Fatal Congenital Toxoplasmosis with Progressive Liver Failure and Genomic Characterization of a Novel Isolate from the United States
Abstract
1. Introduction
2. Case Presentation and Genomic Characterization
2.1. Case Presentation
2.1.1. Maternal and Perinatal History
2.1.2. Neonatal Presentation
2.1.3. Diagnostic Workup
2.1.4. Clinical Course and Treatment
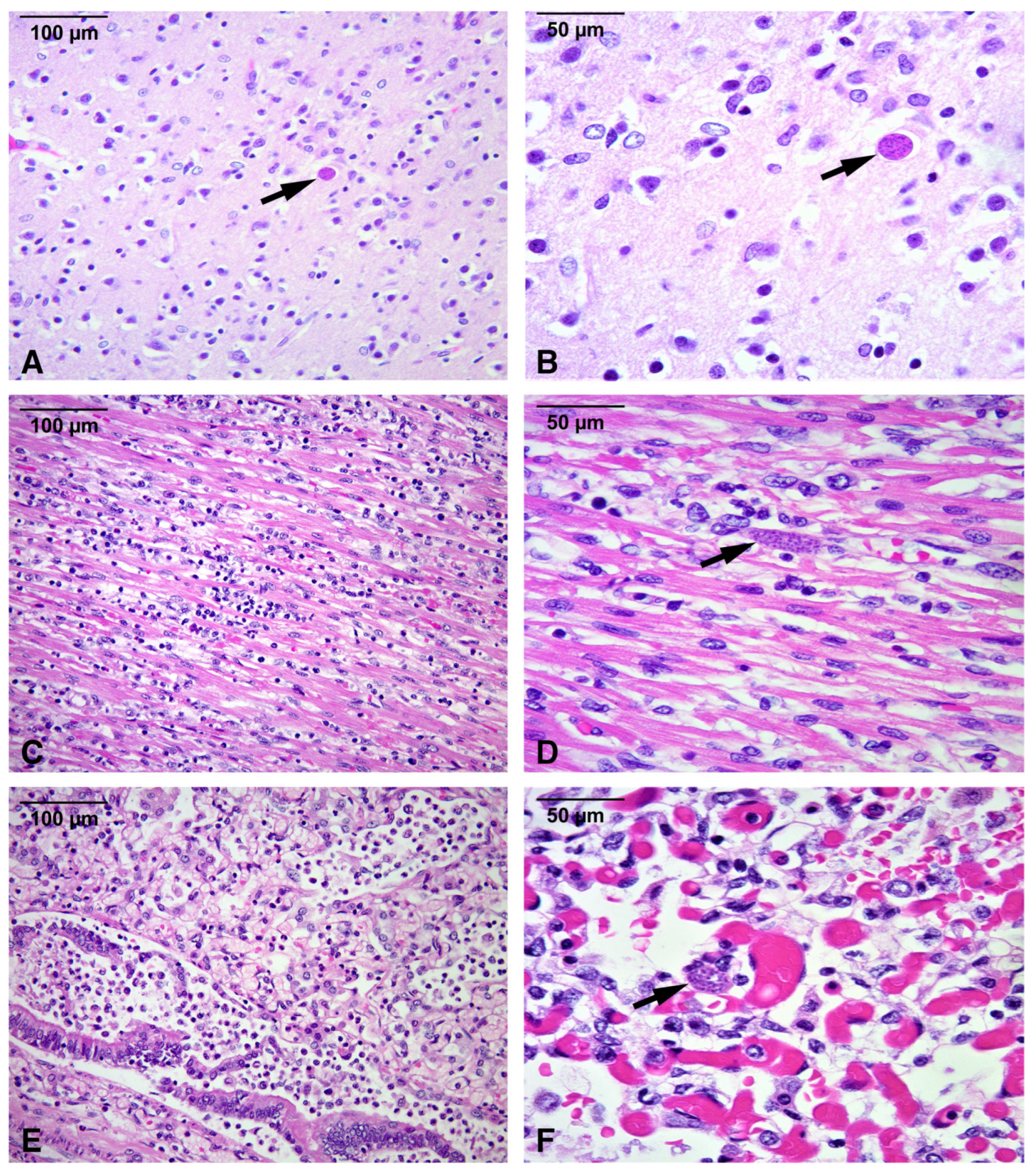
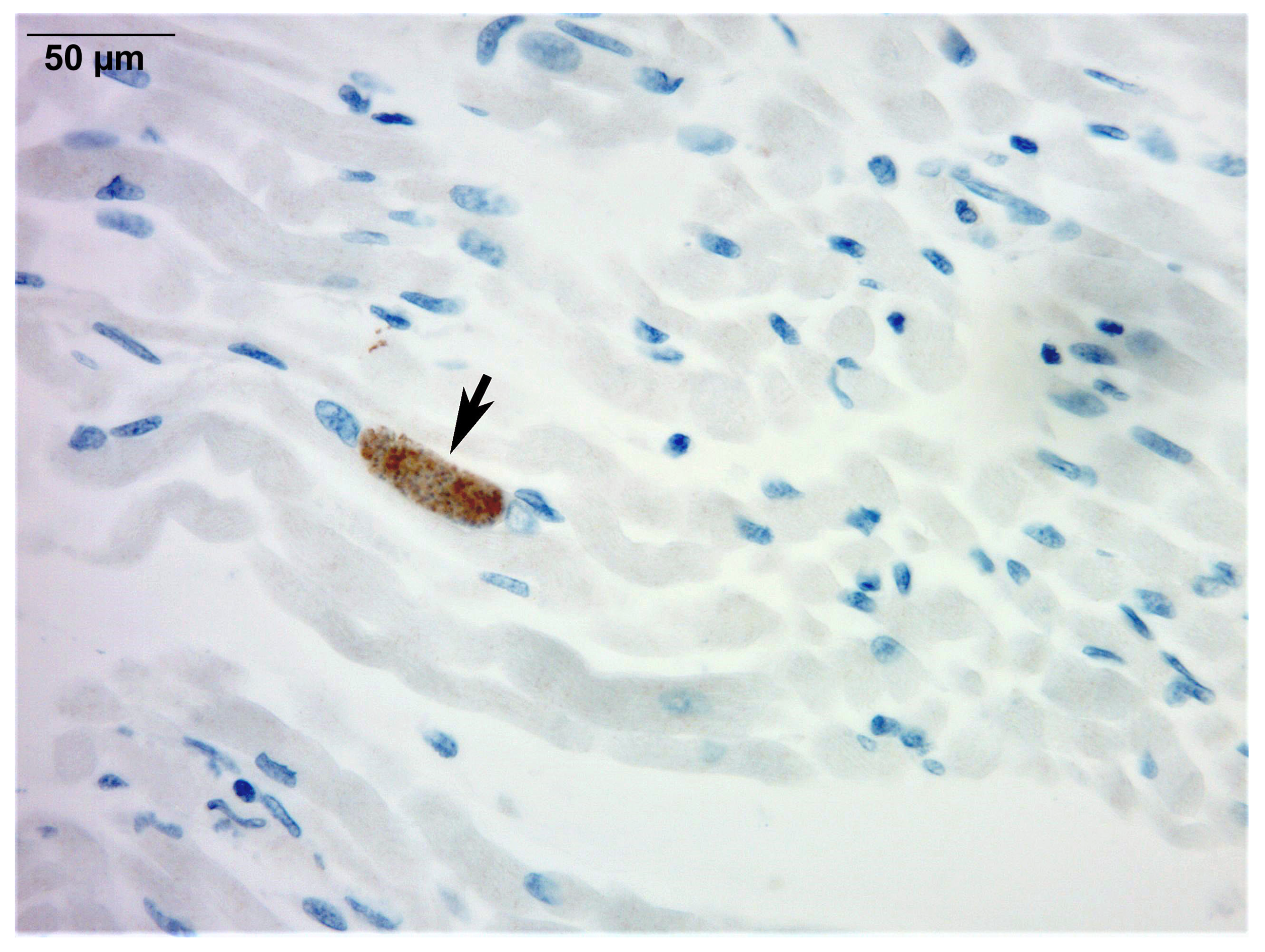
2.1.5. Autopsy Findings
| Postnatal Day | Key Events and Findings | Interventions/Outcomes |
|---|---|---|
| Birth (Day 0) | Male infant born at 38 weeks by emergency cesarean section. Birth weight 2680 g (7th percentile). Apgar 7/8. No hepatosplenomegaly. Labs: hypoglycemia, thrombocytopenia (16,000/mm3), elevated AST and direct bilirubin. | Non-invasive respiratory support. Platelet transfusions. IVIG × 3 for suspected NAIT (later excluded). |
| Day 6 | T. gondii serologies drawn after IVIG: IgG elevated (37 IU/mL), IgM negative. Abdominal/head ultrasound: normal. | Supportive care continued. |
| Day 15 | Fever, abdominal distension, leukocytosis. | Started nafcillin + gentamicin for presumed sepsis. |
| Day 16 | Infectious disease consultation. Maternal/infant paired T. gondii serologies sent. | — |
| Day 17 | Abdominal ultrasound: hepatosplenomegaly, ascites. Paracentesis: lymphocytic fluid. CSF: pleocytosis, elevated protein, PCR negative. Ophthalmology: no chorioretinitis. | Peritoneal fluid sent for T. gondii PCR. |
| Days 18–24 | Progressive liver failure: direct hyperbilirubinemia, thrombocytopenia, coagulopathy, ascites. HLH considered (elevated ferritin, sIL-2R, cytopenias) but excluded by additional testing. | Peritoneal drain placed. Exome sequencing sent. Anti-T. gondii enteral therapy considered but contraindicated due to high lactate. Intravenous trimethoprim-sulfamethoxazole and clindamycin considered but not started due to volume concerns. Supportive care. |
| Day 25 | Reference lab results: infant T. gondii IgG 1:64 (Sabin-Feldman), IgM positive (ISAGA), PCR positive in blood and peritoneal fluid. Maternal IgG 1:8000 (high avidity), IgM negative, IgA positive. Diagnosis: congenital toxoplasmosis. | Began pyrimethamine, sulfadiazine, folinic acid. |
| Day 26 | Acute cardiopulmonary decompensation → mechanical ventilation, nitric oxide, inotropes. | Liver transplantation not feasible. Maximal support. |
| Day 27 | Further deterioration despite intensive care. | Compassionate extubation. Death due to multiorgan failure secondary to disseminated toxoplasmosis. |
| Post-mortem | Autopsy: disseminated T. gondii infection with multi-organ inflammation and necrosis (liver, spleen, brain, heart, lungs, muscle, bladder). T. gondii cysts on hematoxylin and eosin. Immunohistochemistry and PCR confirmed infection. | Cause of death: multiorgan failure from disseminated congenital toxoplasmosis. |
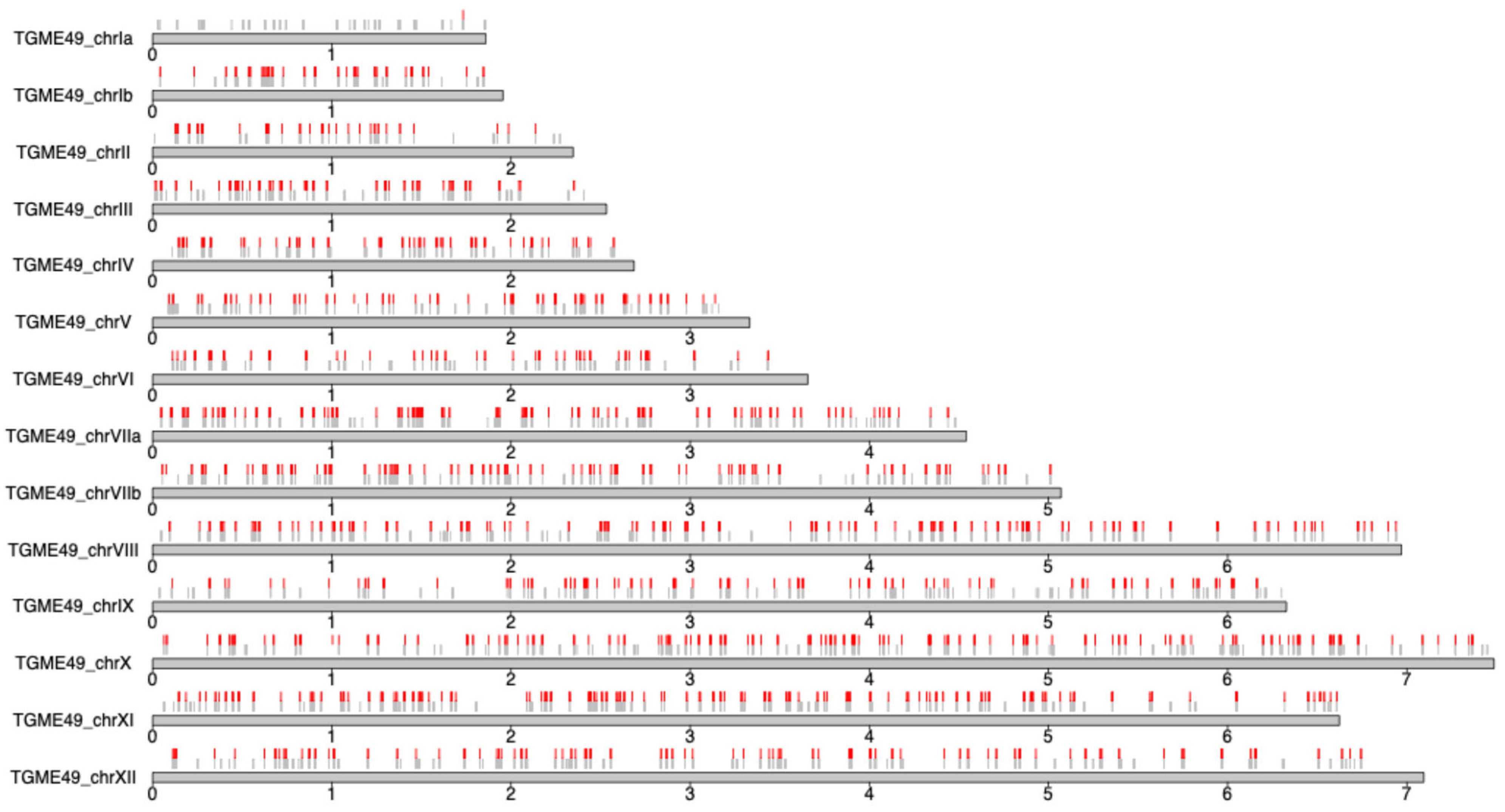
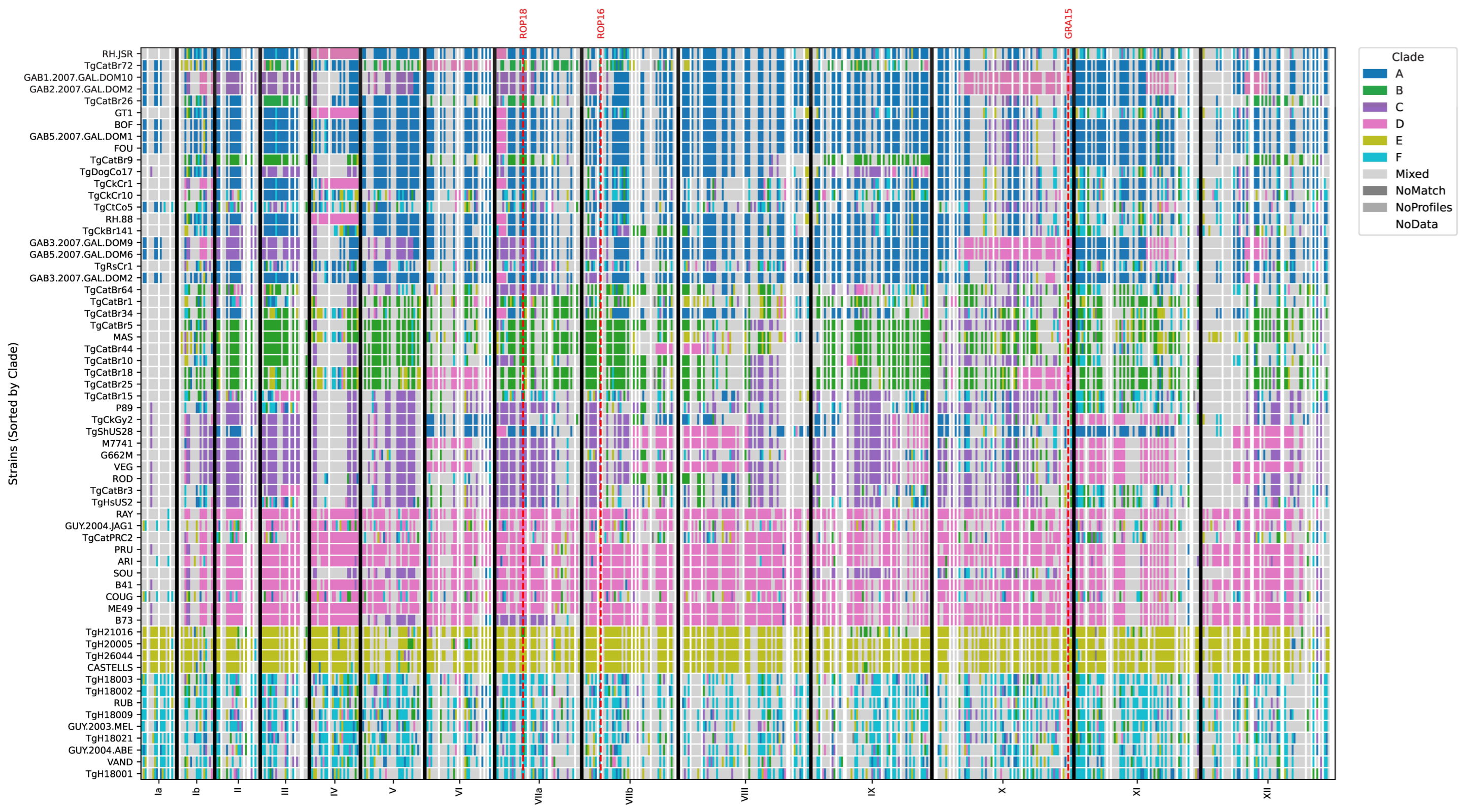
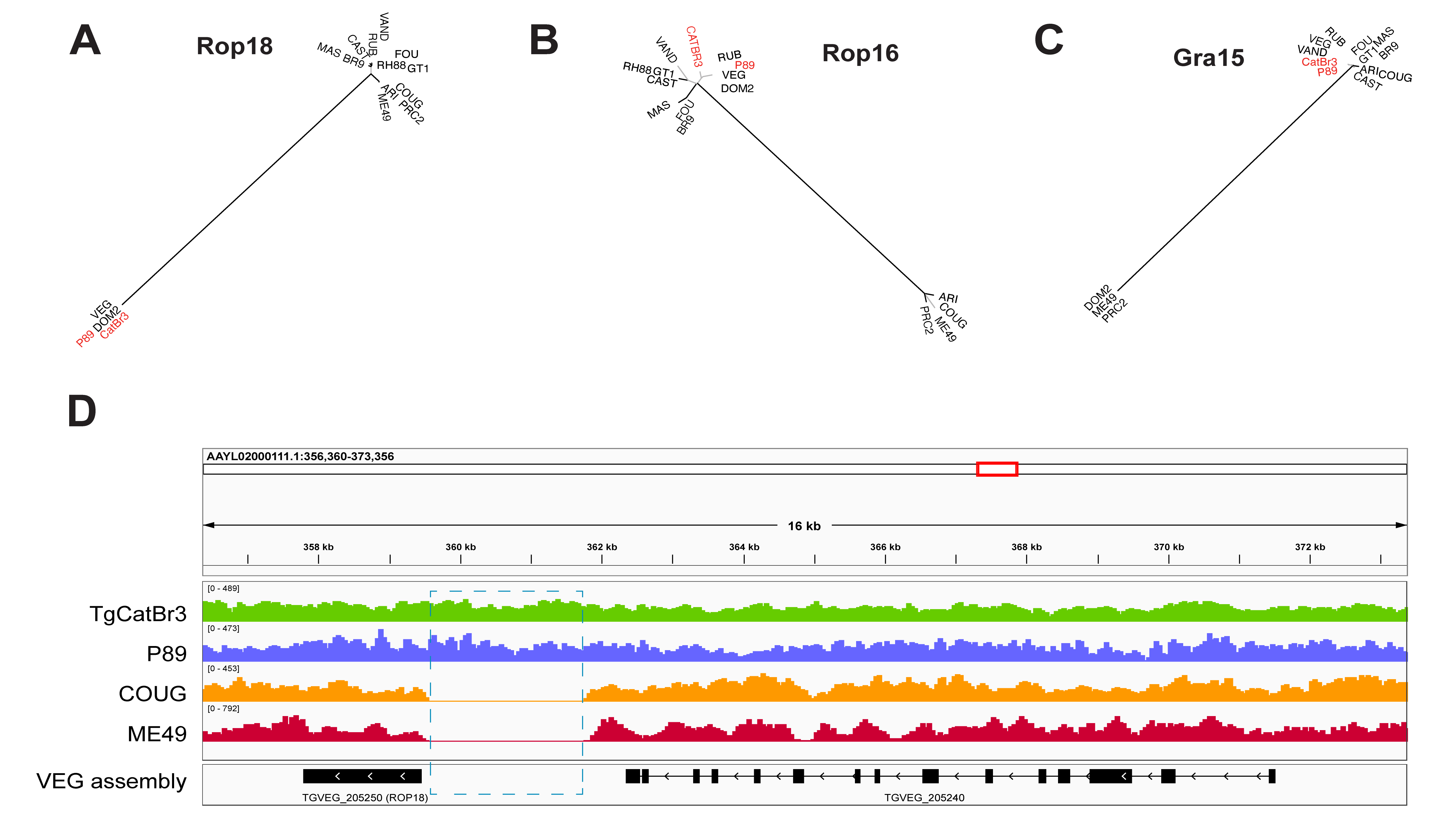
2.2. Genomic Characterization of the Infecting Strain
3. Methods
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HLH | Hemophagocytic lymphohistiocytosis |
| IVIG | Intravenous immunoglobulin |
| NE-II | Non-exclusively serotype II |
| SNV | Single-nucleotide variant |
| urGS | Ultra-rapid genome sequencing |
References
- McAuley, J.B. Congenital Toxoplasmosis. J. Pediatric Infect. Dis. Soc. 2014, 3, S30–S35. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, Y.A.; Read, J.S.; Committee On Infectious. Diagnosis, Treatment, and Prevention of Congenital Toxoplasmosis in the United States. Pediatrics 2017, 139, e20163860. [Google Scholar] [CrossRef]
- Wallon, M.; Peyron, F. Congenital Toxoplasmosis: A Plea for a Neglected Disease. Pathogens 2018, 7, 25. [Google Scholar] [CrossRef]
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Montoya, J.G.; Remington, J.S. Management of Toxoplasma gondii infection during pregnancy. Clin. Infect. Dis. 2008, 47, 554–566. [Google Scholar] [CrossRef]
- Bigna, J.J.; Tochie, J.N.; Tounouga, D.N.; Bekolo, A.O.; Ymele, N.S.; Youda, E.L.; Sime, P.S.; Nansseu, J.R. Global, regional, and country seroprevalence of Toxoplasma gondii in pregnant women: A systematic review, modelling and meta-analysis. Sci. Rep. 2020, 10, 12102. [Google Scholar] [CrossRef]
- Rostami, A.; Riahi, S.M.; Gamble, H.R.; Fakhri, Y.; Nourollahpour Shiadeh, M.; Danesh, M.; Behniafar, H.; Paktinat, S.; Foroutan, M.; Mokdad, A.H.; et al. Global prevalence of latent toxoplasmosis in pregnant women: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2020, 26, 673–683. [Google Scholar] [CrossRef]
- Mizani, A.; Alipour, A.; Sharif, M.; Sarvi, S.; Amouei, A.; Shokri, A.; Rahimi, M.T.; Hosseini, S.A.; Daryani, A. Toxoplasmosis seroprevalence in Iranian women and risk factors of the disease: A systematic review and meta-analysis. Trop. Med. Health 2017, 45, 7. [Google Scholar] [CrossRef]
- Bollani, L.; Auriti, C.; Achille, C.; Garofoli, F.; De Rose, D.U.; Meroni, V.; Salvatori, G.; Tzialla, C. Congenital Toxoplasmosis: The State of the Art. Front. Pediatr. 2022, 10, 894573. [Google Scholar] [CrossRef] [PubMed]
- El Bissati, K.; Levigne, P.; Lykins, J.; Adlaoui, E.B.; Barkat, A.; Berraho, A.; Laboudi, M.; El Mansouri, B.; Ibrahimi, A.; Rhajaoui, M.; et al. Global initiative for congenital toxoplasmosis: An observational and international comparative clinical analysis. Emerg. Microbes Infect. 2018, 7, 165. [Google Scholar] [CrossRef] [PubMed]
- Lykins, J.; Wang, K.; Wheeler, K.; Clouser, F.; Dixon, A.; El Bissati, K.; Zhou, Y.; Lyttle, C.; Rzhetsky, A.; McLeod, R. Understanding Toxoplasmosis in the United States Through “Large Data” Analyses. Clin. Infect. Dis. 2016, 63, 468–475. [Google Scholar] [CrossRef]
- Olariu, T.R.; Remington, J.S.; McLeod, R.; Alam, A.; Montoya, J.G. Severe congenital toxoplasmosis in the United States: Clinical and serologic findings in untreated infants. Pediatr. Infect. Dis. J. 2011, 30, 1056–1061. [Google Scholar] [CrossRef]
- McLeod, R.; Boyer, K.; Karrison, T.; Kasza, K.; Swisher, C.; Roizen, N.; Jalbrzikowski, J.; Remington, J.; Heydemann, P.; Noble, A.G.; et al. Outcome of treatment for congenital toxoplasmosis, 1981-2004: The National Collaborative Chicago-Based, Congenital Toxoplasmosis Study. Clin. Infect. Dis. 2006, 42, 1383–1394. [Google Scholar] [CrossRef]
- McLeod, R.; Boyer, K.M.; Lee, D.; Mui, E.; Wroblewski, K.; Karrison, T.; Noble, A.G.; Withers, S.; Swisher, C.N.; Heydemann, P.T.; et al. Prematurity and severity are associated with Toxoplasma gondii alleles (NCCCTS, 1981-2009). Clin. Infect. Dis. 2012, 54, 1595–1605. [Google Scholar] [CrossRef]
- Mohanty, S.; Shah, I.; Bhatnagar, S. Neonatal hepatitis with toxoplasmosis. J. Clin. Neonatol. 2012, 1, 96–97. [Google Scholar] [CrossRef]
- Paquet, C.; Yudin, M.H.; Allen, V.M.; Bouchard, C.; Boucher, M.; Caddy, S.; Castillo, E.; Money, D.M.; Murphy, K.E.; Ogilvie, G.; et al. Toxoplasmosis in pregnancy: Prevention, screening, and treatment. J. Obstet. Gynaecol. Can. 2013, 35, 78–79. [Google Scholar] [CrossRef] [PubMed]
- Dash, C.A.; Madden, J.A.; Cummings, C.; Rose, M.; Wilson, S.D.; Mori, M.; Agrawal, P.; Chaudhari, B.; Wojcik, M. Perinatal-Lethal Non-Immune Fetal Hydrops Attributed to MECOM-associated Bone Marrow Failure. Cold Spring Harb. Mol. Case Stud. 2023, 9, a006289. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.; Fitch, J.; Lammi, G.; Crist, E.; Cottrell, C.; Magrini, V.; Chaudhari, B.; White, P. Application of constitutional rapid genome sequencing to detect infectious disease in a critically ill neonate. Mol. Genet. Metab. 2021, 132, S273. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Warrenfeltz, S.; Basenko, E.Y.; Crouch, K.; Harb, O.S.; Kissinger, J.C.; Roos, D.S.; Shanmugasundram, A.; Silva-Franco, F. EuPathDB: The Eukaryotic Pathogen Genomics Database Resource. In Eukaryotic Genomic Databases: Methods and Protocols; Kollmar, M., Ed.; Springer: New York, NY, USA, 2018; pp. 69–113. [Google Scholar]
- Lorenzi, H.; Khan, A.; Behnke, M.S.; Namasivayam, S.; Swapna, L.S.; Hadjithomas, M.; Karamycheva, S.; Pinney, D.; Brunk, B.P.; Ajioka, J.W.; et al. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun. 2016, 7, 10147. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Miller, N.; Roos, D.S.; Dubey, J.P.; Ajzenberg, D.; Darde, M.L.; Ajioka, J.W.; Rosenthal, B.; Sibley, L.D. A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii. mBio 2011, 2, e00228–00211. [Google Scholar] [CrossRef]
- Ihara, F.; Kyan, H.; Takashima, Y.; Ono, F.; Hayashi, K.; Matsuo, T.; Igarashi, M.; Nishikawa, Y.; Hikosaka, K.; Sakamoto, H.; et al. Far-East Asian Toxoplasma isolates share ancestry with North and South/Central American recombinant lineages. Nat. Commun. 2024, 15, 4278. [Google Scholar] [CrossRef]
- Kochanowsky, J.A.; Chandrasekaran, S.; Sanchez, J.R.; Thomas, K.K.; Koshy, A.A. ROP16-mediated activation of STAT6 enhances cyst development of type III Toxoplasma gondii in neurons. PLoS Pathog. 2023, 19, e1011347. [Google Scholar] [CrossRef]
- Sanchez, V.; de-la-Torre, A.; Gomez-Marin, J.E. Characterization of ROP18 alleles in human toxoplasmosis. Parasitol. Int. 2014, 63, 463–469. [Google Scholar] [CrossRef]
- Rosowski, E.E.; Lu, D.; Julien, L.; Rodda, L.; Gaiser, R.A.; Jensen, K.D.; Saeij, J.P. Strain-specific activation of the NF-kappaB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 2011, 208, 195–212. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Armstrong, L.; Isaacs, D.; Evans, N. Severe neonatal toxoplasmosis after third trimester maternal infection. Pediatr. Infect. Dis. J. 2004, 23, 968–969. [Google Scholar] [CrossRef]
- Elbez-Rubinstein, A.; Ajzenberg, D.; Darde, M.L.; Cohen, R.; Dumetre, A.; Yera, H.; Gondon, E.; Janaud, J.C.; Thulliez, P. Congenital toxoplasmosis and reinfection during pregnancy: Case report, strain characterization, experimental model of reinfection, and review. J. Infect. Dis. 2009, 199, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Al-Hamod, D.; Vauloup, C.; Goulet, M.; Zupan-Simunek, V.; Castel, C.; Boileau, P. Delayed onset of severe neonatal toxoplasmosis. J. Perinatol. 2010, 30, 231–232. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, F.; Rigourd, V.; Ikounga, P.; Bessieres, B.; Magny, J.F.; Thulliez, P. Disseminated congenital toxoplasma infection with a type II strain. Pediatr. Infect. Dis. J. 2011, 30, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Pomares, C.; Montoya, J.G. Laboratory Diagnosis of Congenital Toxoplasmosis. J. Clin. Microbiol. 2016, 54, 2448–2454. [Google Scholar] [CrossRef] [PubMed]
- Gruter, T.; Ott, A.; Meyer, W.; Jarius, S.; Kinner, M.; Motte, J.; Pitarokoili, K.; Gold, R.; Komorowski, L.; Ayzenberg, I. Effects of IVIg treatment on autoantibody testing in neurological patients: Marked reduction in sensitivity but reliable specificity. J. Neurol. 2020, 267, 715–720. [Google Scholar] [CrossRef]
- Hanson, K.E.; Gabriel, N.; McHardy, I.; Hoffmann, W.; Cohen, S.H.; Welch, R.; Couturier, M.R.; Thompson, G.R. 2031. False-positive Serologic Results attributable to IVIG therapy. Open Forum Infect. Dis. 2018, 5, S591–S592. [Google Scholar] [CrossRef]
- Leveque, M.F.; Albaba, S.; Arrada, N.; Avignon, M.; Sasso, M.; Fillaux, J.; Lachaud, L. Diagnosis of Congenital Toxoplasmosis: No Benefit of IgA Antibody Detection by Platelia ELISA in a Tricentric Evaluation. J. Clin. Microbiol. 2022, 60, e0011622. [Google Scholar] [CrossRef]
- Olariu, T.R.; Blackburn, B.G.; Press, C.; Talucod, J.; Remington, J.S.; Montoya, J.G. Role of Toxoplasma IgA as Part of a Reference Panel for the Diagnosis of Acute Toxoplasmosis during Pregnancy. J. Clin. Microbiol. 2019, 57, e01357-18. [Google Scholar] [CrossRef] [PubMed]
- Teimouri, A.; Mohtasebi, S.; Kazemirad, E.; Keshavarz, H. Role of Toxoplasma gondii IgG Avidity Testing in Discriminating between Acute and Chronic Toxoplasmosis in Pregnancy. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Ribeiro, S.K.; Mariano, I.M.; Cunha, A.C.R.; Pajuaba, A.; Mineo, T.W.P.; Mineo, J.R. Treatment Protocols for Gestational and Congenital Toxoplasmosis: A Systematic Review and Meta-Analysis. Microorganisms 2025, 13, 723. [Google Scholar] [CrossRef]
- Hutson, S.L.; Wheeler, K.M.; McLone, D.; Frim, D.; Penn, R.; Swisher, C.N.; Heydemann, P.T.; Boyer, K.M.; Noble, A.G.; Rabiah, P.; et al. Patterns of Hydrocephalus Caused by Congenital Toxoplasma gondii Infection Associate With Parasite Genetics. Clin. Infect. Dis. 2015, 61, 1831–1834. [Google Scholar] [CrossRef]
- McLeod, R. Human Toxoplasma Infection. In Toxoplasma gondii: The Model Apicomplexan—Perspectives and Methods, 3rd ed.; Weiss, L.M., Kim, K., Eds.; Academic Press: Waltham, MA, USA, 2020. [Google Scholar]
- Carme, B.; Demar, M.; Ajzenberg, D.; Darde, M.L. Severe acquired toxoplasmosis caused by wild cycle of Toxoplasma gondii, French Guiana. Emerg. Infect. Dis. 2009, 15, 656–658. [Google Scholar] [CrossRef]
- Pomares, C.; Devillard, S.; Holmes, T.H.; Olariu, T.R.; Press, C.J.; Ramirez, R.; Talucod, J.; Estran, R.; Su, C.; Dubey, J.P.; et al. Genetic Characterization of Toxoplasma gondii DNA Samples Isolated From Humans Living in North America: An Unexpected High Prevalence of Atypical Genotypes. J. Infect. Dis. 2018, 218, 1783–1791. [Google Scholar] [CrossRef]
- Ajzenberg, D.; Yera, H.; Marty, P.; Paris, L.; Dalle, F.; Menotti, J.; Aubert, D.; Franck, J.; Bessieres, M.H.; Quinio, D.; et al. Genotype of 88 Toxoplasma gondii isolates associated with toxoplasmosis in immunocompromised patients and correlation with clinical findings. J. Infect. Dis. 2009, 199, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Ahmed, M.; Ibrahim, N. The global seroprevalence of Toxoplasma gondii infection in workers occupationally exposed to animals (1972-2023): A systematic review and meta-analysis. Vet. Q. 2024, 44, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, A.C.; Andrade, G.M.; Costa, J.G.; Pinheiro, B.V.; Vasconcelos-Santos, D.V.; Ferreira, A.M.; Su, C.; Januario, J.N.; Vitor, R.W. Genetic characterization of Toxoplasma gondii revealed highly diverse genotypes for isolates from newborns with congenital toxoplasmosis in southeastern Brazil. J. Clin. Microbiol. 2013, 51, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Costa, J.G.L.; Martins-Duarte, E.S.; Pinto, L.V.; Baraviera, R.A.C.; do Rego, W.M.F.; Vitor, R.W.A. Investigation of Virulence-Related Markers in Atypical Strains of Toxoplasma gondii from Brazil. Microorganisms 2025, 13, 301. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, H.; Liu, D.; Huo, X.; Gao, J.; Song, X.; Xu, X.; Huang, K.; Liu, W.; Wang, Y.; et al. Genotypes and mouse virulence of Toxoplasma gondii isolates from animals and humans in China. PLoS ONE 2013, 8, e53483, Erratum in PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Chaichan, P.; Mercier, A.; Galal, L.; Mahittikorn, A.; Ariey, F.; Morand, S.; Boumediene, F.; Udonsom, R.; Hamidovic, A.; Murat, J.B.; et al. Geographical distribution of Toxoplasma gondii genotypes in Asia: A link with neighboring continents. Infect. Genet. Evol. 2017, 53, 227–238. [Google Scholar] [CrossRef]
- Su, C.; Khan, A.; Zhou, P.; Majumdar, D.; Ajzenberg, D.; Darde, M.L.; Zhu, X.Q.; Ajioka, J.W.; Rosenthal, B.M.; Dubey, J.P.; et al. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc. Natl. Acad. Sci. USA 2012, 109, 5844–5849. [Google Scholar] [CrossRef]
- Dubey, J.P.; Velmurugan, G.V.; Rajendran, C.; Yabsley, M.J.; Thomas, N.J.; Beckmen, K.B.; Sinnett, D.; Ruid, D.; Hart, J.; Fair, P.A.; et al. Genetic characterisation of Toxoplasma gondii in wildlife from North America revealed widespread and high prevalence of the fourth clonal type. Int. J. Parasitol. 2011, 41, 1139–1147. [Google Scholar] [CrossRef]
- Dubey, J.P.; Cerqueira-Cezar, C.K.; Murata, F.H.A.; Kwok, O.C.H.; Hill, D.; Yang, Y.; Su, C. All about Toxoplasma gondii infections in pigs: 2009-2020. Vet. Parasitol. 2020, 288, 109185. [Google Scholar] [CrossRef]
- Dubey, J.P.; Sundar, N.; Hill, D.; Velmurugan, G.V.; Bandini, L.A.; Kwok, O.C.; Majumdar, D.; Su, C. High prevalence and abundant atypical genotypes of Toxoplasma gondii isolated from lambs destined for human consumption in the USA. Int. J. Parasitol. 2008, 38, 999–1006. [Google Scholar] [CrossRef]
- Dubey, J.P.; Rajendran, C.; Ferreira, L.R.; Martins, J.; Kwok, O.C.; Hill, D.E.; Villena, I.; Zhou, H.; Su, C.; Jones, J.L. High prevalence and genotypes of Toxoplasma gondii isolated from goats, from a retail meat store, destined for human consumption in the USA. Int. J. Parasitol. 2011, 41, 827–833. [Google Scholar] [CrossRef]
- Taylor, S.; Barragan, A.; Su, C.; Fux, B.; Fentress, S.J.; Tang, K.; Beatty, W.L.; Hajj, H.E.; Jerome, M.; Behnke, M.S.; et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 2006, 314, 1776–1780. [Google Scholar] [CrossRef]
- Mukhopadhyay, D.; Sangare, L.O.; Braun, L.; Hakimi, M.A.; Saeij, J.P. Toxoplasma GRA15 limits parasite growth in IFNgamma-activated fibroblasts through TRAF ubiquitin ligases. EMBO J. 2020, 39, e103758. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, K.M.; Gorfu, G.; Hassan, M.A.; Printz, M.; Crown, D.; Leppla, S.H.; Grigg, M.E.; Saeij, J.P.; Moayeri, M. Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 2014, 10, e1003927. [Google Scholar] [CrossRef]
- Dubey, J.P.; Murata, F.H.A.; Cerqueira-Cezar, C.K.; Kwok, O.C.H.; Yang, Y.; Su, C. Recent epidemiologic, clinical, and genetic diversity of Toxoplasma gondii infections in non-human primates. Res. Vet. Sci. 2021, 136, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Witola, W.H.; Mui, E.; Hargrave, A.; Liu, S.; Hypolite, M.; Montpetit, A.; Cavailles, P.; Bisanz, C.; Cesbron-Delauw, M.F.; Fournie, G.J.; et al. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect. Immun. 2011, 79, 756–766. [Google Scholar] [CrossRef]
- Witola, W.H.; Liu, S.R.; Montpetit, A.; Welti, R.; Hypolite, M.; Roth, M.; Zhou, Y.; Mui, E.; Cesbron-Delauw, M.F.; Fournie, G.J.; et al. ALOX12 in human toxoplasmosis. Infect. Immun. 2014, 82, 2670–2679. [Google Scholar] [CrossRef] [PubMed]
- Karanovic, D.; Michelow, I.C.; Hayward, A.R.; DeRavin, S.S.; Delmonte, O.M.; Grigg, M.E.; Dobbs, A.K.; Niemela, J.E.; Stoddard, J.; Alhinai, Z.; et al. Disseminated and Congenital Toxoplasmosis in a Mother and Child With Activated PI3-Kinase delta Syndrome Type 2 (APDS2): Case Report and a Literature Review of Toxoplasma Infections in Primary Immunodeficiencies. Front. Immunol. 2019, 10, 77. [Google Scholar] [CrossRef]
- Bonnet Ducrot, S.; Plantaz, D.; Mathieu, N.; Debillon, T.; Bost Bru, C.; Brenier-Pinchart, M.P.; Fricker-Hidalgo, H.; Chevallier, M. Neonatal fever: A puzzling case. Arch. Pediatr. 2018, 25, 435–438. [Google Scholar] [CrossRef]
- Campos, F.A.; Andrade, G.M.; Lanna Ade, P.; Lage, B.F.; Assumpcao, M.V.; Pinto, J.A. Incidence of congenital toxoplasmosis among infants born to HIV-coinfected mothers: Case series and literature review. Braz. J. Infect. Dis. 2014, 18, 609–617. [Google Scholar] [CrossRef]
- Phan, L.; Kasza, K.; Jalbrzikowski, J.; Noble, A.G.; Latkany, P.; Kuo, A.; Mieler, W.; Meyers, S.; Rabiah, P.; Boyer, K.; et al. Longitudinal study of new eye lesions in treated congenital toxoplasmosis. Ophthalmology 2008, 115, 553–559.E8. [Google Scholar] [CrossRef] [PubMed]
- Felin, M.S.; Wang, K.; Moreira, A.; Grose, A.; Leahy, K.; Zhou, Y.; Clouser, F.A.; Siddiqui, M.; Leong, N.; Goodall, P.; et al. Building Programs to Eradicate Toxoplasmosis Part I: Introduction and Overview. Curr. Pediatr. Rep. 2022, 10, 57–92. [Google Scholar] [CrossRef]
- Cortina-Borja, M.; Tan, H.K.; Wallon, M.; Paul, M.; Prusa, A.; Buffolano, W.; Malm, G.; Salt, A.; Freeman, K.; Petersen, E.; et al. Prenatal treatment for serious neurological sequelae of congenital toxoplasmosis: An observational prospective cohort study. PLoS Med. 2010, 7, e1000351. [Google Scholar] [CrossRef]
- Wallon, M.; Peyron, F.; Cornu, C.; Vinault, S.; Abrahamowicz, M.; Kopp, C.B.; Binquet, C. Congenital toxoplasma infection: Monthly prenatal screening decreases transmission rate and improves clinical outcome at age 3 years. Clin. Infect. Dis. 2013, 56, 1223–1231. [Google Scholar] [CrossRef]
- Prusa, A.R.; Kasper, D.C.; Pollak, A.; Gleiss, A.; Waldhoer, T.; Hayde, M. The Austrian Toxoplasmosis Register, 1992-2008. Clin. Infect. Dis. 2015, 60, e4–e10. [Google Scholar] [CrossRef] [PubMed]
- Hotop, A.; Hlobil, H.; Gross, U. Efficacy of rapid treatment initiation following primary Toxoplasma gondii infection during pregnancy. Clin. Infect. Dis. 2012, 54, 1545–1552. [Google Scholar] [CrossRef]
- Abraham, S.; Piarroux, R.; Zhou, Y.; Tesic, V.; Abeleda, A.; Houhou-Fidouh, N.; Nicaise-Rolland, P.; Landraud, L.; McLeod, R.; Houze, S. Performances of ICT Toxoplasma IgG-IgM test in comparison with Vidas(R) toxo competition to determine the immune status against Toxoplasma gondii. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1327–1335. [Google Scholar] [CrossRef]
- Zhou, Y.; Leahy, K.; Grose, A.; Lykins, J.; Siddiqui, M.; Leong, N.; Goodall, P.; Withers, S.; Ashi, K.; Schrantz, S.; et al. Novel paradigm enables accurate monthly gestational screening to prevent congenital toxoplasmosis and more. PLoS Negl. Trop. Dis. 2024, 18, e0011335. [Google Scholar] [CrossRef] [PubMed]
- Prusa, A.R.; Kasper, D.C.; Sawers, L.; Walter, E.; Hayde, M.; Stillwaggon, E. Congenital toxoplasmosis in Austria: Prenatal screening for prevention is cost-saving. PLoS Negl. Trop. Dis. 2017, 11, e0005648. [Google Scholar] [CrossRef]
- Sawers, L.; Wallon, M.; Mandelbrot, L.; Villena, I.; Stillwaggon, E.; Kieffer, F. Prevention of congenital toxoplasmosis in France using prenatal screening: A decision-analytic economic model. PLoS ONE 2022, 17, e0273781. [Google Scholar] [CrossRef]
- Sagel, U.; Kramer, A.; Mikolajczyk, R.T. Incidence of maternal Toxoplasma infections in pregnancy in Upper Austria, 2000-2007. BMC Infect. Dis. 2011, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Benard, A.; Petersen, E.; Salamon, R.; Chene, G.; Gilbert, R.; Salmi, L.R.; European Toxo Prevention Study Group. Survey of European programmes for the epidemiological surveillance of congenital toxoplasmosis. Euro Surveill. 2008, 13, 18834. [Google Scholar] [CrossRef] [PubMed]
- Mandelbrot, L. Congenital toxoplasmosis: What is the evidence for chemoprophylaxis to prevent fetal infection? Prenat. Diagn. 2020, 40, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Thiebaut, R.; Leproust, S.; Chene, G.; Gilbert, R. Effectiveness of prenatal treatment for congenital toxoplasmosis: A meta-analysis of individual patients’ data. Lancet 2007, 369, 115–122. [Google Scholar] [CrossRef]
- Gomes Ferrari Strang, A.G.; Ferrar, R.G.; Falavigna-Guilherme, A.L. Gestational toxoplasmosis treatment changes the child’s prognosis: A cohort study in southern Brazil. PLoS Negl. Trop. Dis. 2023, 17, e0011544. [Google Scholar] [CrossRef]
- Guarch-Ibanez, B.; Carreras-Abad, C.; Frick, M.A.; Blazquez-Gamero, D.; Baquero-Artigao, F.; Fuentes, I.; Spanish Research Network of Congenital Toxoplasmosis Group; Soler-Palacin, P. REIV-TOXO Project: Results from a Spanish cohort of congenital toxoplasmosis (2015-2022). The beneficial effects of prenatal treatment on clinical outcomes of infected newborns. PLoS Negl. Trop. Dis. 2024, 18, e0012619. [Google Scholar] [CrossRef]


| Patient | Age at Diagnosis | Diagnostic Testing of the Infant | Maternal Serologies | Clinical Presentation | Outcome | Year, Source | T. gondii Strain Genotype (if Reported) |
|---|---|---|---|---|---|---|---|
| 1 | Day 2 | IgM+ | IgG-, IgM- | Thrombocytopenia, coagulopathy, liver failure, respiratory failure, pulmonary hypertension | Survived | 2004 [29] | Not reported |
| 2 | Day 8 | IgM+, blood PCR+ | IgG+, IgM- | Thrombocytopenia, hepatosplenomegaly, respiratory distress, chorioretinitis | Survived | 2009 [30] | Atypical, South American-like microsatellite genotype (non-type II), isolated from newborn blood |
| 3 | Day 6 | IgM+ | IgG+, IgM+ | Thrombocytopenia, coagulopathy, transaminitis, cardiac arrest, respiratory failure, pulmonary hemorrhage, pulmonary hypertension, chorioretinitis, intracranial calcification | Survived | 2010 [31] | Not reported |
| 4 | Day 3 | IgM+, PCR+ (blood, tracheal aspirate, urine, ascites) | IgG+, IgM+ | Thrombocytopenia, coagulopathy, respiratory failure, pulmonary hypertension, hepatosplenomegaly, ascites, macular bleeding and edema | Death on day 10 | 2011 [32] | Type II strain on genotyping of infant samples |
| 5 | Day 25 | IgM+, blood PCR+ | IgG+, IgM- | Thrombocytopenia, liver failure, ascites, respiratory distress | Death on day 27 | 2020, present case | Clade C (haplogroup 9) isolate TgHsUS2, closely related to P89/TgCatBr3, with type III-like ROP18/ROP16/GRA15 alleles |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kojima, K.; Varier, I.; Sayegh, R.; Shimamura, M.; Chaudhari, B.P.; Bernieh, A.; Schulz, M.J.; White, P.; Fitch, J.; Medoro, A.K.; et al. Fatal Congenital Toxoplasmosis with Progressive Liver Failure and Genomic Characterization of a Novel Isolate from the United States. Microorganisms 2025, 13, 2865. https://doi.org/10.3390/microorganisms13122865
Kojima K, Varier I, Sayegh R, Shimamura M, Chaudhari BP, Bernieh A, Schulz MJ, White P, Fitch J, Medoro AK, et al. Fatal Congenital Toxoplasmosis with Progressive Liver Failure and Genomic Characterization of a Novel Isolate from the United States. Microorganisms. 2025; 13(12):2865. https://doi.org/10.3390/microorganisms13122865
Chicago/Turabian StyleKojima, Katsuaki, Indu Varier, Rouba Sayegh, Masako Shimamura, Bimal P. Chaudhari, Anas Bernieh, Matthew J. Schulz, Peter White, James Fitch, Alexandra K. Medoro, and et al. 2025. "Fatal Congenital Toxoplasmosis with Progressive Liver Failure and Genomic Characterization of a Novel Isolate from the United States" Microorganisms 13, no. 12: 2865. https://doi.org/10.3390/microorganisms13122865
APA StyleKojima, K., Varier, I., Sayegh, R., Shimamura, M., Chaudhari, B. P., Bernieh, A., Schulz, M. J., White, P., Fitch, J., Medoro, A. K., Lorenzi, H. A., & McLeod, R. (2025). Fatal Congenital Toxoplasmosis with Progressive Liver Failure and Genomic Characterization of a Novel Isolate from the United States. Microorganisms, 13(12), 2865. https://doi.org/10.3390/microorganisms13122865






