Different Land Use Systems in the Brazilian Cerrado and Their Effects on Soil Bacterial Communities
Abstract
1. Introduction
2. Materials and Methods
2.1. General Description of the Areas and Soil Sampling
2.2. Physicochemical and Soil Enzyme Analysis
2.3. Amplicon Sequencing
2.4. Statistical Analyses
3. Results
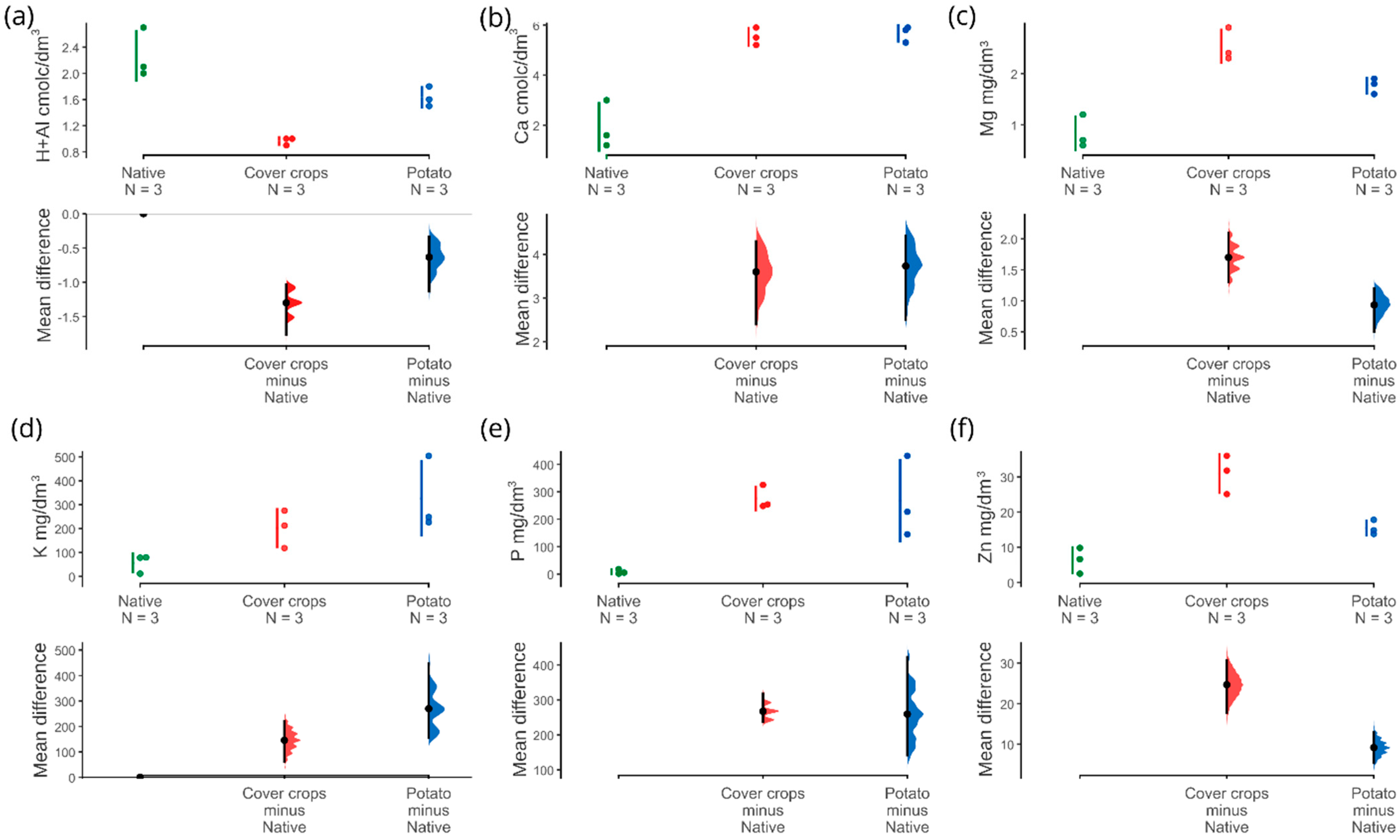
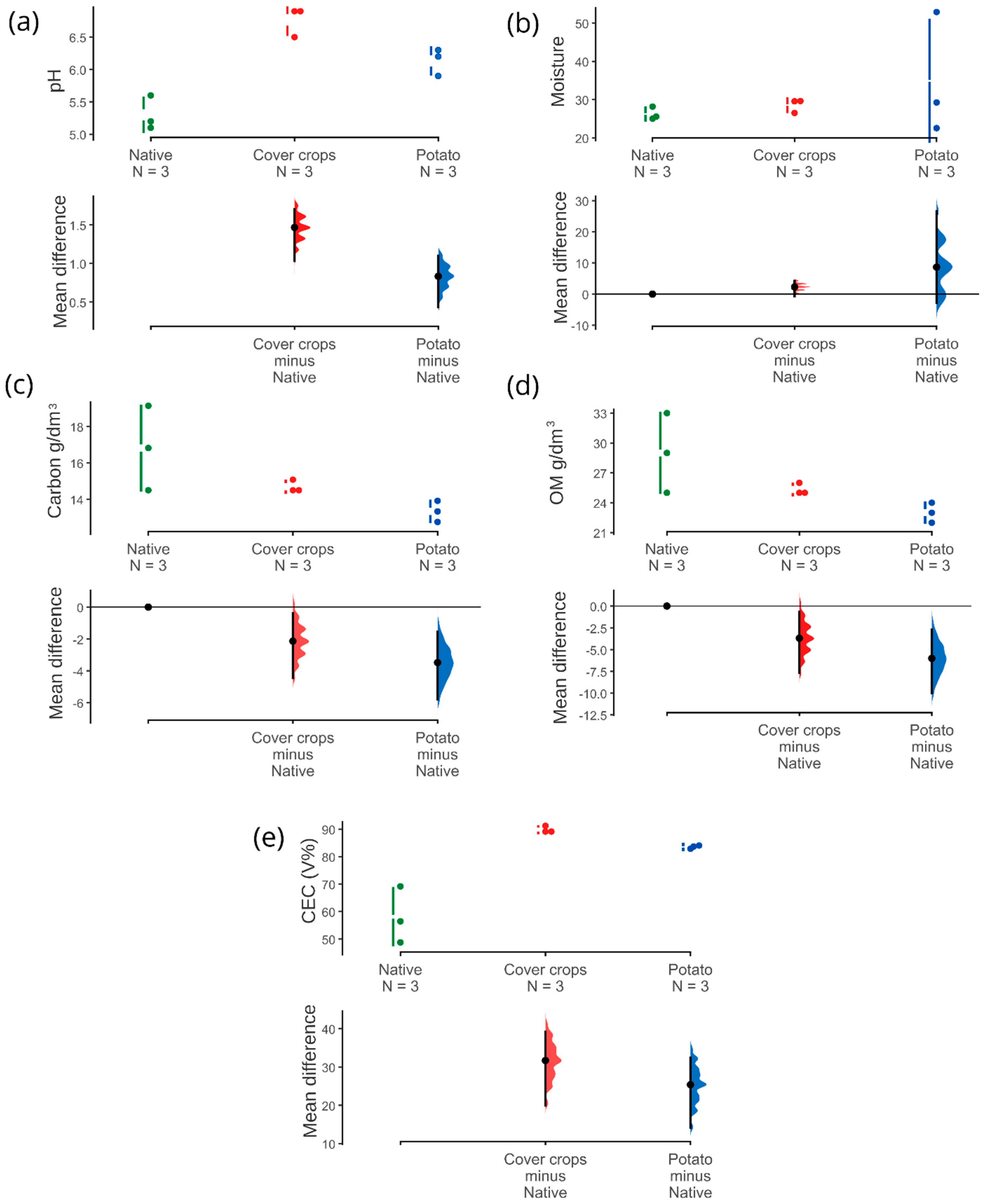
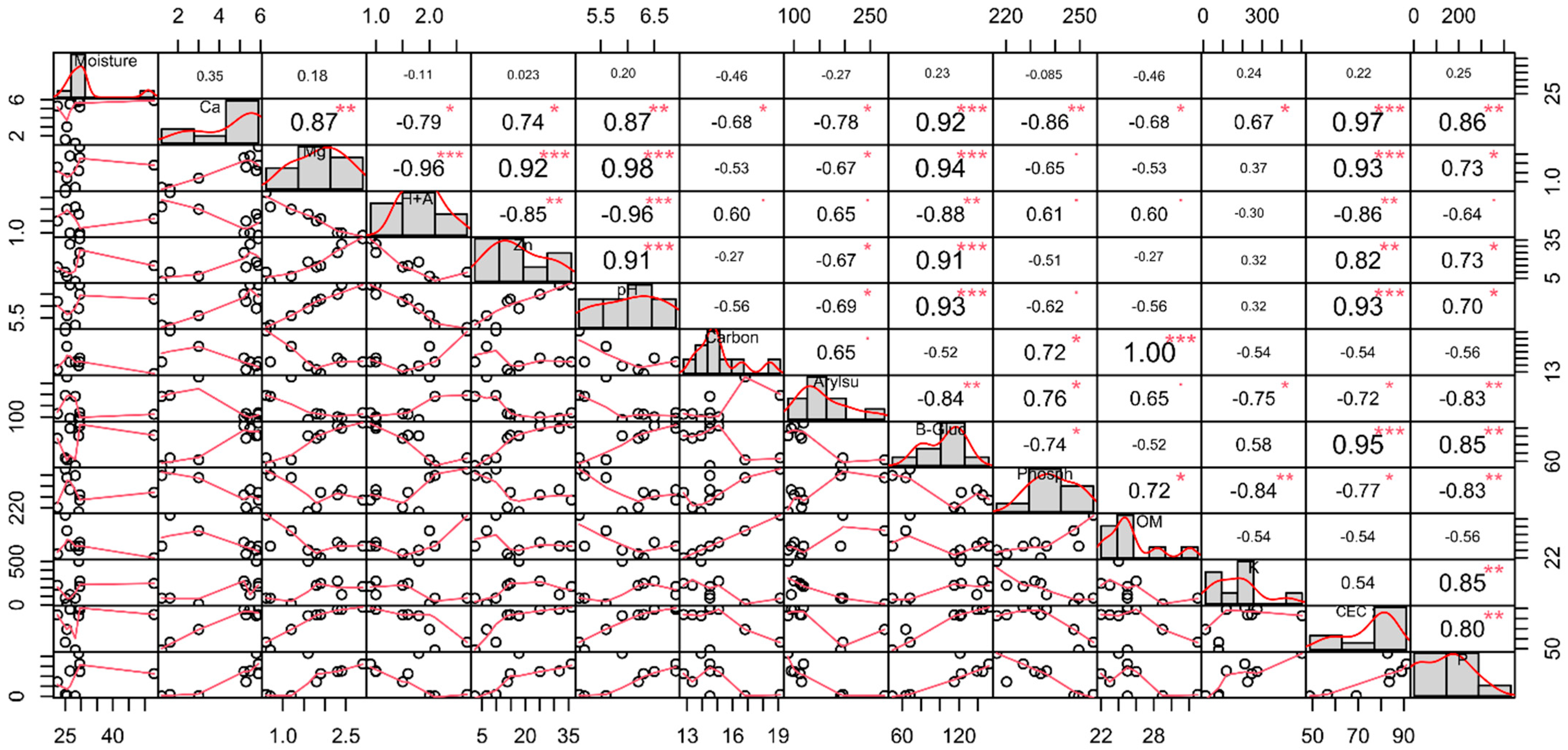
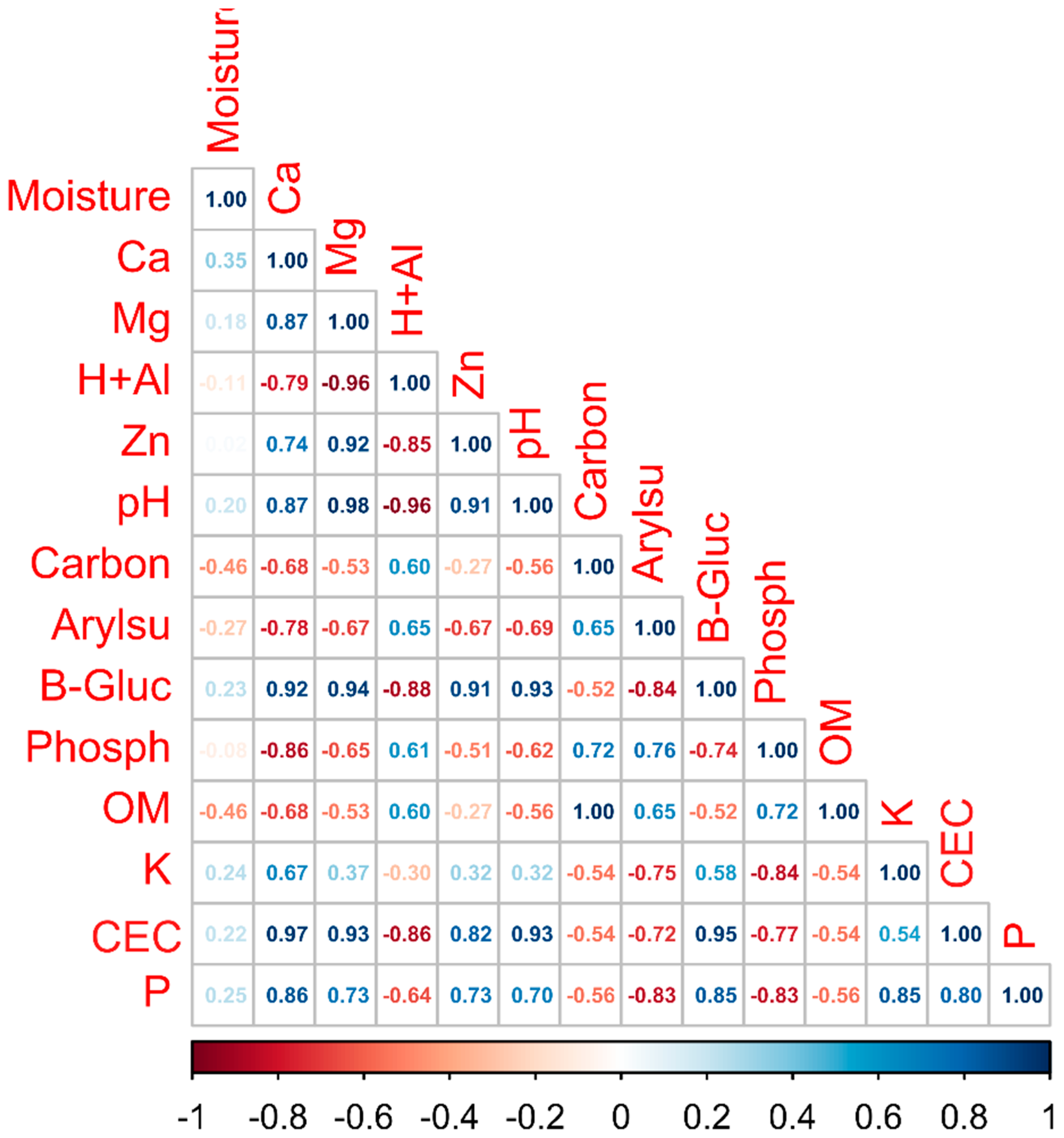
3.1. Physicochemical and Soil Enzyme Analysis
3.2. Relative Abundance
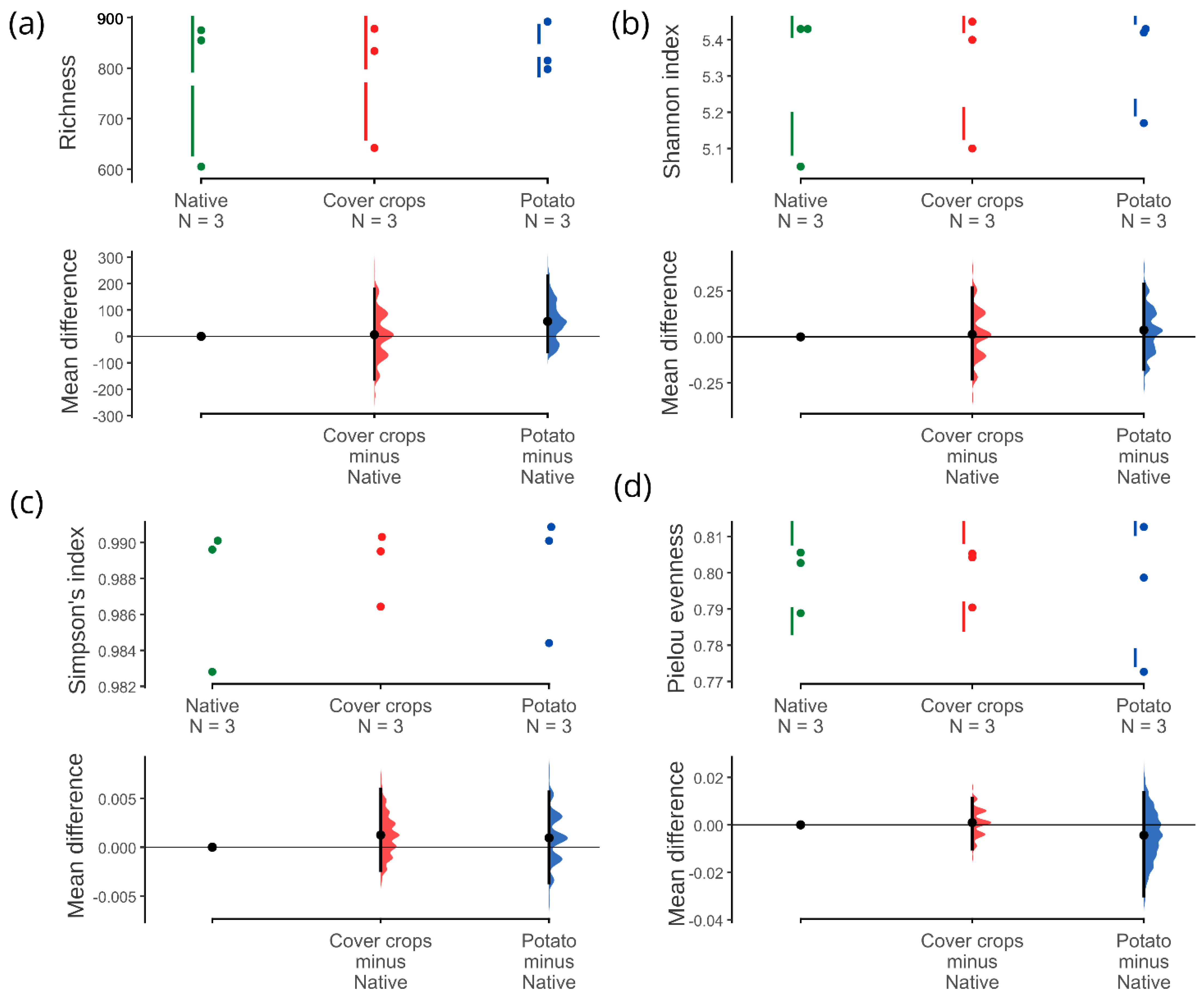
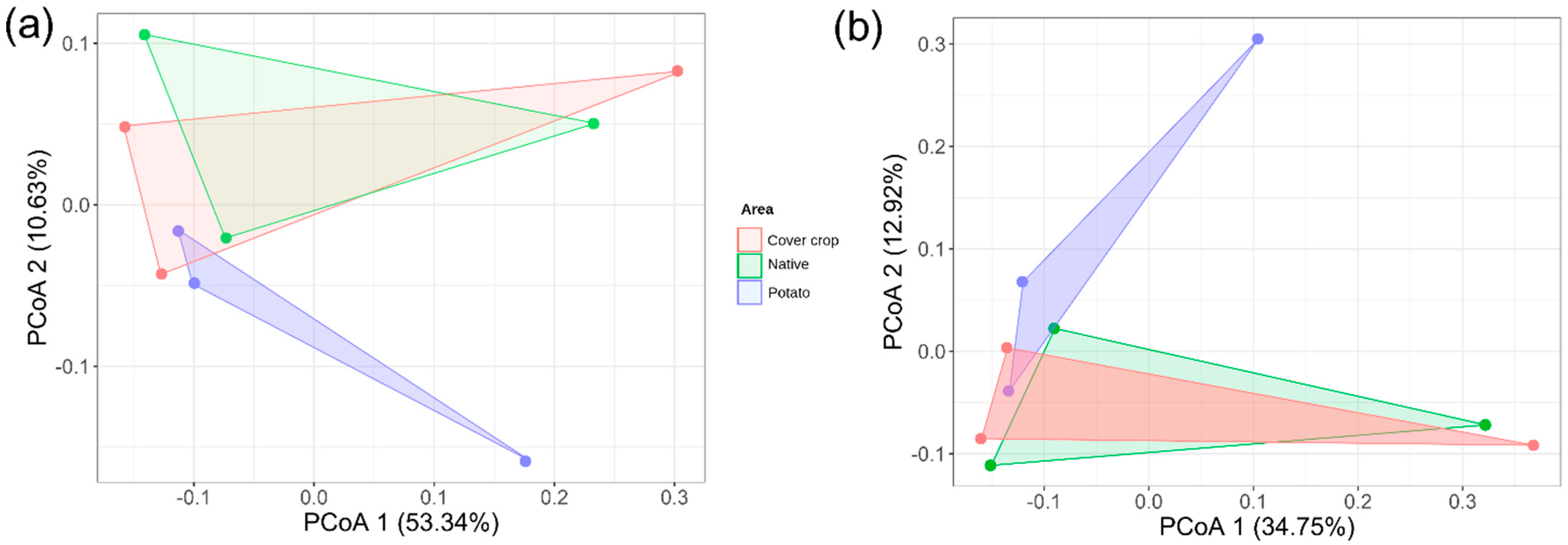
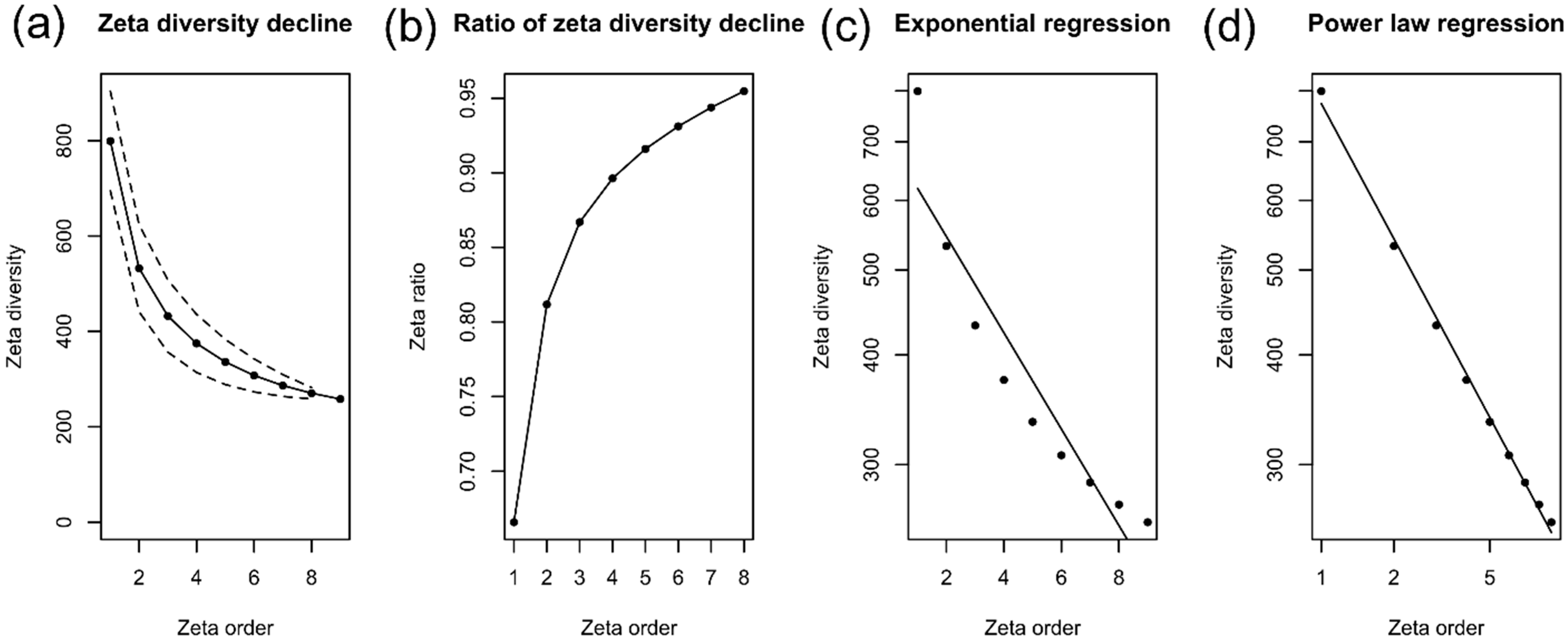
3.3. Alpha, Beta, and Zeta Diversity
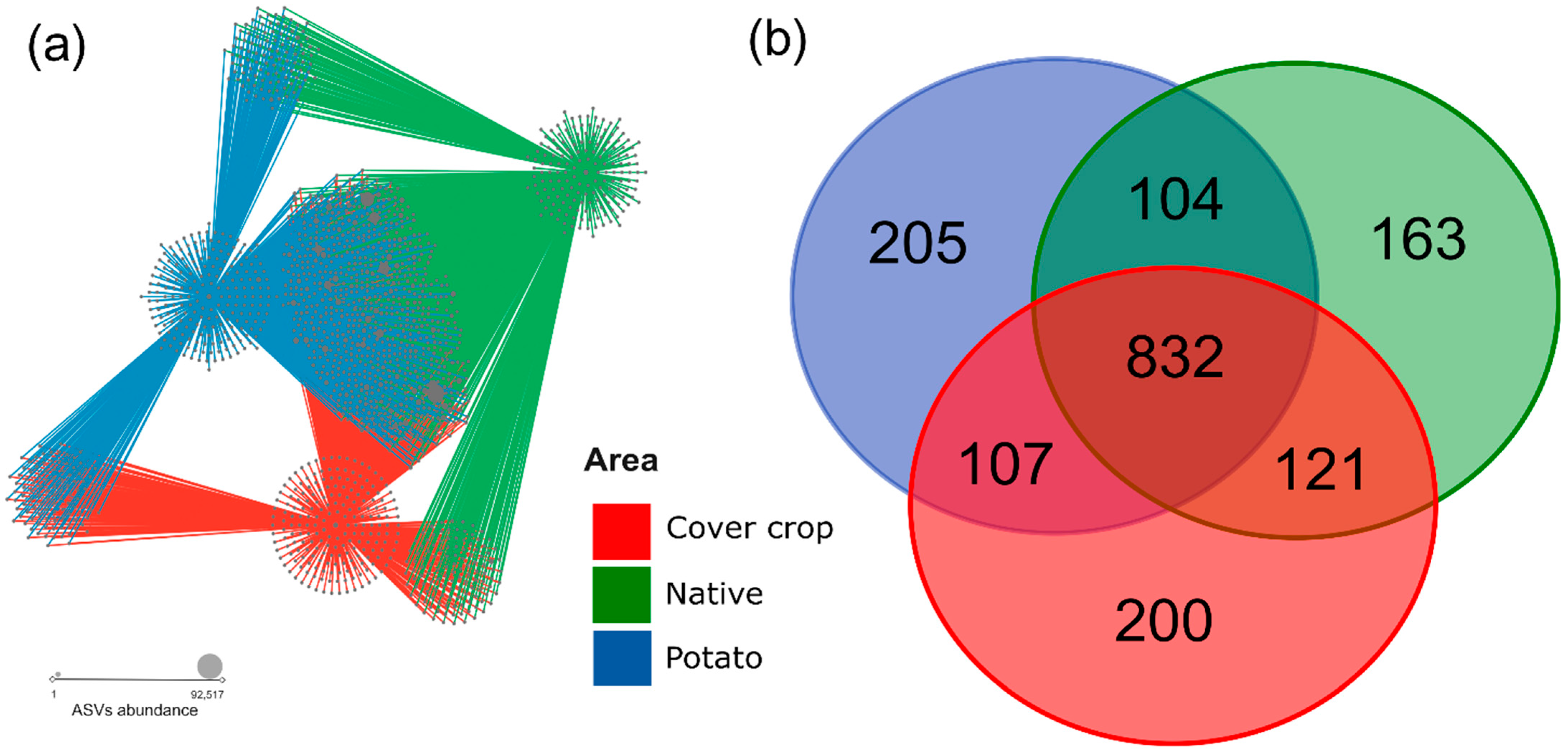
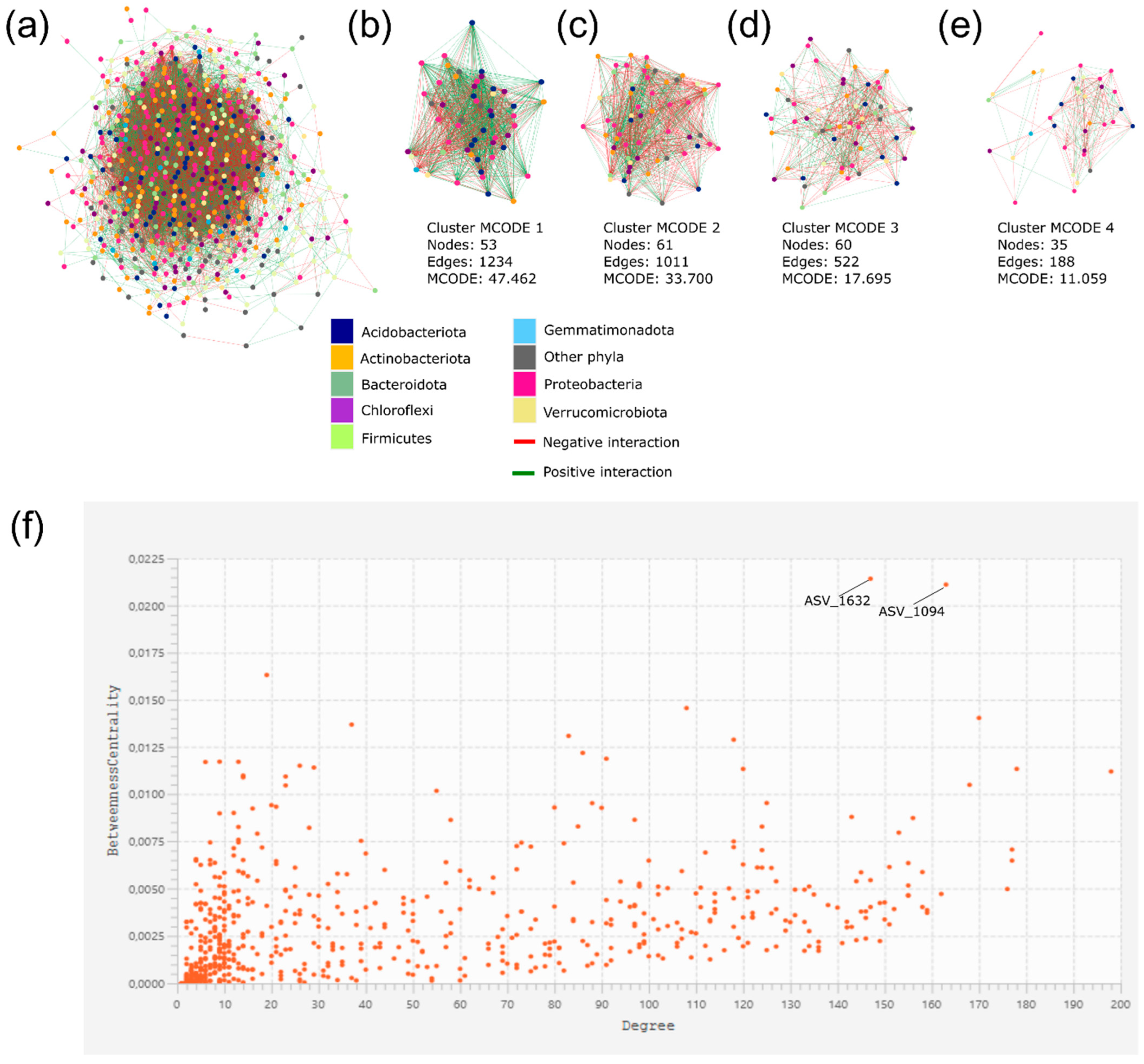
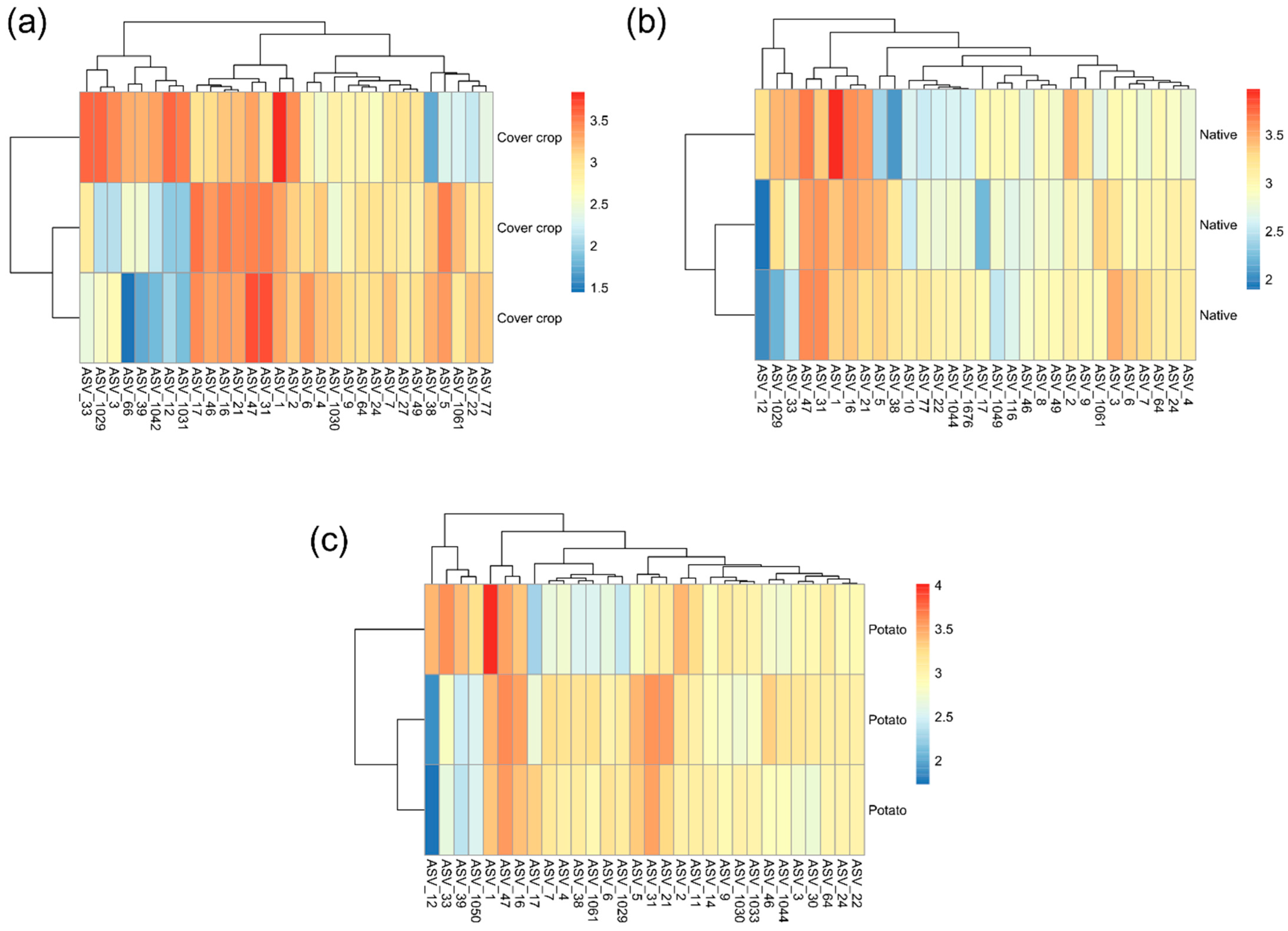
3.4. ASVs Co-Occurrence and Sharing Networks
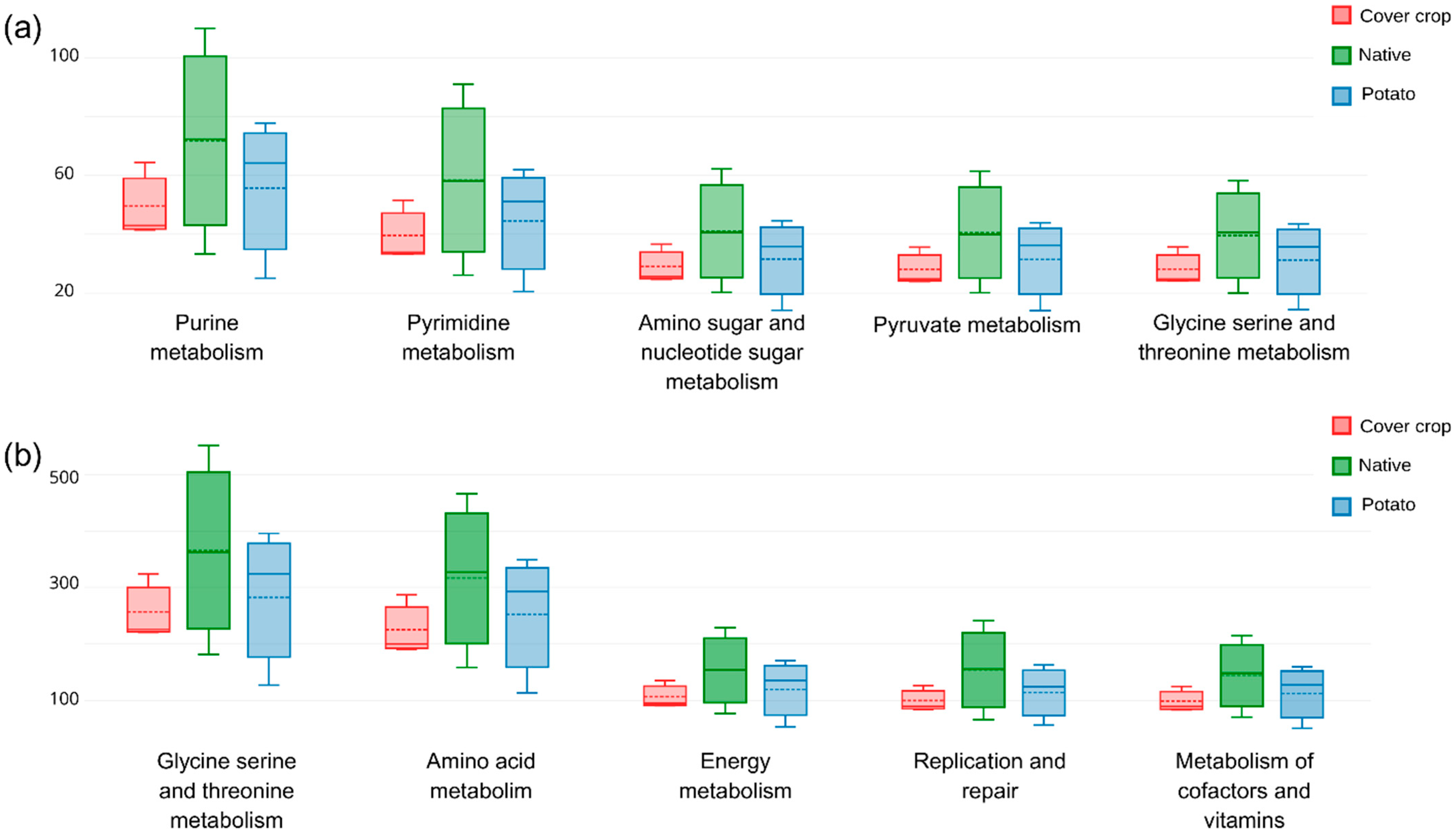
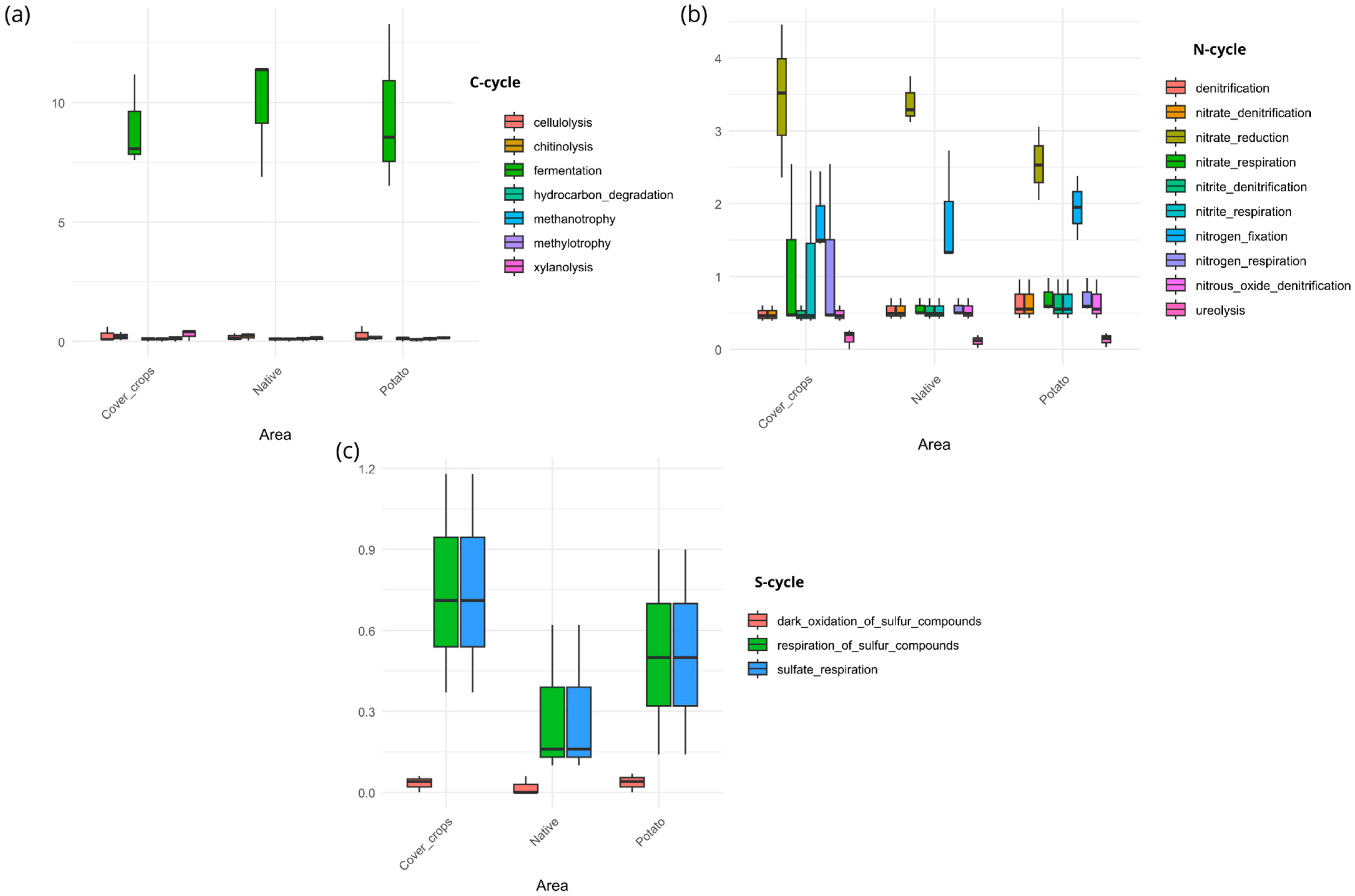
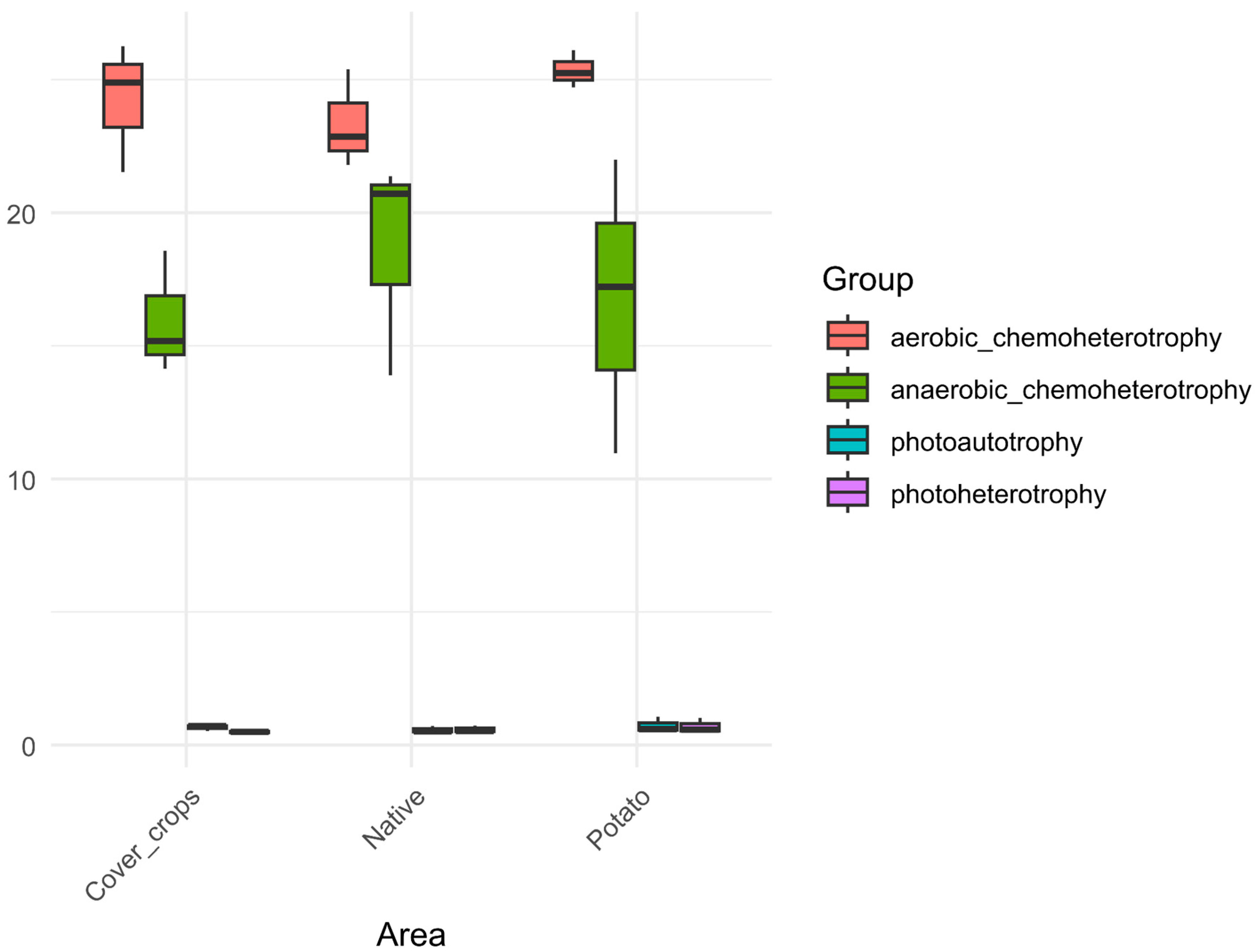
3.5. Habitat Preference and Functional Capacity of the Soil Bacterial Community
4. Discussion
4.1. Physicochemical Properties and Soil Enzymes
4.2. Community of Soil Bacteria
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Appendix A


References
- Curtis, T.P.; Sloan, W.T.; Scannell, J.W. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 2002, 99, 10494–10499. [Google Scholar] [CrossRef] [PubMed]
- Blum, W.E.H. Functions of soil for society and the environment. Rev. Environ. Sci. Technol. 2005, 4, 75–79. [Google Scholar] [CrossRef]
- Orwin, K.H.; Stevenson, B.A.; Smaill, S.J.; Kirschbaum, M.U.; Dickie, I.A.; Clothier, B.E.; Garrett, L.G.; van der Weerden, T.J.; Beare, M.H.; Curtin, D.; et al. Effects of climate change on the delivery of soil-mediated ecosystem services within the primary sector in temperate ecosystems: A review and New Zealand case study. Glob. Change Biol. 2015, 21, 2844–2860. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Torres, O.; Lugo-Melchor, O.Y.; de Anda, J.; Pacheco, A.; Yebra-Montes, C.; Gradilla-Hernandez, M.S.; Senes-Guerrero, C. Bacterial Dynamics and Their Influence on the Biogeochemical Cycles in a Subtropical Hypereutrophic Lake During the Rainy Season. Front. Microbiol. 2022, 13, 832477. [Google Scholar] [CrossRef]
- Ma, B.; Stirling, E.; Liu, Y.; Zhao, K.; Zhou, J.; Singh, B.K.; Tang, C.; Dahlgren, R.A.; Xu, J. Soil Biogeochemical Cycle Couplings Inferred from a Function-Taxon Network. Research 2021, 2021, 7102769. [Google Scholar] [CrossRef]
- Lori, M.; Symnaczik, S.; Mader, P.; De Deyn, G.; Gattinger, A. Organic farming enhances soil microbial abundance and activity-A meta-analysis and meta-regression. PLoS ONE 2017, 12, e0180442. [Google Scholar] [CrossRef]
- Pastuszczak, M.; Stanek-Tarkowska, J.; Kačaniova, M. Impact of Soil Fertilized with Biomass Ash on Depth-Related Variability of Culturable Bacterial Diversity and Selected Physicochemical Parameters in Spring Barley Cultivation. Int. J. Envron. Res. Public Health 2022, 19, 13721. [Google Scholar] [CrossRef]
- Finley, B.K.; Mau, R.L.; Hayer, M.; Stone, B.W.; Morrissey, E.M.; Koch, B.J.; Rasmussen, C.; Dijkstra, P.; Schwartz, E.; Hungate, B.A. Soil minerals affect taxon-specific bacterial growth. ISME J. 2022, 16, 1318–1326. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Zaborowska, M.; Kucharski, J. Sensitivity of Zea mays and Soil Microorganisms to the Toxic Effect of Chromium (VI). Int. J. Mol. Sci. 2022, 24, 178. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Effects of Tebuconazole Application on Soil Microbiota and Enzymes. Molecules 2022, 27, 7501. [Google Scholar] [CrossRef]
- Zhang, Z.; Kaye, J.P.; Bradley, B.A.; Amsili, J.P.; Suseela, V. Cover crop functional types differentially alter the content and composition of soil organic carbon in particulate and mineral-associated fractions. Glob. Change Biol. 2022, 28, 5831–5848. [Google Scholar] [CrossRef]
- Tsiafouli, M.A.; Thebault, E.; Sgardelis, S.P.; de Ruiter, P.C.; van der Putten, W.H.; Birkhofer, K.; Hemerik, L.; de Vries, F.T.; Bardgett, R.D.; Brady, M.V.; et al. Intensive agriculture reduces soil biodiversity across Europe. Glob. Change Biol. 2015, 21, 973–985. [Google Scholar] [CrossRef] [PubMed]
- van Rijssel, S.Q.; Veen, G.F.C.; Koorneef, G.J.; Bakx-Schotman, J.M.T.T.; Ten Hooven, F.C.; Geisen, S.; van der Putten, W.H. Soil microbial diversity and community composition during conversion from conventional to organic agriculture. Mol. Ecol. 2022, 31, 4017–4030. [Google Scholar] [CrossRef]
- Zhou, Z.; Krashevska, V.; Widyastuti, R.; Scheu, S.; Potapov, A. Tropical land use alters functional diversity of soil food webs and leads to monopolization of the detrital energy channel. eLife 2022, 11, e75428. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.; Mittermeier, C.; da Fonseca, G.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Sloan, S.; Jenkins, C.N.; Joppa, L.N.; Gaveau, D.L.; Laurance, W.F. Remaining natural vegetation in the global biodiversity hotspots. Biol. Conserv. 2014, 177, 12–24. [Google Scholar] [CrossRef]
- Vieira, D.L.M.; Sano, E.E.; Silva, T.R. A classification of cultivated pastures in the Brazilian Cerrado for sustainable intensification and savanna restoration. Ambio 2022, 51, 1219–1226. [Google Scholar] [CrossRef]
- de Carvalho, A.M.; de Jesus, D.R.; de Sousa, T.R.; Ramos, M.L.G.; de Figueiredo, C.C.; de Oliveira, A.D.; Marchao, R.L.; Ribeiro, F.P.; Dantas, R.A.; Borges, L.A.B. Soil Carbon Stocks and Greenhouse Gas Mitigation of Agriculture in the Brazilian Cerrado—A Review. Plants 2023, 12, 2449. [Google Scholar] [CrossRef]
- Klink, C.A.; Sato, M.N.; Cordeiro, G.G.; Ramos, M.I.M. The Role of Vegetation on the Dynamics of Water and Fire in the Cerrado Ecosystems: Implications for Management and Conservation. Plants 2020, 9, 1803. [Google Scholar] [CrossRef]
- Peixoto, R.S.; Chaer, G.M.; Franco, N.; Reis Junior, F.B.; Mendes, I.C.; Rosado, A.S. A decade of land use contributes to changes in the chemistry, biochemistry and bacterial community structures of soils in the Cerrado. Antonie Leeuwenhoek 2010, 98, 403–413. [Google Scholar] [CrossRef]
- Rampelotto, P.H.; de Siqueira, F.A.; Barboza, A.D.M.; Roesch, L.F.W. Changes in diversity, abundance, and structure of soil bacterial communities in Brazilian Savanna under different land use systems. Microb. Ecol. 2013, 66, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.C.; Mendes, I.C.; Reis-Junior, F.B.; Carvalho, F.M.; Nogueira, M.A.; Vasconcelos, A.T.; Vicente, V.A.; Hungria, M. Shifts in taxonomic and functional microbial diversity with agriculture: How fragile is the Brazilian Cerrado? BMC Microbiol. 2016, 16, 42. [Google Scholar] [CrossRef]
- Moreira, F.M.S.; Huising, E.J.; Bignell, D.E. A Handbook of Tropical Soil Biology: Sampling & Characterization of Below-Ground Biodiversity; Routledge: London, UK, 2008. [Google Scholar]
- Embrapa—Brazilian Company of Agricultural Research. Brazilian System of Soil Classification, 3rd ed.; Embrapa Solos: Rio de Janeiro, RJ, USA, 2013; 353p. [Google Scholar]
- Department of Agriculture, Soil Survey Staff. Keys to Soil Taxonomy, 11th ed.; USDA-NRCS: Washington, DC, USA, 2010; 346p. [Google Scholar]
- Silva, F.C. Manual de Analises Quimicas de Solos, plantas e Fertilizantes, 2nd ed.; Embrapa Informacao Tecnologica: Brasilia, DF, Brazil; Embrapa Solos: Rio de Janeiro, Brazil, 2009; 627p. [Google Scholar]
- Tabatabai, M.A. Soil Enzymes. In Methods of Soil Analysis: Part 2 Microbiological and Biochemical Properties; Page, A.L., Ed.; The American Society of Agronomy: Madison, WI, USA, 1994; pp. 775–833. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, Interactive, Scalable, and Extensible Microbiome Data Science. PeerJ Prepr. 2018, 6, e27295v2. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar]
- Ho, J.; Tumkaya, T.; Aryal, S.; Choi, H.; Claridge-Chang, A. Moving beyond P values: Data analysis with estimation graphics. Nat. Methods 2019, 16, 565–566. [Google Scholar] [CrossRef]
- Latombe, G.; McGeoch, M.; Nipperess, D.; Hui, C. Zetadiv: Functions to Compute Compositional Turnover Using Zeta Diversity. R Package Version 1.0. 2017. Available online: https://cran.r-project.org/package=zetadiv (accessed on 10 September 2018).
- Hui, C.; McGeoch, M.A. Zeta diversity as a concept and metric that unifies incidence-based biodiversity patterns. Am. Nat. 2014, 184, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Latombe, G.; Hui, C.; McGeoch, M.A. Multi-site generalised dissimilarity modelling: Using zeta diversity to differentiate drivers of turnover in rare and widespread species. Methods Ecol. Evol. 2017, 8, 431–442. [Google Scholar] [CrossRef]
- Feng, K.; Peng, X.; Zhang, Z.; Gu, S.; He, Q.; Shen, W.; Wang, Z.; Wang, D.; Hu, Q.; Li, Y.; et al. iNAP: An integrated network analysis pipeline for microbiome studies. iMeta 2022, 1, e13. [Google Scholar] [CrossRef]
- Zuo, Y.; Li, X.; Yang, J.; Liu, J.; Zhao, L.; He, X. Fungal Endophytic Community and Diversity Associated with Desert Shrubs Driven by Plant Identity and Organ Differentiation in Extremely Arid Desert Ecosystem. J. Fungi 2021, 7, 578. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, F.; Luo, S.; Yin, X.; He, D.; Liu, J.; Yue, Z.; Song, J. Co-expression of key gene modules and pathways of human breast cancer cell lines. Biosci. Rep. 2019, 39, BSR20181925. [Google Scholar] [CrossRef]
- Chazdon, R.L.; Chao, A.; Colwell, R.K.; Lin, S.Y.; Norden, N.; Letcher, S.G.; Clark, D.B.; Finegan, B.; Arroyo, J.P. A Novel Statistical Method for Classifying Habitat Generalists and Specialists. Ecology 2011, 92, 1332–1343. [Google Scholar] [CrossRef]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and Standard Workflow for Inferring, Analyzing, Comparing, and Visualizing the Functional Potential of Microbial Communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef]
- Chen, D.; Duan, Y.; Jin, Y.; Yang, Y.; Yuan, L. Soil quality and microbiology in response to fertilizations in a paddy-upland rotation with multiple crops and frequent tillage. Exp. Agric. 2020, 56, 227–238. [Google Scholar] [CrossRef]
- Lopes, A.S.; Guimaraes, G.L.R. A Career Perspective on Soil Management in the Cerrado Region of Brazil. Adv. Agron. 2016, 137, 1–72. [Google Scholar] [CrossRef]
- Babujia, L.; Hungria, M.; Franchini, J.; Brookes, P. Microbial biomass and activity at various soil depths in a Brazilian oxisol after two decades of no-tillage and conventional tillage. Soil. Biol. Biochem. 2010, 42, 2174–2181. [Google Scholar] [CrossRef]
- Mathew, R.P.; Feng, Y.; Githinji, L.; Ankumah, R.; Balkcom, K.S. Impact of no-tillage and conventional tillage systems on soil microbial communities. Appl. Environ. Soil Sci. 2012, 2012, 548620. [Google Scholar] [CrossRef]
- Hills, K.; Collins, H.; Yorgey, G.; McGuire, A.; Kruger, C. Improving Soil Health in Pacific Northwest Potato Production: A Review. Am. J. Potato Res. 2020, 97, 1–22. [Google Scholar] [CrossRef]
- Maas, A.; Fuller, K.B.; Hatzenbuehler, P.; McIntosh, C. An exploration of preferences for soil health practices in potato production. Farming Syst. 2023, 1, 100054. [Google Scholar] [CrossRef]
- Brito, A.C.C.; Boechat, C.L.; de Sena, A.F.S.; Duarte, L.d.S.L.; Nascimento, C.W.A.D.; da Silva, Y.J.A.B.; Saraiva, P.C. Assessing the Distribution and Concentration of Heavy Metals in Soils of an Agricultural Frontier in the Brazilian Cerrado. Water Air Soil. Pollut. 2020, 231, 388. [Google Scholar] [CrossRef]
- Shi, W.-M.; Yao, J.; Yan, F. Vegetable cultivation under greenhouse conditions leads to rapid accumulation of nutrients, acidification and salinity of soils and groundwater contamination in South-Eastern China. Nutr. Cycl. Agroecosyst. 2009, 83, 73–84. [Google Scholar] [CrossRef]
- Khan, A.R.; Reichmann, L.G.; Ibal, J.C.; Shin, J.H.; Liu, Y.; Collins, H.; LePage, B.; Terry, N. Variation in pickleweed root-associated microbial communities at different locations of a saline solid waste management unit contaminated with petroleum hydrocarbons. PLoS ONE 2019, 14, e0222901. [Google Scholar] [CrossRef]
- Szoboszlay, M.; Nather, A.; Liu, B.; Carrillo, A.; Castellanos, T.; Smalla, K.; Jia, Z.; Tebbe, C.C. Contrasting microbial community responses to salinization and straw amendment in a semiarid bare soil and its wheat rhizosphere. Sci. Rep. 2019, 9, 9795. [Google Scholar] [CrossRef]
- Lv, H.; Ji, C.; Ding, J.; Yu, L.; Cai, H. High Levels of Zinc Affect Nitrogen and Phosphorus Transformation in Rice Rhizosphere Soil by Modifying Microbial Communities. Plants 2022, 11, 2271. [Google Scholar] [CrossRef]
- Santos-Goulart, P.F.; Schulman, P.; Macedo, R.; Mendes, L.W.; Costa-Coelho, G.R.; Rodrigues, L.A.; Mello, R.N.; Lobo, M. Biodiversity and community structures of fungi and bacteria in soils under a crop-livestock integration system. Appl. Soil. Ecol. 2024, 202, 105565. [Google Scholar] [CrossRef]
- Gschwendtner, S.; Leberecht, M.; Engel, M.; Kublik, S.; Dannenmann, M.; Polle, A.; Schloter, M. Effects of Elevated Atmospheric CO2 on Microbial Community Structure at the Plant-Soil Interface of Young Beech Trees (Fagus sylvatica L.) Grown at Two Sites with Contrasting Climatic Conditions. Microb. Ecol. 2015, 69, 867–878. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y.; Zhang, P.; Yu, F.; Perez-Moreno, J. Macrofungi Cultivation in Shady Forest Areas Significantly Increases Microbiome Diversity, Abundance and Functional Capacity in Soil Furrows. J. Fungi 2021, 7, 775. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, Y.; Ji, S.; Dong, D.; Li, Y.; Wang, M.; Han, L.; Chen, X. Effects of Elevation and Distance from Highway on the Abundance and Community Structure of Bacteria in Soil along Qinghai-Tibet Highway. Int. J. Environ. Res. Public Health 2021, 18, 13137. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Yang, X.; Li, Q.; Sun, X. Effects of planting of two common crops, Allium fistulosum and Brassica napus, on soil properties and microbial communities of ginseng cultivation in northeast China. BMC Microbiol. 2022, 22, 182. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, H.; Li, Y.; Ge, C.; Yao, H. Elevated CO2 altered the nano-ZnO-induced influence on bacterial and fungal composition in tomato (Solanum lycopersicum L.) rhizosphere soils. Env. Sci. Pollut. Res. Int. 2023, 30, 75894–75907. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Miksch, S.; Meiners, M.; Meyerdierks, A.; Probandt, D.; Wegener, G.; Titschack, J.; Jensen, M.A.; Ellrott, A.; Amann, R.; Knittel, K. Bacterial communities in temperate and polar coastal sands are seasonally stable. ISME Commun. 2021, 1, 29. [Google Scholar] [CrossRef]
- Somee, M.R.; Amoozegar, M.A.; Dastgheib, S.M.M.; Shavandi, M.; Maman, L.G.; Bertilsson, S.; Mehrshad, M. Genome-resolved analyses show an extensive diversification in key aerobic hydrocarbon-degrading enzymes across bacteria and archaea. BMC Genom. 2022, 23, 690. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Terrat, S.; Horrigue, W.; Dequiedt, S.; Saby, N.P.A.; Lelievre, M.; Nowak, V.; Tripied, J.; Regnier, T.; Jolivet, C.; Arrouays, D.; et al. Mapping and predictive variations of soil bacterial richness across France. PLoS ONE 2017, 12, e0186766, Erratum in PLoS ONE 2017, 12, e0190128; Erratum in PLoS ONE 2022, 17, e0268101. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Oliverio, A.M.; Brewer, T.E.; Benavent-Gonzalez, A.; Eldridge, D.J.; Bardgett, R.D.; Maestre, F.T.; Singh, B.K.; Fierer, N. A global atlas of the dominant bacteria found in soil. Science 2018, 359, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Karimi, B.; Terrat, S.; Dequiedt, S.; Saby, N.P.A.; Horrigue, W.; Lelievre, M.; Nowak, V.; Jolivet, C.; Arrouays, D.; Wincker, P.; et al. Biogeography of soil bacteria and archaea across France. Sci. Adv. 2018, 4, eaat1808. [Google Scholar] [CrossRef] [PubMed]
- Greening, C.; Grinter, R.; Chiri, E. Uncovering the Metabolic Strategies of the Dormant Microbial Majority: Towards Integrative Approaches. Msystems 2019, 4, e00107-19. [Google Scholar] [CrossRef] [PubMed]
- Locey, K.J.; Muscarella, M.E.; Larsen, M.L.; Bray, S.R.; Jones, S.E.; Lennon, J.T. Dormancy dampens the microbial distance-decay relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190243. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef]
- Woodcock, S.; Curtis, T.P.; Head, I.M.; Lunn, M.; Sloan, W.T. Taxa-area relationships for microbes: The unsampled and the unseen. Ecol. Lett. 2006, 9, 805–812. [Google Scholar] [CrossRef]
- Meyer, K.M.; Memiaghe, H.; Korte, L.; Kenfack, D.; Alonso, A.; Bohannan, B.J.M. Why do microbes exhibit weak biogeographic patterns? ISME J. 2018, 12, 1404–1413. [Google Scholar] [CrossRef]
- Bent, S.J.; Forney, L.J. The tragedy of the uncommon: Understanding limitations in the analysis of microbial diversity. ISME J. 2008, 2, 689–695. [Google Scholar] [CrossRef]
- Singh, I.; Hussain, M.; Manjunath, G.; Chandra, N.; Ravikanth, G. Regenerative agriculture augments bacterial community structure for a healthier soil and agriculture. Front. Agron. 2023, 5, 1134514. [Google Scholar] [CrossRef]
- Ramkumar, D.; Marty, A.; Ramkumar, J.; Rosencranz, H.; Vedantham, R.; Goldman, M.; Meyer, E.; Steinmetz, J.; Weckle, A.; Bloedorn, K.; et al. Food for thought: Making the case for food produced via regenerative agriculture in the battle against noncommunicable chronic diseases (NCDs). One Health 2024, 18, 100734. [Google Scholar] [CrossRef]
- Tang, Q.; Lin, T.; Sun, Z.; Yan, A.; Zhang, J.; Jiang, P.; Wu, F.; Zhang, H. Effects of mulching film on soil microbial diversity and community of cotton. AMB Express 2022, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, P.G.C.; Batalha, M.A.; Pivello, V.R.; Meirelles, S.T. Soil-vegetation relationships in cerrado (Brazilian savanna) and semideciduous forest. Southeast. Braz. Plant Ecol. 2002, 160, 1–16. [Google Scholar] [CrossRef]
- Lock, K.; Janssen, C.R. Influence of soil zinc concentrations on zinc sensitivity and functional diversity of microbial communities. Environ. Pollut. 2005, 136, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Mertens, J.; Springael, D.; De Troyer, I.; Cheyns, K.; Wattiau, P.; Smolders, E. Long-term exposure to elevated zinc concentrations induced structural changes and zinc tolerance of the nitrifying community in soil. Environ. Microbiol. 2006, 8, 2170–2178. [Google Scholar] [CrossRef]
- Witzgall, K.; Vidal, A.; Schubert, D.I.; Hoschen, C.; Schweizer, S.A.; Buegger, F.; Pouteau, V.; Chenu, C.; Mueller, C.W. Particulate organic matter as a functional soil component for persistent soil organic carbon. Nat. Commun. 2021, 12, 4115. [Google Scholar] [CrossRef]



| Distance Method | Variable | R2 | p Value |
|---|---|---|---|
| Area | 0.16350 | 0.678 | |
| Zn | 0.29698 | 0.041 | |
| Bray–Curtis | OM | 0.07801 | 0.601 |
| Residual | 0.46152 | NA | |
| Total | 1 | NA | |
| Area | 0.20946 | 0.604 | |
| Al | 0.22323 | 0.033 | |
| Jaccard | Carbon | 0.09898 | 0.568 |
| Residual | 0.46833 | NA | |
| Total | 1 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reis, J.B.A.d.; Oliveira, T.M.R.d.; Sartori da Silva, M.R.S.; Lopes, F.A.C.; de Paula, A.M.; Pontes, N.d.C.; Vale, H.M.M.d. Different Land Use Systems in the Brazilian Cerrado and Their Effects on Soil Bacterial Communities. Microorganisms 2025, 13, 804. https://doi.org/10.3390/microorganisms13040804
Reis JBAd, Oliveira TMRd, Sartori da Silva MRS, Lopes FAC, de Paula AM, Pontes NdC, Vale HMMd. Different Land Use Systems in the Brazilian Cerrado and Their Effects on Soil Bacterial Communities. Microorganisms. 2025; 13(4):804. https://doi.org/10.3390/microorganisms13040804
Chicago/Turabian StyleReis, Jefferson Brendon Almeida dos, Thayssa Monize Rosa de Oliveira, Maria Regina Silveira Sartori da Silva, Fabyano Alvares Cardoso Lopes, Alessandra Monteiro de Paula, Nadson de Carvalho Pontes, and Helson Mario Martins do Vale. 2025. "Different Land Use Systems in the Brazilian Cerrado and Their Effects on Soil Bacterial Communities" Microorganisms 13, no. 4: 804. https://doi.org/10.3390/microorganisms13040804
APA StyleReis, J. B. A. d., Oliveira, T. M. R. d., Sartori da Silva, M. R. S., Lopes, F. A. C., de Paula, A. M., Pontes, N. d. C., & Vale, H. M. M. d. (2025). Different Land Use Systems in the Brazilian Cerrado and Their Effects on Soil Bacterial Communities. Microorganisms, 13(4), 804. https://doi.org/10.3390/microorganisms13040804








