Biofilm Formation in Arcobacter butzleri and Arcobacter cryaerophilus: Phenotypic and Genotypic Characterization of Food and Environmental Isolates
Abstract
1. Introduction
2. Materials and Methods
2.1. A. butzleri Strains Identification by mPCR and 16S rRNA-RFLP
2.2. Biofilm-Associated Gene Detection
2.3. Motility Assay
2.4. Congo-Red Assay
2.5. Biofilm Formation Assay
2.6. Statistical Analysis
3. Results
3.1. Detection of Biofilm-Associated Genes
3.2. Bacterial Motility Assessed by Motility Assay
3.3. Biofilm Formation Assessed by Congo Red Agar (CRA) Assay
3.4. Quantitative Biofilm Formation Assessed by Christensen Method
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramees, T.P.; Dhama, K.; Karthik, K.; Rathore, R.S.; Kumar, A.; Saminathan, M.; Tiwari, R.; Malik, Y.S.; Singh, R.K. Arcobacter: An emerging food-borne zoonotic pathogen, its public health concerns and advances in diagnosis and control—A comprehensive review. Vet. Q. 2017, 37, 136–161. [Google Scholar] [CrossRef] [PubMed]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef] [PubMed]
- Levican, A.; Collado, L.; Figueras, M.J. Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 2013, 36, 22–27. [Google Scholar] [CrossRef]
- Ferreira, S.; Queiroz, J.A.; Oleastro, M.; Domingues, F.C. Insights in the pathogenesis and resistance of Arcobacter: A review. Crit. Rev. Microbiol. 2016, 42, 364–383. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, E.; Buzzanca, D.; Chiesa, F.; Botta, C.; Rantsiou, K.; Houf, K.; Alessandria, V. Exploring multi-antibiotic resistance in Arcobacter butzleri isolates from a poultry processing plant in northern Italy: An in-depth inquiry. Food Control 2024, 163, 110500. [Google Scholar] [CrossRef]
- Couto, F.; Martins, I.; Vale, F.; Domingues, F.; Oleastro, M.; Ferreira, S. Insights into macrolide resistance in Arcobacter butzleri: Potential resistance mechanisms and impact on bacterial fitness and virulence. J. Antimicrob. Chemother. 2024, 79, 2708–2717. [Google Scholar] [CrossRef]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Fanelli, F.; Di Pinto, A.; Mottola, A.; Mule, G.; Chieffi, D.; Baruzzi, F.; Tantillo, G.; Fusco, V. Genomic Characterization of Arcobacter butzleri Isolated from Shellfish: Novel Insight into Antibiotic Resistance and Virulence Determinants. Front. Microbiol. 2019, 10, 670. [Google Scholar] [CrossRef]
- Assanta, M.A.; Roy, D.; Lemay, M.-J.; Montpetit, D. Attachment of Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii to abiotic surfaces and survival in water at 4 °C. J. Food Prot. 2002, 65, 1240–1247. [Google Scholar] [CrossRef]
- Šilha, D.; Sirotková, S.; Švarcová, K.; Hofmeisterová, L.; Koryčanová, K.; Šilhová, L. Biofilm formation ability of Arcobacter-like and Campylobacter strains under different conditions and on food processing materials. Microorganisms 2021, 9, 2017. [Google Scholar] [CrossRef]
- Martins, R.; Mateus, C.; Domingues, F.; Bücker, R.; Oleastro, M.; Ferreira, S. Effect of atmospheric conditions on pathogenic phenotypes of Arcobacter butzleri. Microorganisms 2022, 10, 2409. [Google Scholar] [CrossRef] [PubMed]
- Mateus, C.; Maia, C.J.; Domingues, F.; Bücker, R.; Oleastro, M.; Ferreira, S. Evaluation of bile salts on the survival and modulation of virulence of Aliarcobacter butzleri. Antibiotics 2023, 12, 1387. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Sánchez, A.; Baztarrika, I.; Alonso, R.; Fernández-Astorga, A.; Martínez-Ballesteros, I.; Martinez-Malaxetxebarria, I. Arcobacter butzleri Biofilms: Insights into the Genes Beneath Their Formation. Microorganisms 2022, 10, 1280. [Google Scholar] [CrossRef] [PubMed]
- Guerry, P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007, 15, 456–461. [Google Scholar] [CrossRef]
- Martínez-Malaxetxebarria, I.; Girbau, C.; Salazar-Sánchez, A.; Baztarrika, I.; Martínez-Ballesteros, I.; Laorden, L.; Alonso, R.; Fernández-Astorga, A. Genetic characterization and biofilm formation of potentially pathogenic foodborne Arcobacter isolates. Int. J. Food Microbiol. 2022, 373, 109712. [Google Scholar] [CrossRef]
- Reeser, R.J.; Medler, R.T.; Billington, S.J.; Jost, B.H.; Joens, L.A. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 2007, 73, 1908–1913. [Google Scholar] [CrossRef]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef]
- Šilha, D.; Šilhová-Hrušková, L.; Vytřasová, J. Modified isolation method of Arcobacter spp. from different environmental and food samples. Folia Microbiol. 2015, 60, 515–521. [Google Scholar] [CrossRef]
- Douidah, L.; De Zutter, L.; Vandamme, P.; Houf, K. Identification of five human and mammal associated Arcobacter species by a novel multiplex-PCR assay. J. Microbiol. Methods 2010, 80, 281–286. [Google Scholar] [CrossRef]
- Figueras, M.J.; Levican, A.; Collado, L. Updated 16S rRNA-RFLP method for the identification of all currently characterised Arcobacter spp. BMC Microbiol. 2012, 12, 292. [Google Scholar] [CrossRef]
- Marshall, S.M.; Melito, P.L.; Woodward, D.L.; Johnson, W.M.; Rodgers, F.G.; Mulvey, M.R. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 1999, 37, 4158–4160. [Google Scholar] [CrossRef] [PubMed]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989, 42, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, T.D.L.; Pereira, E.M.; dos Santos, K.R.N.; Maciel, E.L.N.; Schuenck, R.P.; Nunes, A.P.F. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Diagn. Microbiol. Infect. Dis. 2013, 75, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Høiby, N.; Ciofu, O.; Bjarnsholt, T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010, 5, 1663–1674. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Ferreira, S.; Fraqueza, M.J.; Queiroz, J.A.; Domingues, F.C.; Oleastro, M. Genetic diversity, antibiotic resistance and biofilm-forming ability of Arcobacter butzleri isolated from poultry and environment from a Portuguese slaughterhouse. Int. J. Food Microbiol. 2013, 162, 82–88. [Google Scholar] [CrossRef]
- Kalmokoff, M.; Lanthier, P.; Tremblay, T.-L.; Foss, M.; Lau, P.C.; Sanders, G.; Austin, J.; Kelly, J.; Szymanski, C.M. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 2006, 188, 4312–4320. [Google Scholar] [CrossRef]
- Harshey, R.M. Bacterial motility on a surface: Many ways to a common goal. Annu. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.T.; Lipman, L.J.A.; Wösten, M.M.S.M.; van Asten, A.J.; Gaastra, W. Arcobacter spp. possess two very short flagellins of which FlaA is essential for motility. FEMS Immunol. Med. Microbiol. 2008, 53, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Neves, P.; Flores-Martin, S.; Manosalva, C.; Andaur, M.; Otth, C.; Lincopan, N.; Fernández, H. Transcriptional analysis of flagellar and putative virulence genes of Arcobacter butzleri as an endocytobiont of Acanthamoeba castellanii. Arch. Microbiol. 2019, 201, 1075–1083. [Google Scholar] [CrossRef]
- Lee, J.-S.; Bae, Y.-M.; Han, A.; Lee, S.-Y. Development of Congo red broth method for the detection of biofilm-forming or slime-producing Staphylococcus sp. LWT 2016, 73, 707–714. [Google Scholar] [CrossRef]
- Moreno, X.; Ventura, M.; Panizo, M.M.; Garcés, M.F. Evaluación de la formación de biopelículas en aislamientos bacterianos y fúngicos por el método semicuantitativo de microtitulación con cristal violeta y el cualitativo de agar con rojo Congo. Biomédica 2023, 43, 77–88. [Google Scholar] [CrossRef]
- Girbau, C.; Martinez-Malaxetxebarria, I.; Muruaga, G.; Carmona, S.; Alonso, R.; Fernández-Astorga, A. Study of biofilm formation ability of foodborne Arcobacter butzleri under different conditions. J. Food Prot. 2017, 80, 758–762. [Google Scholar] [CrossRef]
- Disli, H.B.; Hizlisoy, H.; Gungor, C.; Barel, M.; Dishan, A.; Gundog, D.A.; Al, S.; Onmaz, N.E.; Yildirim, Y.; Gonulalan, Z. Investigation and characterization of Aliarcobacter spp. isolated from cattle slaughterhouse in Türkiye. Int. Microbiol. 2024, 27, 1321–1332. [Google Scholar] [CrossRef]
- Stepanović, S.; Cirković, I.; Mijac, V.; Švabić-Vlahović, M. Influence of the incubation temperature, atmosphere and dynamic conditions on biofilm formation by Salmonella spp. Food Microbiol. 2003, 20, 339–343. [Google Scholar] [CrossRef]
- Wolska, K.I.; Grudniak, A.M.; Rudnicka, Z.; Markowska, K. Genetic control of bacterial biofilms. J. Appl. Genet. 2016, 57, 225–238. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p)ppGpp and its role in bacterial persistence: New challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A brief recap of microbial adhesion and biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef]
- Schütze, A.; Benndorf, D.; Püttker, S.; Kohrs, F.; Bettenbrock, K. The impact of ackA, pta, and ackA-pta mutations on growth, gene expression and protein acetylation in Escherichia coli K-12. Front. Microbiol. 2020, 11, 233. [Google Scholar] [CrossRef]
- Ju, X.; Li, J.; Zhu, M.; Lu, Z.; Lv, F.; Zhu, X.; Bie, X. Effect of the luxS gene on biofilm formation and antibiotic resistance by Salmonella serovar Dublin. Food Res. Int. 2018, 107, 385–393. [Google Scholar] [CrossRef]
- Han, X.; Bai, H.; Liu, L.; Dong, H.; Liu, R.; Song, J.; Ding, C.; Qi, K.; Liu, H.; Yu, S. The luxS gene functions in the pathogenesis of avian pathogenic Escherichia coli. Microb. Pathog. 2013, 55, 21–27. [Google Scholar] [CrossRef]
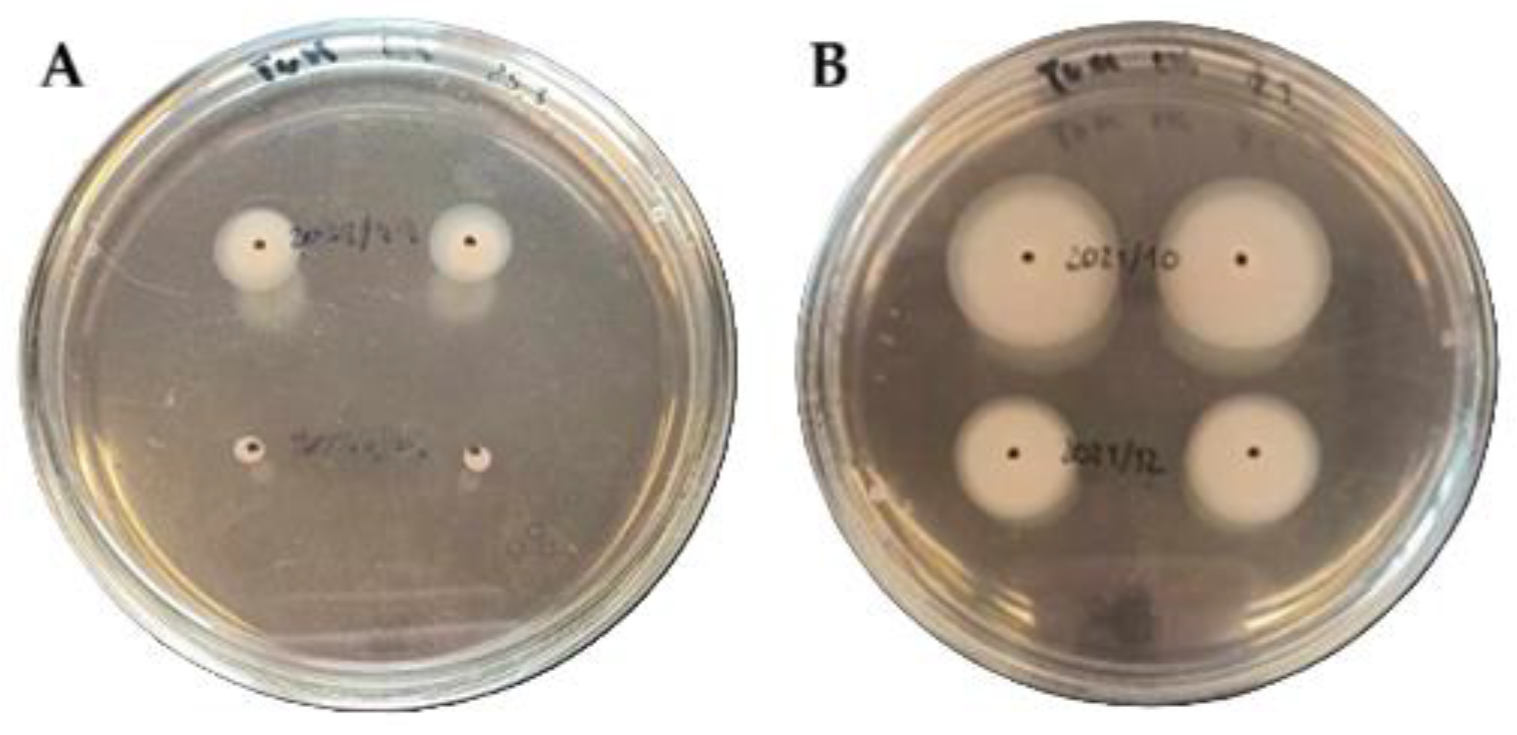
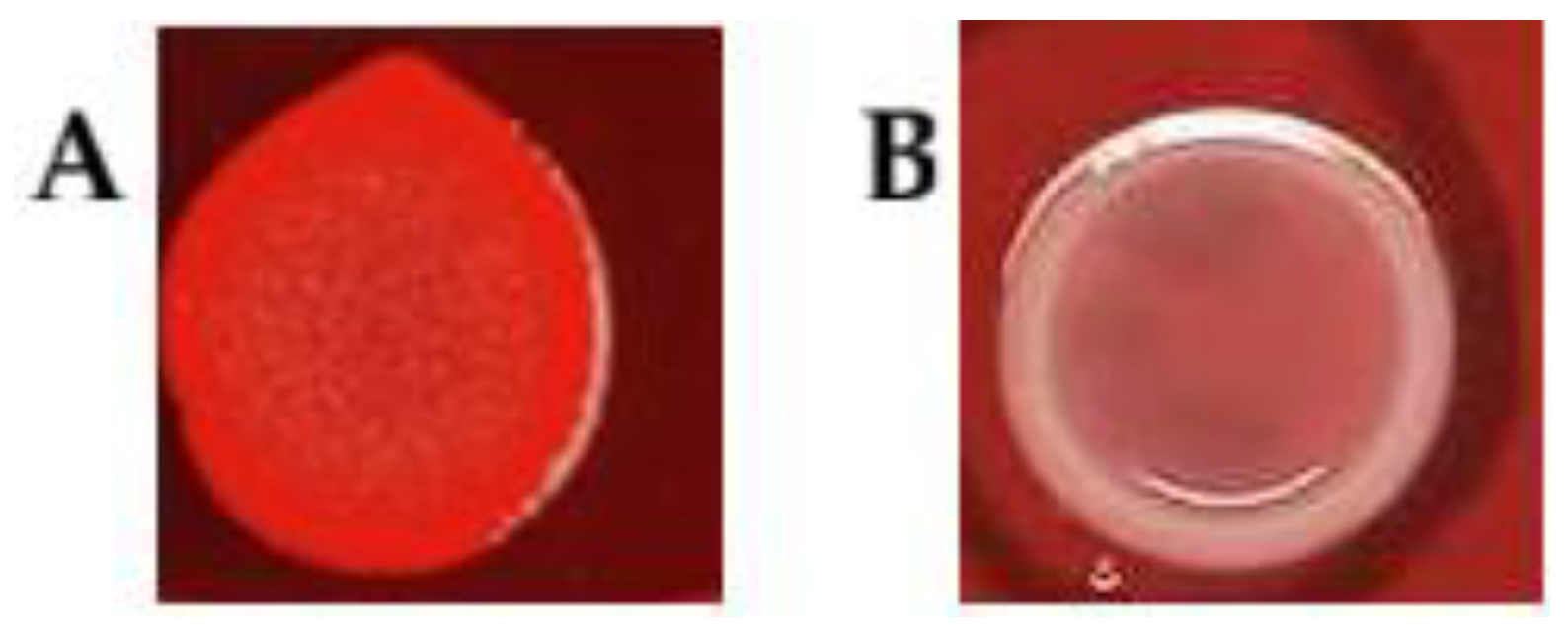
| Bacterial Strain | Static Cultivation | Dynamic Cultivation | Motility | CRA | Biofilm-Associated Genes | |||
|---|---|---|---|---|---|---|---|---|
| Absorbance | Category | Absorbance | Category | Mean (cm) | Category | |||
| LMG 10828 C | 0.1261 ± 0.0097 | ** | 0.1372 ± 0.0147 | ** | 1.55 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2013/KK B | 0.2134 ± 0.0552 | *** | 0.1683 ± 0.0433 | *** | 0.35 ± 0.05 | # | White | fliS, luxS, pta, spoT |
| UPCE 2013/23 A | 0.1297 ± 0.0055 | ** | 0.1281 ± 0.0073 | ** | 0.35 ± 0.05 | # | Red | fliS, luxS, pta, spoT |
| UPCE 2013/30 B | 0.3380 ± 0.0931 | *** | 0.2796 ± 0.0736 | *** | 0.65 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/1 B | 0.1107 ± 0.0018 | – | 0.1095 ± 0.0030 | – | 0.40 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/5 A | 0.1213 ± 0.0041 | ** | 0.1478 ± 0.0052 | ** | 0.30 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/10 B | 0.3119 ± 0.0219 | *** | 0.2110 ± 0.1163 | *** | 0.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/11 B | 0.1091 ± 0.0020 | – | 0.1100 ± 0.0052 | – | 0.45 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/13 B | 0.2096 ± 0.0050 | *** | 0.1775 ± 0.0091 | *** | 0.30 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/14 B | 0.1135 ± 0.1086 | * | 0.1100 ± 0.0345 | – | 0.45 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/15 B | 0.2783 ± 0.0200 | *** | 0.3099 ± 0.0165 | *** | 0.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/16 B | 0.2771 ± 0.0422 | *** | 0.2677 ± 0.0306 | *** | 0.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/18 B | 0.1992 ± 0.0652 | *** | 0.1742 ± 0.0072 | *** | 0.30 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/19 B | 0.1227 ± 0.0022 | ** | 0.1219 ± 0.0022 | ** | 0.55 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2019/20 B | 0.1110 ± 0.0029 | * | 0.1101 ± 0.0284 | – | 0.50 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/4 B | 0.1169 ± 0.0037 | * | 0.1149 ± 0.0024 | * | 1.60 ± 0.2 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/10 B | 0.1238 ± 0.0078 | ** | 0.1478 ± 0.0148 | ** | 2.15 ± 0.05 | ## | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/11 B | 0.1955 ± 0.0233 | *** | 0.2754 ± 0.0116 | *** | 1.90 ± 0.0 | # | Red | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/12 B | 0.2449 ± 0.0180 | *** | 0.1335 ± 0.0071 | ** | 1.45 ± 0.05 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/13 B | 0.1286 ± 0.0061 | ** | 0.1285 ± 0.0035 | ** | 1.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/15 B | 0.1635 ± 0.0082 | *** | 0.1530 ± 0.0055 | *** | 1.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/32 B | 0.1279 ± 0.0035 | ** | 0.1240 ± 0.0015 | ** | 0.20 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/33 B | 0.1174 ± 0.0042 | * | 0.1190 ± 0.0027 | * | 0.20 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/41 B | 0.1233 ± 0.0015 | ** | 0.1170 ± 0.0023 | * | 0.40 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/42 B | 0.3435 ± 0.1163 | *** | 0.2830 ± 0.0351 | *** | 0.45 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/43 B | 0.4109 ± 0.1386 | *** | 0.2900 ± 0.0796 | *** | 1.10 ± 0.10 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/44 B | 0.1246 ± 0.0021 | ** | 0.1216 ± 0.0027 | ** | 0.45 ± 0.05 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/45 B | 0.1265 ± 0.0020 | ** | 0.1240 ± 0.0035 | ** | 0.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/46 B | 0.1332 ± 0.0033 | ** | 0.1290 ± 0.0038 | ** | 0.45 ± 0.05 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/47 B | 0.4073 ± 0.0141 | *** | 0.3724 ± 0.2063 | *** | 1.15 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2021/48 B | 0.1341 ± 0.0505 | ** | 0.1306 ± 0.0040 | ** | 0.40 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 2021/49 B | 0.1263 ± 0.0028 | ** | 0.1180 ± 0.0019 | * | 1.10 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 24B B | 0.3058 ± 0.0137 | *** | 0.2978 ± 0.0266 | *** | 0.45 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 26 B | 0.1200 ± 0.0034 | ** | 0.1181 ± 0.0033 | * | 0.95 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 30 B | 0.1199 ± 0.0025 | * | 0.1163 ± 0.0046 | * | 0.40 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 43 B | 0.1371 ± 0.0080 | ** | 0.1230 ± 0.0074 | ** | 0.40 ± 0.0 | # | Red | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 48 B | 0.1141 ± 0.0303 | * | 0.1174 ± 0.0076 | * | 0.80 ± 0.1 | # | White | fliS, luxS, pta, spoT |
| UPCE 49 B | 0.1183 ± 0.0047 | * | 0.1180 ± 0.0028 | * | 0.35 ± 0.05 | # | White | fliS, luxS, pta, spoT |
| UPCE 65 B | 0.2509 ± 0.0192 | *** | 0.2243 ± 0.0387 | *** | 0.45 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 69 B | 0.3351 ± 0.0202 | *** | 0.2846 ± 0.0252 | *** | 2.10 ± 0.0 | ## | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 89 B | 0.1237 ± 0.0013 | ** | 0.1176 ± 0.0033 | * | 0.50 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 93 B | 0.1169 ± 0.0031 | * | 0.1118 ± 0.0021 | * | 1.10 ± 0.0 | # | Red | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 94 B | 0.1208 ± 0.0030 | ** | 0.1172 ± 0.0029 | * | 1.30 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 107 B | 0.1240 ± 0.0035 | ** | 0.1132 ± 0.0030 | * | 0.40 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 132 B | 0.1184 ± 0.0034 | * | 0.1129 ± 0.0036 | * | 0.35 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 134 B | 0.3333 ± 0.0261 | *** | 0.2767 ± 0.0449 | *** | 1.0 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 138 B | 0.1606 ± 0.0105 | *** | 0.1569 ± 0.0129 | *** | 1.70 ± 0.0 | # | White | fliS, luxS, pta, spoT |
| UPCE 141 B | 0.1192 ± 0.0053 | * | 0.1211 ± 0.0052 | ** | 0.85 ± 0.05 | # | Red | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 160 B | 0.1178 ± 0.0039 | * | 0.1209 ± 0.0030 | ** | 0.30 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 161 B | 0.1707 ± 0.0119 | *** | 0.1279 ± 0.0057 | ** | 0.50 ± 0.0 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| Bacterial Strain | Static Cultivation | Dynamic Cultivation | Motility | CRA | Biofilm-Associated Genes | |||
|---|---|---|---|---|---|---|---|---|
| Absorbance | Category | Absorbance | Category | Mean (cm) | Category | |||
| CCM 3933 C | 0.1280 ± 0.0088 | ** | 0.1198 ± 0.0058 | * | 0.45 ± 0.08 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2013/13 A | 0.1412 ± 0.0109 | ** | 0.1614 ± 0.0325 | *** | 1.05 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/58 A | 0.1262 ± 0.0099 | ** | 0.1285 ± 0.0085 | ** | 0.40 ± 0.07 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/59 B | 0.1812 ± 0.0242 | *** | 0.2015 ± 0.0514 | *** | 0.50 ± 0.04 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/51 B | 0.1638 ± 0.0143 | *** | 0.1252 ± 0.0041 | ** | 0.85 ± 0.10 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/52 A | 0.1586 ± 0.0229 | *** | 0.1410 ± 0.0075 | ** | 0.40 ± 0.10 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/54 B | 0.1460 ± 0.0101 | ** | 0.1614 ± 0.0785 | *** | 0.85 ± 0.05 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/55 B | 0.1144 ± 0.0049 | * | 0.1100 ± 0.0085 | – | 0.75 ± 0.08 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/56 B | 0.1117 ± 0.0034 | * | 0.1215 ± 0.0043 | ** | 0.60 ± 0.07 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
| UPCE 2015/57 B | 0.1251 ± 0.0047 | ** | 0.1158 ± 0.0074 | * | 1.50 ± 0.07 | # | White | flaA, flaB, fliS, luxS, pta, spoT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Musilová, I.; Kozlová, K.; Šilha, D. Biofilm Formation in Arcobacter butzleri and Arcobacter cryaerophilus: Phenotypic and Genotypic Characterization of Food and Environmental Isolates. Microorganisms 2025, 13, 2835. https://doi.org/10.3390/microorganisms13122835
Musilová I, Kozlová K, Šilha D. Biofilm Formation in Arcobacter butzleri and Arcobacter cryaerophilus: Phenotypic and Genotypic Characterization of Food and Environmental Isolates. Microorganisms. 2025; 13(12):2835. https://doi.org/10.3390/microorganisms13122835
Chicago/Turabian StyleMusilová, Irena, Kateřina Kozlová, and David Šilha. 2025. "Biofilm Formation in Arcobacter butzleri and Arcobacter cryaerophilus: Phenotypic and Genotypic Characterization of Food and Environmental Isolates" Microorganisms 13, no. 12: 2835. https://doi.org/10.3390/microorganisms13122835
APA StyleMusilová, I., Kozlová, K., & Šilha, D. (2025). Biofilm Formation in Arcobacter butzleri and Arcobacter cryaerophilus: Phenotypic and Genotypic Characterization of Food and Environmental Isolates. Microorganisms, 13(12), 2835. https://doi.org/10.3390/microorganisms13122835







