Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Specimen
2.2. Process of Sanger Sequencing
2.3. Process of mNGS
2.4. Criteria for Identification and Discrepant Analysis
2.5. Statistical Analysis
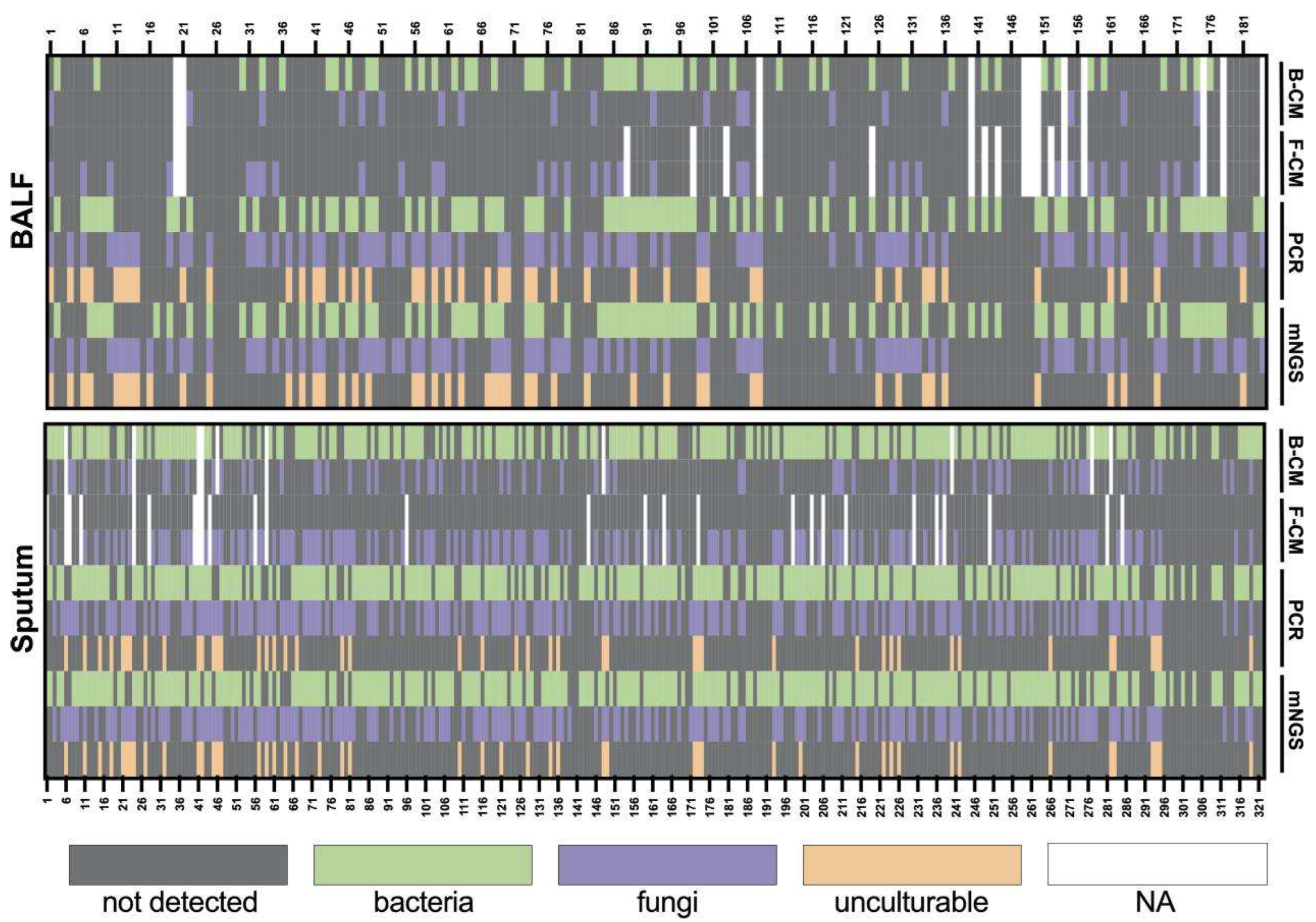
3. Results
3.1. Patient Characteristics
3.2. Test Performance of Culture, Sanger Sequencing, and mNGS
3.3. Co-Infection Detected in BALF
3.4. Comparison of Culture, Sanger Sequencing, and mNGS in Certain Microorganisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mNGS | Metagenomic next-generation sequencing |
| LRTI | Lower Respiratory Tract Infections |
| BALF | bronchoalveolar lavage fluid |
| PCR | polymerase chain reaction |
References
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 7 August 2024).
- Abbafati, C.; Abbas, K.M.; Abbasi, M.; Abbasifard, M.; Abbasi-Kangevari, M.; Abbastabar, H.; Abd-Allah, F.; Abdelalim, A.; Abdollahi, M.; Abdollahpour, I.; et al. Global Burden of 369 Diseases and Injuries in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Waterer, G. The Global Burden of Respiratory Infectious Diseases Before and Beyond COVID. Respirology 2023, 28, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gilligan, P.H.; Gonzalez, M.D.; Jerris, R.C.; Kehl, S.C.; Patel, R.; et al. A Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2018 Update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin. Infect. Dis. 2018, 67, e1–e94. [Google Scholar] [CrossRef] [PubMed]
- Baldassarri, R.J.; Kumar, D.; Baldassarri, S.; Cai, G. Diagnosis of Infectious Diseases in the Lower Respiratory Tract: A Cytopathologist’s Perspective. Arch. Pathol. Lab. Med. 2019, 143, 683–694. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef]
- Wilson, M.R.; Naccache, S.N.; Samayoa, E.; Biagtan, M.; Bashir, H.; Yu, G.; Salamat, S.M.; Somasekar, S.; Federman, S.; Miller, S.; et al. Actionable Diagnosis of Neuroleptospirosis by Next-Generation Sequencing. N. Engl. J. Med. 2014, 370, 2408–2417. [Google Scholar] [CrossRef]
- Mitchell, S.L.; Simner, P.J. Next-Generation Sequencing in Clinical Microbiology: Are We There Yet? Clin. Lab. Med. 2019, 39, 405–418. [Google Scholar] [CrossRef]
- Diao, Z.; Han, D.; Zhang, R.; Li, J. Metagenomics Next-Generation Sequencing Tests Take the Stage in the Diagnosis of Lower Respiratory Tract Infections. J. Adv. Res. 2022, 38, 201–212. [Google Scholar] [CrossRef]
- Diao, Z.; Zhang, Y.; Chen, Y.; Han, Y.; Chang, L.; Ma, Y.; Feng, L.; Huang, T.; Zhang, R.; Li, J. Assessing the Quality of Metagenomic Next-Generation Sequencing for Pathogen Detection in Lower Respiratory Infections. Clin. Chem. 2023, 69, 1038–1049. [Google Scholar] [CrossRef]
- Han, D.; Li, Z.; Li, R.; Tan, P.; Zhang, R.; Li, J. MNGS in Clinical Microbiology Laboratories: On the Road to Maturity. Crit. Rev. Microbiol. 2019, 45, 668–685. [Google Scholar] [CrossRef]
- Simner, P.J.; Miller, S.; Carroll, K.C. Understanding the Promises and Hurdles of Metagenomic Next-Generation Sequencing as a Diagnostic Tool for Infectious Diseases. Clin. Infect. Dis. 2018, 66, 778–788. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Zhao, Q.; Liu, J.; Wang, S.; Wu, W.; Miao, L.; Zhan, P.; Chen, X.; Huang, M.; Ye, M.; et al. Utilizing Metagenomic Next-Generation Sequencing for Pathogen Detection and Diagnosis in Lower Respiratory Tract Infections in Real-World Clinical Practice. Infection 2024, 52, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Qiu, X.; Wang, T.; Zhang, J. The Diagnostic Value of Metagenomic Next–Generation Sequencing in Lower Respiratory Tract Infection. Front. Cell Infect. Microbiol. 2021, 11, 694756. [Google Scholar] [CrossRef] [PubMed]
- Hogan, C.A.; Yang, S.; Garner, O.B.; Green, D.A.; Gomez, C.A.; DIen Bard, J.; Pinsky, B.A.; Banaei, N. Clinical Impact of Metagenomic Next-Generation Sequencing of Plasma Cell-Free DNA for the Diagnosis of Infectious Diseases: A Multicenter Retrospective Cohort Study. Clin. Infect. Dis. 2021, 72, 239–245. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Xu, J.; Yang, Q.; Zhu, H.; Yang, J. Diagnostic Value of Metagenomic Next-Generation Sequencing of Lower Respiratory Tract Specimen for the Diagnosis of Suspected Pneumocystis jirovecii Pneumonia. Ann. Med. 2023, 55, 2232358. [Google Scholar] [CrossRef]
- Carr, V.R.; Chaguza, C. Metagenomics for Surveillance of Respiratory Pathogens. Nat. Rev. Microbiol. 2021, 19, 285. [Google Scholar] [CrossRef]
- Vijayvargiya, P.; Thoendel, M.J. Sequencing for Infection Diagnostics: Is It Time to Embrace the Next Generation? Clin. Infect. Dis. 2022, 74, 1669–1670. [Google Scholar] [CrossRef]
- Buchan, B.W.; Windham, S.; Balada-Llasat, J.M.; Leber, A.; Harrington, A.; Relich, R.; Murphy, C.; Bard, J.D.; Naccache, S.; Ronen, S.; et al. Practical Comparison of the BioFire FilmArray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J. Clin. Microbiol. 2020, 58, 10-1128. [Google Scholar] [CrossRef]
- Bender, R.G.; Sirota, S.B.; Swetschinski, L.R.; Dominguez, R.M.V.; Novotney, A.; Wool, E.E.; Ikuta, K.S.; Vongpradith, A.; Rogowski, E.L.B.; Doxey, M.; et al. Global, Regional, and National Incidence and Mortality Burden of Non-COVID-19 Lower Respiratory Infections and Aetiologies, 1990–2021: A Systematic Analysis from the Global Burden of Disease Study 2021. Lancet Infect. Dis. 2024, 24, 974–1002. [Google Scholar] [CrossRef]
- Langelier, C.; Kalantar, K.L.; Moazed, F.; Wilson, M.R.; Crawford, E.D.; Deiss, T.; Belzer, A.; Bolourchi, S.; Caldera, S.; Fung, M.; et al. Integrating Host Response and Unbiased Microbe Detection for Lower Respiratory Tract Infection Diagnosis in Critically Ill Adults. Proc. Natl. Acad. Sci. USA 2018, 115, E12353–E12362. [Google Scholar] [CrossRef]
- Miller, J.M.; Binnicker, M.J.; Campbell, S.; Carroll, K.C.; Chapin, K.C.; Gonzalez, M.D.; Harrington, A.; Jerris, R.C.; Kehl, S.C.; Leal, S.M.; et al. Guide to Utilization of the Microbiology Laboratory for Diagnosis of Infectious Diseases: 2024 Update by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM). Clin. Infect. Dis. 2024, ciae104. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, K.; Johnson, B.; Klimowich, K.; Chiotos, K.; Jensen, E.A.; Planet, P.; Phinizy, P.; Piccione, J. Comparison of Tracheal Aspirate and Bronchoalveolar Lavage Samples in the Microbiological Diagnosis of Lower Respiratory Tract Infection in Pediatric Patients. Pediatr. Pulmonol. 2022, 57, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Diao, Z.; Lai, H.; Han, Y.; Xie, J.; Zhang, R.; Li, J. Multilaboratory Assessment of Metagenomic Next-Generation Sequencing for Unbiased Microbe Detection. J. Adv. Res. 2022, 38, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Edward, P.; Handel, A.S. Metagenomic Next-Generation Sequencing for Infectious Disease Diagnosis: A Review of the Literature with a Focus on Pediatrics. J. Pediatr. Infect. Dis. Soc. 2021, 10, S71–S77. [Google Scholar] [CrossRef]
- Benoit, P.; Brazer, N.; de Lorenzi-Tognon, M.; Kelly, E.; Servellita, V.; Oseguera, M.; Nguyen, J.; Tang, J.; Omura, C.; Streithorst, J.; et al. Seven-Year Performance of a Clinical Metagenomic next-Generation Sequencing Test for Diagnosis of Central Nervous System Infections. Nat. Med. 2024, 30, 3522–3533. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical Metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef]
- Rader, T.S.; Stevens, M.P.; Bearman, G. Syndromic Multiplex Polymerase Chain Reaction (MPCR) Testing and Antimicrobial Stewardship: Current Practice and Future Directions. Curr. Infect. Dis. Rep. 2021, 23, 5. [Google Scholar] [CrossRef]
- Clark, T.W.; Lindsley, K.; Wigmosta, T.B.; Bhagat, A.; Hemmert, R.B.; Uyei, J.; Timbrook, T.T. Rapid Multiplex PCR for Respiratory Viruses Reduces Time to Result and Improves Clinical Care: Results of a Systematic Review and Meta-Analysis. J. Infect. 2023, 86, 462–475. [Google Scholar] [CrossRef]
| Issues | BALF (n = 184) | Sputum (n = 322) |
|---|---|---|
| Sex, male, n (%) | 116 (63.04%) | 207 (64.29%) |
| Age at onset, y, median (range) | 57.43 ± 16.703, 59 (13–90) | 60.61 ± 16.517, 64 (10–100) |
| Clinical classification | ||
| Patients with severe respiratory conditions | 121 (65.76%) | 117 (36.34%) |
| Immunocompromised patients with respiratory-related symptoms | 12 (6.52%) | 18 (5.59%) |
| Special patients with respiratory-related symptoms | 2 (1.09%) | 2 (0.62%) |
| Critically ill patients with respiratory-related symptoms due to infections | 6 (3.26%) | 23 (7.14%) |
| Critically ill patients with respiratory-related symptoms in settings | 6 (3.26%) | 105 (32.61%) |
| Patients with underlying diseases presenting respiratory symptoms | 21 (11.41%) | 34 (10.56%) |
| Patients with other respiratory symptoms | 16 (8.70%) | 23 (7.14%) |
| Organism | TP | FP | FN | TN | Sensitivity [95% CI] | Specificity [95% CI] | PPV [95% CI] | NPV [95% CI] | PLR | NLR | Accuracy [95% CI] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BALF | |||||||||||
| Aspergillus fumigatus | 8 | 10 | 0 | 147 | 1.00 [0.6306, 1] | 0.94 [0.886, 0.969] | 0.44 [0.2153, 0.6924] | 1.00 [0.9752, 1] | 15.70 | 0.00 | 0.94 [0.8914, 0.9706] |
| Talaromyces marneffei | 0 | 0 | 0 | 165 | 0.00 [0, 0] | 1.00 [0.9779, 1] | 0.00 [0, 0] | 1.00 [0.9779, 1] | NA | 0.00 | 1.00 [0.9779, 1] |
| Cryptococcus neoformans | 1 | 0 | 0 | 164 | 1.00 [0.025, 1] | 1.00 [0.9778, 1] | 1.00 [0.025, 1] | 1.00 [0.9778, 1] | Inf | 0.00 | 1.00 [0.9779, 1] |
| Candida albicans | 23 | 4 | 1 | 137 | 0.96 [0.7888, 0.9989] | 0.97 [0.929, 0.9922] | 0.85 [0.6627, 0.9581] | 0.99 [0.9603, 0.9998] | 33.78 | 0.04 | 0.97 [0.9307, 0.9901] |
| Candida tropicalis | 4 | 1 | 0 | 160 | 1.00 [0.3976, 1] | 0.99 [0.9659, 0.9998] | 0.80 [0.2836, 0.9949] | 1.00 [0.9772, 1] | 161.00 | 0.00 | 0.99 [0.9667, 0.9998] |
| Candida glabrata | 5 | 1 | 0 | 159 | 1.00 [0.4782, 1] | 0.99 [0.9657, 0.9998] | 0.83 [0.3588, 0.9958] | 1.00 [0.9771, 1] | 160.00 | 0.00 | 0.99 [0.9667, 0.9998] |
| Haemophilus influenzae | 0 | 0 | 0 | 172 | 0.00 [0, 0] | 1.00 [0.9788, 1] | 0.00 [0, 0] | 1.00 [0.9788, 1] | NA | 0.00 | 1.00 [0.9788, 1] |
| Streptococcus pneumoniae | 0 | 1 | 0 | 171 | 0.00 [0, 0] | 0.99 [0.968, 0.9999] | 0.00 [0, 0.975] | 1.00 [0.9787, 1] | 0.00 | 0.00 | 0.99 [0.968, 0.9999] |
| Moraxella catarrhalis | 0 | 0 | 0 | 172 | 0.00 [0, 0] | 1.00 [0.9788, 1] | 0.00 [0, 0] | 1.00 [0.9788, 1] | NA | 0.00 | 1.00 [0.9788, 1] |
| Nocardia asteroids | 0 | 0 | 0 | 172 | 0.00 [0, 0] | 1.00 [0.9788, 1] | 0.00 [0, 0] | 1.00 [0.9788, 1] | NA | 0.00 | 1.00 [0.9788, 1] |
| Klebsiella pneumoniae | 10 | 10 | 0 | 152 | 1.00 [0.6915, 1] | 0.94 [0.8894, 0.97] | 0.50 [0.272, 0.728] | 1.00 [0.976, 1] | 16.20 | 0.00 | 0.94 [0.8957, 0.9718] |
| Acinetobacter baumannii | 27 | 17 | 0 | 128 | 1.00 [0.8723, 1] | 0.88 [0.8189, 0.9302] | 0.61 [0.455, 0.7564] | 1.00 [0.9716, 1] | 8.53 | 0.00 | 0.90 [0.8465, 0.9414] |
| Pseudomonas aeruginosa | 16 | 10 | 0 | 146 | 1.00 [0.7941, 1] | 0.94 [0.8853, 0.9688] | 0.62 [0.4057, 0.7977] | 1.00 [0.9751, 1] | 15.60 | 0.00 | 0.94 [0.8957, 0.9718] |
| Staphylococcus aureus | 5 | 8 | 0 | 159 | 1.00 [0.4782, 1] | 0.95 [0.9078, 0.9791] | 0.38 [0.1386, 0.6842] | 1.00 [0.9771, 1] | 20.88 | 0.00 | 0.95 [0.9104, 0.9797] |
| Sputum | |||||||||||
| Aspergillus fumigatus | 29 | 9 | 1 | 256 | 0.97 [0.8278, 0.9992] | 0.97 [0.9365, 0.9844] | 0.76 [0.5976, 0.8856] | 1.00 [0.9785, 0.9999] | 28.46 | 0.03 | 0.97 [0.9385, 0.9836] |
| Talaromyces marneffei | 0 | 0 | 0 | 295 | 0.00 [0, 0] | 1.00 [0.9876, 1] | 0.00 [0, 0] | 1.00 [0.9876, 1] | NA | 0.00 | 1.00 [0.9876, 1] |
| Cryptococcus neoformans | 1 | 4 | 0 | 290 | 1.00 [0.025, 1] | 0.99 [0.9655, 0.9963] | 0.20 [0.0051, 0.7164] | 1.00 [0.9874, 1] | 73.50 | 0.00 | 0.99 [0.9656, 0.9963] |
| Candida albicans | 85 | 15 | 3 | 196 | 0.97 [0.9036, 0.9929] | 0.93 [0.8855, 0.9597] | 0.85 [0.7647, 0.9135] | 0.98 [0.9566, 0.9969] | 13.59 | 0.04 | 0.94 [0.9065, 0.9639] |
| Candida tropicalis | 18 | 8 | 0 | 269 | 1.00 [0.8147, 1] | 0.97 [0.9439, 0.9875] | 0.69 [0.4821, 0.8567] | 1.00 [0.9864, 1] | 34.63 | 0.00 | 0.97 [0.9473, 0.9882] |
| Candida glabrata | 27 | 4 | 0 | 265 | 1.00 [0.8723, 1] | 0.99 [0.9624, 0.9959] | 0.87 [0.7017, 0.9637] | 1.00 [0.9862, 1] | 67.25 | 0.00 | 0.99 [0.9658, 0.9963] |
| Haemophilus influenzae | 19 | 13 | 0 | 280 | 1.00 [0.8235, 1] | 0.96 [0.9253, 0.9762] | 0.59 [0.4064, 0.763] | 1.00 [0.9869, 1] | 22.54 | 0.00 | 0.96 [0.9298, 0.9776] |
| Streptococcus pneumoniae | 9 | 12 | 0 | 291 | 1.00 [0.6637, 1] | 0.96 [0.9318, 0.9794] | 0.43 [0.2182, 0.6598] | 1.00 [0.9874, 1] | 25.25 | 0.00 | 0.96 [0.9338, 0.98] |
| Moraxella catarrhalis | 5 | 8 | 0 | 299 | 1.00 [0.4782, 1] | 0.97 [0.9493, 0.9887] | 0.38 [0.1386, 0.6842] | 1.00 [0.9877, 1] | 38.38 | 0.00 | 0.97 [0.9501, 0.9889] |
| Nocardia asteroids | 0 | 0 | 0 | 312 | 0.00 [0, 0] | 1.00 [0.9882, 1] | 0.00 [0, 0] | 1.00 [0.9882, 1] | NA | 0.00 | 1.00 [0.9882, 1] |
| Klebsiella pneumoniae | 46 | 16 | 1 | 250 | 0.98 [0.8871, 0.9995] | 0.94 [0.9042, 0.9652] | 0.74 [0.615, 0.8447] | 1.00 [0.978, 0.9999] | 16.27 | 0.02 | 0.95 [0.9145, 0.968] |
| Acinetobacter baumannii | 50 | 19 | 1 | 242 | 0.98 [0.8955, 0.9995] | 0.93 [0.8887, 0.9556] | 0.72 [0.6038, 0.8254] | 1.00 [0.9773, 0.9999] | 13.47 | 0.02 | 0.94 [0.9027, 0.9604] |
| Pseudomonas aeruginosa | 46 | 25 | 0 | 241 | 1.00 [0.9229, 1] | 0.91 [0.8644, 0.9383] | 0.65 [0.5254, 0.7576] | 1.00 [0.9848, 1] | 10.64 | 0.00 | 0.92 [0.884, 0.9475] |
| Staphylococcus aureus | 40 | 27 | 0 | 245 | 1.00 [0.9119, 1] | 0.90 [0.8589, 0.9336] | 0.60 [0.47, 0.7151] | 1.00 [0.9851, 1] | 10.07 | 0.00 | 0.91 [0.8766, 0.9422] |
| Organism | TP | FP | FN | TN | Sensitivity [95% CI] | Specificity [95% CI] | PPV [95% CI] | NPV [95% CI] | PLR | NLR | Accuracy [95% CI] |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BALF | |||||||||||
| Aspergillus fumigatus | 17 | 1 | 0 | 166 | 1.00 [0.8049, 1] | 0.99 [0.9671, 0.9998] | 0.94 [0.7271, 0.9986] | 1.00 [0.978, 1] | 167.00 | 0.00 | 0.99 [0.9701, 0.9999] |
| Pneumocystis jirovecii | 40 | 2 | 0 | 142 | 1.00 [0.9119, 1] | 0.99 [0.9507, 0.9983] | 0.95 [0.8384, 0.9942] | 1.00 [0.9744, 1] | 72.00 | 0.00 | 0.99 [0.9613, 0.9987] |
| Talaromyces marneffei | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| Cryptococcus neoformans | 1 | 0 | 0 | 183 | 1.00 [0.025, 1] | 1.00 [0.98, 1] | 1.00 [0.025, 1] | 1.00 [0.98, 1] | Inf | 0.00 | 1.00 [0.9802, 1] |
| Nocardia asteroids | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| Candida albicans | 29 | 2 | 1 | 152 | 0.97 [0.8278, 0.9992] | 0.99 [0.9539, 0.9984] | 0.94 [0.7858, 0.9921] | 0.99 [0.9641, 0.9998] | 74.43 | 0.03 | 0.98 [0.9531, 0.9966] |
| Candida tropicalis | 5 | 1 | 0 | 178 | 1.00 [0.4782, 1] | 0.99 [0.9693, 0.9999] | 0.83 [0.3588, 0.9958] | 1.00 [0.9795, 1] | 179.00 | 0.00 | 0.99 [0.9701, 0.9999] |
| Candida glabrata | 6 | 0 | 0 | 178 | 1.00 [0.5407, 1] | 1.00 [0.9795, 1] | 1.00 [0.5407, 1] | 1.00 [0.9795, 1] | Inf | 0.00 | 1.00 [0.9802, 1] |
| Haemophilus influenzae | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| Streptococcus pneumoniae | 1 | 0 | 0 | 183 | 1.00 [0.025, 1] | 1.00 [0.98, 1] | 1.00 [0.025, 1] | 1.00 [0.98, 1] | Inf | 0.00 | 1.00 [0.9802, 1] |
| Moraxella catarrhalis | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| Legionella pneumophila | 5 | 1 | 0 | 178 | 1.00 [0.4782, 1] | 0.99 [0.9693, 0.9999] | 0.83 [0.3588, 0.9958] | 1.00 [0.9795, 1] | 179.00 | 0.00 | 0.99 [0.9701, 0.9999] |
| Klebsiella pneumoniae | 20 | 1 | 0 | 163 | 1.00 [0.8316, 1] | 0.99 [0.9665, 0.9998] | 0.95 [0.7618, 0.9988] | 1.00 [0.9776, 1] | 164.00 | 0.00 | 0.99 [0.9701, 0.9999] |
| Acinetobacter baumannii | 44 | 5 | 0 | 135 | 1.00 [0.9196, 1] | 0.96 [0.9186, 0.9883] | 0.90 [0.7777, 0.966] | 1.00 [0.973, 1] | 28.00 | 0.00 | 0.97 [0.9377, 0.9911] |
| Pseudomonas aeruginosa | 25 | 2 | 1 | 156 | 0.96 [0.8036, 0.999] | 0.99 [0.955, 0.9985] | 0.93 [0.7571, 0.9909] | 0.99 [0.965, 0.9998] | 75.96 | 0.04 | 0.98 [0.9531, 0.9966] |
| Staphylococcus aureus | 12 | 1 | 0 | 171 | 1.00 [0.7354, 1] | 0.99 [0.968, 0.9999] | 0.92 [0.6397, 0.9981] | 1.00 [0.9787, 1] | 172.00 | 0.00 | 0.99 [0.9701, 0.9999] |
| Mycoplasma pneumoniae | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| human adenoviruses | 0 | 0 | 0 | 184 | 0.00 [0, 0] | 1.00 [0.9802, 1] | 0.00 [0, 0] | 1.00 [0.9802, 1] | NA | 0.00 | 1.00 [0.9802, 1] |
| Sputum | |||||||||||
| Aspergillus fumigatus | 38 | 3 | 1 | 280 | 0.97 [0.8652, 0.9994] | 0.99 [0.9693, 0.9978] | 0.93 [0.8008, 0.9846] | 1.00 [0.9803, 0.9999] | 91.91 | 0.03 | 0.99 [0.9685, 0.9966] |
| Pneumocystis jirovecii | 40 | 3 | 0 | 279 | 1.00 [0.9119, 1] | 0.99 [0.9692, 0.9978] | 0.93 [0.8094, 0.9854] | 1.00 [0.9869, 1] | 94.00 | 0.00 | 0.99 [0.973, 0.9981] |
| Talaromyces marneffei | 0 | 0 | 0 | 322 | 0.00 [0, 0] | 1.00 [0.9886, 1] | 0.00 [0, 0] | 1.00 [0.9886, 1] | NA | 0.00 | 1.00 [0.9886, 1] |
| Cryptococcus neoformans | 5 | 0 | 0 | 317 | 1.00 [0.4782, 1] | 1.00 [0.9884, 1] | 1.00 [0.4782, 1] | 1.00 [0.9884, 1] | Inf | 0.00 | 1.00 [0.9886, 1] |
| Nocardia asteroids | 0 | 0 | 0 | 322 | 0.00 [0, 0] | 1.00 [0.9886, 1] | 0.00 [0, 0] | 1.00 [0.9886, 1] | NA | 0.00 | 1.00 [0.9886, 1] |
| Candida albicans | 99 | 2 | 2 | 219 | 0.98 [0.9303, 0.9976] | 0.99 [0.9677, 0.9989] | 0.98 [0.9303, 0.9976] | 0.99 [0.9677, 0.9989] | 108.31 | 0.02 | 0.99 [0.9685, 0.9966] |
| Candida tropicalis | 26 | 1 | 0 | 295 | 1.00 [0.8677, 1] | 1.00 [0.9813, 0.9999] | 0.96 [0.8103, 0.9991] | 1.00 [0.9876, 1] | 296.00 | 0.00 | 1.00 [0.9828, 0.9999] |
| Candida glabrata | 31 | 0 | 0 | 291 | 1.00 [0.8878, 1] | 1.00 [0.9874, 1] | 1.00 [0.8878, 1] | 1.00 [0.9874, 1] | Inf | 0.00 | 1.00 [0.9886, 1] |
| Haemophilus influenzae | 30 | 2 | 0 | 290 | 1.00 [0.8843, 1] | 0.99 [0.9755, 0.9992] | 0.94 [0.7919, 0.9923] | 1.00 [0.9874, 1] | 146.00 | 0.00 | 0.99 [0.9777, 0.9992] |
| Streptococcus pneumoniae | 21 | 2 | 0 | 299 | 1.00 [0.8389, 1] | 0.99 [0.9762, 0.9992] | 0.91 [0.7196, 0.9893] | 1.00 [0.9877, 1] | 150.50 | 0.00 | 0.99 [0.9777, 0.9992] |
| Moraxella catarrhalis | 13 | 1 | 0 | 308 | 1.00 [0.7529, 1] | 1.00 [0.9821, 0.9999] | 0.93 [0.6613, 0.9982] | 1.00 [0.9881, 1] | 309.00 | 0.00 | 1.00 [0.9828, 0.9999] |
| Legionella pneumophila | 3 | 0 | 0 | 319 | 1.00 [0.2924, 1] | 1.00 [0.9885, 1] | 1.00 [0.2924, 1] | 1.00 [0.9885, 1] | Inf | 0.00 | 1.00 [0.9886, 1] |
| Klebsiella pneumoniae | 58 | 4 | 2 | 258 | 0.97 [0.8847, 0.9959] | 0.98 [0.9614, 0.9958] | 0.94 [0.843, 0.9821] | 0.99 [0.9725, 0.9991] | 63.32 | 0.03 | 0.98 [0.9599, 0.9931] |
| Acinetobacter baumannii | 66 | 5 | 2 | 249 | 0.97 [0.8978, 0.9964] | 0.98 [0.9547, 0.9936] | 0.93 [0.8433, 0.9767] | 0.99 [0.9715, 0.999] | 49.31 | 0.03 | 0.98 [0.9557, 0.9912] |
| Pseudomonas aeruginosa | 68 | 3 | 1 | 250 | 0.99 [0.9219, 0.9996] | 0.99 [0.9657, 0.9975] | 0.96 [0.8814, 0.9912] | 1.00 [0.978, 0.9999] | 83.11 | 0.01 | 0.99 [0.9685, 0.9966] |
| Staphylococcus aureus | 61 | 7 | 0 | 254 | 1.00 [0.9413, 1] | 0.97 [0.9455, 0.9892] | 0.90 [0.7993, 0.9576] | 1.00 [0.9856, 1] | 37.29 | 0.00 | 0.98 [0.9557, 0.9912] |
| Mycoplasma pneumoniae | 0 | 0 | 0 | 322 | 0.00 [0, 0] | 1.00 [0.9886, 1] | 0.00 [0, 0] | 1.00 [0.9886, 1] | NA | 0.00 | 1.00 [0.9886, 1] |
| human adenoviruses | 2 | 1 | 1 | 318 | 0.67 [0.0943, 0.9916] | 1.00 [0.9827, 0.9999] | 0.67 [0.0943, 0.9916] | 1.00 [0.9827, 0.9999] | 212.67 | 0.33 | 0.99 [0.9777, 0.9992] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, Q.; Zhang, G.; Wang, T.; Li, J.; Kang, W.; Zhang, J.; Liu, Y.; Xu, Y. Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples. Microorganisms 2025, 13, 682. https://doi.org/10.3390/microorganisms13030682
Yi Q, Zhang G, Wang T, Li J, Kang W, Zhang J, Liu Y, Xu Y. Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples. Microorganisms. 2025; 13(3):682. https://doi.org/10.3390/microorganisms13030682
Chicago/Turabian StyleYi, Qiaolian, Ge Zhang, Tong Wang, Jin Li, Wei Kang, Jingjia Zhang, Yali Liu, and Yingchun Xu. 2025. "Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples" Microorganisms 13, no. 3: 682. https://doi.org/10.3390/microorganisms13030682
APA StyleYi, Q., Zhang, G., Wang, T., Li, J., Kang, W., Zhang, J., Liu, Y., & Xu, Y. (2025). Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples. Microorganisms, 13(3), 682. https://doi.org/10.3390/microorganisms13030682





