. 2002, 93, 976–985. [Google Scholar] [CrossRef]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, 606–612. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Juliao, F.C.; Tonani, K.A.A.; Fregonesi, B.M.; Zagui, G.S.; Machado, C.S.; da Silva, T.V.; Machado, G.P.; Della Torre, G.C.; Muñoz, S.S. Storage Tanks for Household Water Usage in Brazil: Microbiological and Chemical Quality, and Maintenance of Sanitary Conditions. Arq. Ciên. Saud. 2021, 28, 11–15. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing. Data from the EUCAST MIC Distribution Website. Available online: https://www.eucast.org (accessed on 8 April 2025).
- Kahlmeter, G.; Turnidge, J. Wild-type distributions of minimum inhibitory concentrations and epidemiological cut-off values-laboratory and clinical utility. Clin. Microbiol. Rev. 2023, 36, e0010022. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, H.; Liu, Y.; Liu, X.; Qiao, J.; Ge, H.; Zhao, J.; Ma, X.; Chen, M.; Liu, R. Genetic characterization and virulence determinants of multidrug-resistant NDM-1-producing Aeromonas caviae. Front. Microbiol. 2023, 13, 1055654. [Google Scholar] [CrossRef]
- Luo, X.; Yin, Z.; Yu, L.; Zhang, J.; Hu, D.; Xu, M.; Wang, P.; Wang, F.; Feng, J. Genomic analysis of chromosomal cointegrated blaNDM-1-carrying ICE and blaRSA-1-carrying IME from clinical multidrug resistant Aeromonas caviae. Front. Cell. Infect. Microbiol. 2023, 13, 1131059. [Google Scholar] [CrossRef] [PubMed]
- BRCAST (Brazilian Committee on Antimicrobial Susceptibility Testing) Clinical Breakpoints Table. Version 13.0 [Internet] 2023. Available online: https://brcast.org.br (accessed on 10 December 2024).
- Cardoso, A.M.; Flores, V.R.; do Rosario, G.G.; Succar, J.B.; Berbert, L.C.; Oliveira, M.C.F.; Canellas, A.L.B.; Laport, M.S.; Souza, C.R.V.M.; Chagas, T.P.G.; et al. Antimicrobial Susceptibility of Escherichia coli Isolates Causing Community-Acquired Urinary Tract Infections: Comparison of Methods. Microorganisms 2025, 13, 231. [Google Scholar] [CrossRef]
- CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Supplement M100, 30th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Moriel, B.; Cruz, L.M.; Dallagassa, C.B.; Faoro, H.; de Souza, E.M.; Pedrosa, F.O.; Rego, F.G.; Picheth, G.; Fadel-Picheth, C.M. Draft Genome Sequence of Aeromonas caviae BLM, Isolated from Stool culture of a child with diarrhea. Genome Announc. 2015, 3, e00524-15. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Russell, N.J. Mechanisms of thermal adaptation in bacteria: Blueprints for survival. Trends Biochem. Sci. 1984, 9, 108–112. [Google Scholar] [CrossRef]
- Kaneda, T. Iso- and anteiso-fatty acids in bacteria: Biosynthesis, function, and taxonomic significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef]
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J Bacteriol. 2006, 188, 8272–8282. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Zhou, Z.; Huang, H.; Ren, Y.; Zhang, Y.; Li, G.; Zhou, Z.; Wang, L. Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 2011, 193, 3389–3390. [Google Scholar] [CrossRef]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef]
- Tomás, J.M. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012, 2012, 256261. [Google Scholar] [CrossRef]
- Figueira, V.; Vaz-Moreira, I.; Silva, M.; Manaia, C.M. Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 2011, 45, 5599–5611. [Google Scholar] [CrossRef] [PubMed]
- Picao, R.C.; Cardoso, J.P.; Campana, E.H.; Nicoletti, A.G.; Petrolini, F.V.B.; Assis, D.M.; Juliano, L.; Galesa, A.C. The route of antimicrobial resistance from the hospital effluent to the environment: Focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn. Microbiol. Infect. Dis. 2013, 76, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H.; Igumbor, E.U.; Aghdasi, F.; Tom, M.; Okoh, A.I. Emerging Aeromonas Species Infections and Their Significance in Public Health. Sci. World J. 2012, 2012, 625023. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.L.; Ko, W.C.; Wu, C.J. Complexity of β-lactamases among clinical Aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 2012, 45, 398–403. [Google Scholar] [CrossRef]
- Wu, C.J.; Chen, P.L.; Wu, J.J.; Yan, J.J.; Lee, C.C.; Lee, H.C.; Lee, N.Y.; Chang, C.M.; Lin, Y.T.; Chiu, Y.C.; et al. Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic Aeromonas isolates. J. Med. Microbiol. 2012, 61, 712–719. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Wu, C.J.; Chuang, Y.C.; Lee, M.F.; Lee, C.C.; Lee, H.C.; Lee, N.Y.; Chang, C.M.; Chen, P.L.; Lin, Y.T.; Yan, J.J.; et al. Bacteremia due to extended-spectrum-β-lactamase-producing Aeromonas spp. at a medical center in Southern Taiwan. Antimicrob. Agents Chemother. 2011, 55, 5813–5818. [Google Scholar] [CrossRef]
- Ko, W.C.; Lee, H.C.; Chuang, Y.C.; Liu, C.C.; Wu, J.J. Clinical features and therapeutic implications of 104 episodes of monomicrobial Aeromonas bacteraemia. J. Infect. 2000, 40, 267–273. [Google Scholar] [CrossRef]
- Bogaerts, P.; Naas, T.; Saegeman, V.; Bonnin, R.A.; Schuermans, A.; Evrard, S.; Bouchahrouf, W.; Jove, T.; Tande, D.; Bolle, X.; et al. OXA-427, a new plasmid-borne carbapenem-hydrolysing class D β-lactamase in Enterobacteriaceae. J. Antimicrob. Chemother. 2017, 72, 2469–2477. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.P.; Llabrés, S.; Neuberger, A.; Blaza, J.N.; Bai, X.C.; Okada, U.; Murakami, S.; van Veen, H.W.; Zachariae, U.; Scheres, S.H.W.; et al. Structure of the MacAB-ToIC ABC-type tripartite multidrug efflux pump. Nat. Microbiol. 2017, 2, 17070. [Google Scholar] [CrossRef] [PubMed]
- Valdes, N.; Espinoza, C.; Sanhueza, L.; Gonzalez, A.; Corsini, G.; Tello, M. Draft genome sequence of the Chilean isolate Aeromonas salmonicida strain CBA100. FEMS Microbiol. Lett. 2015, 362, frnu062. [Google Scholar] [CrossRef]
- Gofil-Urriza, M.; Arpin, C.; Capdepuy, M.; Dubois, V.; Caumette, P.; Quentin, C. Type II topoisomerase quinolone resistance-determining regions of Aeromonas caviae, A. hydrophila, and A. sobria complexes and mutations associated with quinolone resistance. Antimicrob. Agents Chemother. 2002, 46, 350–359. [Google Scholar] [CrossRef]
- Alcaide, E.; Blasco, M.D.; Esteve, C. Mechanisms of quinolone resistance in Aeromonas species isolated from humans, water and eels. Res. Microbiol. 2010, 161, 40–45. [Google Scholar] [CrossRef]
- Sanchez-Cespedes, J.; Marti, S.; Soto, S.M.; Alba, V.; Melción, C.; Almela, M.; Marco, F.; Vila, J. Two chromosomally located qnrB variants, qnrB6 and the new qnrB16, in Citrobacter spp. isolates causing bacteraemia. Clin. Microbiol. Infect. 2009, 15, 1132–1138. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2014, 2, 1–42. [Google Scholar] [CrossRef]
- Hadi, N.; Yang, Q.; Barnett, T.C.; Tabei, S.M.; Kirov, S.M.; Shaw, J.G. Bundle-forming pilus locus of Aeromonas veronii bv. Sobria. Infect. Immun. 2012, 80, 1351–1360. [Google Scholar] [CrossRef]
- Barnett, T.C.; Kirov, S.M. The type IV Aeromonas pilus (Tap) gene cluster is widely conserved in Aeromonas species. Microb. Pathog. 1999, 26, 77–84. [Google Scholar] [CrossRef]
- Kirov, S.M.; Barnett, T.C.; Pepe, C.M.; Strom, M.S.; Albert, M.J. Investigation of the Role of Type IV Aeromonas Pilus (Tap) in the Pathogenesis of Aeromonas Gastrointestinal Infection. Infect. Immun. 2000, 68, 4040–4048. [Google Scholar] [CrossRef]
- Jiang, X.; Qin, Y.X.; Lin, G.F.; Huang, L.; Huang, B.; Huang, W.S.; Yan, Q.P. FigN plays important roles in the adhesion of Aeromonas hydrophila to host mucus. Genet. Mol. Res. 2015, 14, 6376–6386. [Google Scholar] [CrossRef] [PubMed]
- Canals, R.; Altarriba, M.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomas, J.M.; Merino, S. Analysis of the lateral flagellar gene system of Aeromonas hydrophila AH-3. J. Bacteriol. 2006, 188, 852–862. [Google Scholar] [CrossRef]
- Mohammed, A.N.; Attia, A.S. Control of biofilm-producing Aeromonas bacteria in the water tanks and drinkers of broiler poultry farms using chitosan nanoparticle-based coating thyme oil. Iraqi J. Vet. Sci. 2022, 36, 659–669. [Google Scholar] [CrossRef]
- Albert, M.J.; Ansaruzzaman, M.; Talukder, K.A.; Chopra, A.K.; Kuhn, I.; Rahman, M.; Faruque, A.S.; Islam, M.S.; Sack, R.B.; Mollby, R. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 2000, 38, 3785–3790. [Google Scholar] [CrossRef] [PubMed]
- Erova, T.E.; Sha, J.; Horneman, A.J.; Borchardt, M.A.; Khajanchi, B.K.; Fadl, A.A.; Chopra, A.K. Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol. Lett. 2007, 275, 301–311. [Google Scholar] [CrossRef]
- Erova, T.E.; Kosykh, V.G.; Fadl, A.A.; Sha, J.; Horneman, A.J.; Chopra, A.K. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 2008, 190, 3467–3474. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Lu, M.; Chopra, A.K. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: Characterization of an iron uptake regulator. Infect. Immun. 2001, 69, 6370–6381. [Google Scholar] [CrossRef]
- Sha, J.; Rosenzweig, J.A.; Kozlova, E.V.; Wang, S.; Erova, T.E.; Kirtley, M.L.; van Lier, C.J.; Chopra, A.K. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology 2013, 159, 1120–1135. [Google Scholar] [CrossRef]

| A. caviae Strain | AMK | AMP | ATM | CAZ | TGC | FEP | CIP | LEV | IPM | MEM | SXT | TZP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V15 | 4 (S) | ≥64 (R) | 32 (R) | ≥64 (R) | ≤0.5 (S) | 128 (R) | ≥4 (R) | ≥8 (R) | 8 (R) | ≥16 (R) | ≥150 (R) | ≥128 (R) |
| FAHZZU2447 | 4 (S) | ND | 32 (R) | ≥128 (R) | ≤0.5 (S) | 128 (R) | 16 (R) | ND | 8 (R) | ND | ≥150 (R) | ≥128 (R) |
| 211703 | 16 (S) | ND | ≤1 (S) | ≥64 (R) | ≤0.5 (S) | ND | ≥4 (R) | ≥8 (R) | ≥16 (R) | ≥16 (R) | ≥320 (R) | ≥128 (R) |
| V123 | 4 (S) | ≥64 (R) | 32 (R) | ≥8 (R) | ≤0.5 (S) | 128 (R) | ≥4 (R) | ≥8 (R) | 8 (R) | ≥16 (R) | ≥150 (R) | ≥128 (R) |
| V77 | 4 (S) | ≥64 (R) | 32 (R) | ≥8 (R) | ≤0.5 (S) | 128 (R) | ≥4 (R) | ≥8 (R) | 8 (R) | ≥16 (R) | ≥150 (R) | ≥128 (R) |
| F10 | 4 (S) | ≥64 (R) | ≥8 (R) | ≥8 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥8 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| AE27 | 4 (S) | ≥64 (R) | ≥8 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V2 | 4 (S) | ≥64 (R) | ≥8 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V51 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V13 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V7 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V5 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V44 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| AE21 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| AE33 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| F15 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| F17 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| F18 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V1 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V3 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V4 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V11 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V87 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V55 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V23 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V14 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V61 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V89 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V42 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V71 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V78 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V93 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≥8 (R) |
| V75 | 4 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≤4 (S) |
| V73 | 16 (S) | ≥64 (R) | ≥4 (R) | ≥4 (R) | ≤0.5 (S) | ≥8 (R) | ≥4 (R) | ≥4 (R) | 8 (R) | ≥16 (R) | ≥8 (R) | ≤4 (S) |
| V32 | 16 (S) | ≥64 (R) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤0.25 (S) | ≤2 (S) | ≤0.25 (S) | ≤1 (S) | ≤1 (S) | ≤4 (S) |
| V88 | 4 (S) | ≥64 (R) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤0.25 (S) | ≤2 (S) | ≤0.25 (S) | ≤1 (S) | ≤1 (S) | ≤4 (S) |
| V49 | 4 (S) | ≥64 (R) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤1 (S) | ≤4 (S) |
| V59 | 4 (S) | ≥64 (R) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤1 (S) | ≤4 (S) |
| V53 | 16 (S) | ≥64 (R) | ≤1 (S) | ≤1 (S) | ≤0.5 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤0.25 (S) | ≤1 (S) | ≤1 (S) | ≤4 (S) |
| Subject Strain | dDDH (%) | Base Pairs | G + C Content (%) |
|---|---|---|---|
| Aeromonas caviae NCTC 12244 | 91.70 | 4,586,140 | 61.60 |
| Aeromonas hydrophila subsp. anaerogenes CECT 4221 | 80.80 | 4,576,209 | 61.08 |
| Aeromonas sanarellii LMG 24682 | 78.60 | 4,186,421 | 63.13 |
| Aeromonas taiwanensis LMG 24683 | 76.80 | 4,230,588 | 62.83 |
| Aeromonas media CECT 4232 | 71.50 | 4,467,324 | 61.15 |
| Aeromonas eucrenophila CECT 4224 | 66.50 | 4,534,044 | 61.17 |
| Aeromonas hydrophila ATCC 7966 | 62.70 | 4,744,448 | 61.55 |
| Aeromonas dhakensis CIP 107500 | 61.80 | 4,711,264 | 61.78 |
| Aeromonas aquariorum CECT 7289 | 61.60 | 4,677,943 | 61.95 |
| Aeromonas hydrophila subsp. ranae CIP 107985 | 61.00 | 4,681,175 | 61.56 |
| Aeromonas trota CECT 4255 | 50.40 | 4,332,624 | 60.07 |
| Aeromonas enteropelogenes CECT 4487 | 48.90 | 4,461,928 | 59.70 |
| Functions | Virulence Factors | Related Genes | ||
|---|---|---|---|---|
| A. caviae V15 | A. caviae FAHZZU2447 | A. hydrophila subsp. hydrophila | ||
| Adherence | Lateral flagela | flgCEIJ, fliFGP, lafBCEFKSTUX, lfgABFGHKLMN, lfhAB, lfiEHIJMNQR, maf-5 | flgCEIJ, fliFGP, lafBCEFKSTUX, lfgABFGHKLMN, lfhAB, lfiEHIJMNQR, maf-5 | - |
| Mannose-sensitive hemagglutinin (Msh) pilus, type IV pili | mshABCDEFGI1IJKLMNOPQ | mshABCDEFG1IJKLMNOP | mshBCDEFGI1IJLMNO, mshQ | |
| Polar flagella | cheA-2B-2R-3VWYZ, flaBHJ, flgABCDEFGHIJKLMN, flhABFG, fliAEFGHIJKLMNOPQR, flrABC, maf-1, motXY, pomA2AB2B | cheABRVWYZ, flaBHJ, flgABCDEFGHIJKLMN, flhABFG, fliAEFGHIJKLNOPQR, flrABC, maf-1, motXY, nueB, pomA2AB2B | cheA-2B-2R-3VWYZ, flaABGHJ, flgABCDEFGHIJKLMN, flhABF, fliAEFGHIJKLMNOPQR, flmDH, flrABC, maf-1-2, motXY, nueAB, pmA2AB2B | |
| Tap type IV pili | tapBCDFMNOPQTVW, tapY1, tppABCDEF | tapBCDFMNOPQTUVWY1, tppABCDE | tapABCDFMNOPQTUVW1, tppABEF | |
| Secretion system | T2SS | exeABCDEFGHIJKLMN | exeABCDEFGHIJKLMN | exeABCDEFGHIJKLMN, tapD |
| T6SS | atsDGHIJKLPQS, clpV1, dtU, hcp, vasHK/atsR, vgrG2G3, vipAB | atsD | atsABCDGHIJKLPQS, clpV1, dotU, hcp1, hcp, vasHK/atsR, vgrG1G2G3, vipAB | |
| Toxin | Hemolysin HlyA | hlyA | hlyA | hlyA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, V.H.; Berbert, L.C.; Adesoji, A.T.; Bianco, K.; Cavalcante, J.J.V.; Pellegrino, F.L.P.C.; Albano, R.M.; Clementino, M.M.; Cardoso, A.M. Aeromonas caviae subsp. aquatica subsp. nov., a New Multidrug-Resistant Subspecies Isolated from a Drinking Water Storage Tank. Microorganisms 2025, 13, 897. https://doi.org/10.3390/microorganisms13040897
Moreira VH, Berbert LC, Adesoji AT, Bianco K, Cavalcante JJV, Pellegrino FLPC, Albano RM, Clementino MM, Cardoso AM. Aeromonas caviae subsp. aquatica subsp. nov., a New Multidrug-Resistant Subspecies Isolated from a Drinking Water Storage Tank. Microorganisms. 2025; 13(4):897. https://doi.org/10.3390/microorganisms13040897
Chicago/Turabian StyleMoreira, Victor Hugo, Lidiane Coelho Berbert, Ayodele Timilehin Adesoji, Kayo Bianco, Janaina Japiassu Vasconcelos Cavalcante, Flávia Lúcia Piffano Costa Pellegrino, Rodolpho Mattos Albano, Maysa Mandetta Clementino, and Alexander Machado Cardoso. 2025. "Aeromonas caviae subsp. aquatica subsp. nov., a New Multidrug-Resistant Subspecies Isolated from a Drinking Water Storage Tank" Microorganisms 13, no. 4: 897. https://doi.org/10.3390/microorganisms13040897
APA StyleMoreira, V. H., Berbert, L. C., Adesoji, A. T., Bianco, K., Cavalcante, J. J. V., Pellegrino, F. L. P. C., Albano, R. M., Clementino, M. M., & Cardoso, A. M. (2025). Aeromonas caviae subsp. aquatica subsp. nov., a New Multidrug-Resistant Subspecies Isolated from a Drinking Water Storage Tank. Microorganisms, 13(4), 897. https://doi.org/10.3390/microorganisms13040897









