Engineering Dual-Input Glucose- and Temperature-Sensitive Lysis Circuits in Corynebacterium glutamicum for Efficient Intracellular Product Recovery
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Construction of Plasmids and Transformation
2.3. Analysis Methods
2.4. Statistical Analysis
3. Results
3.1. Construction and Characterization of a Glucose-Responsive Genetic Element
3.2. Construction and Functional Evaluation of Phage-Derived Lysis Elements
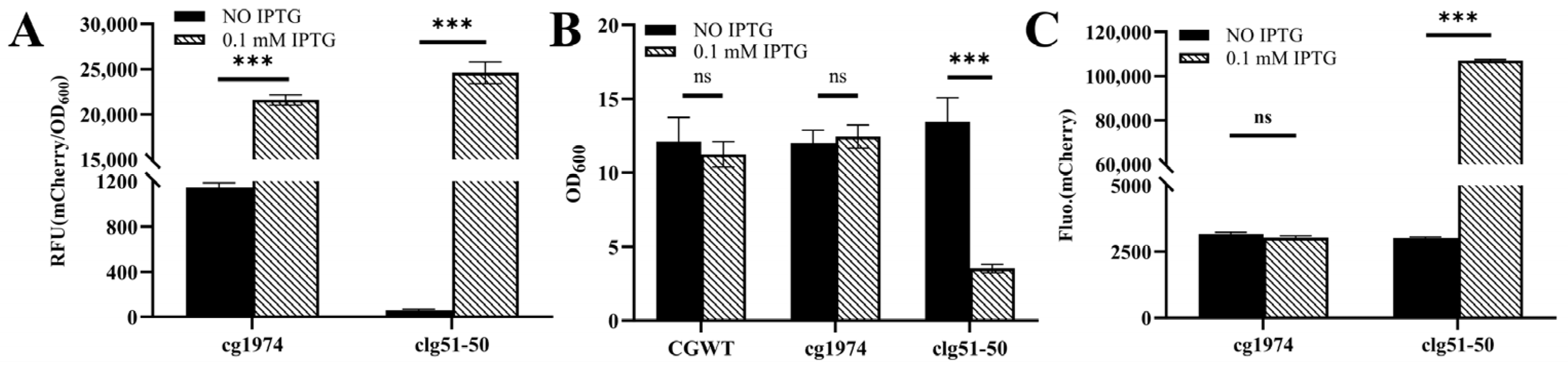
3.3. Construction and Validation of a Controllable Glucose-Responsive Lysis System
3.4. Construction and Validation of a Temperature-Responsive Lysis System
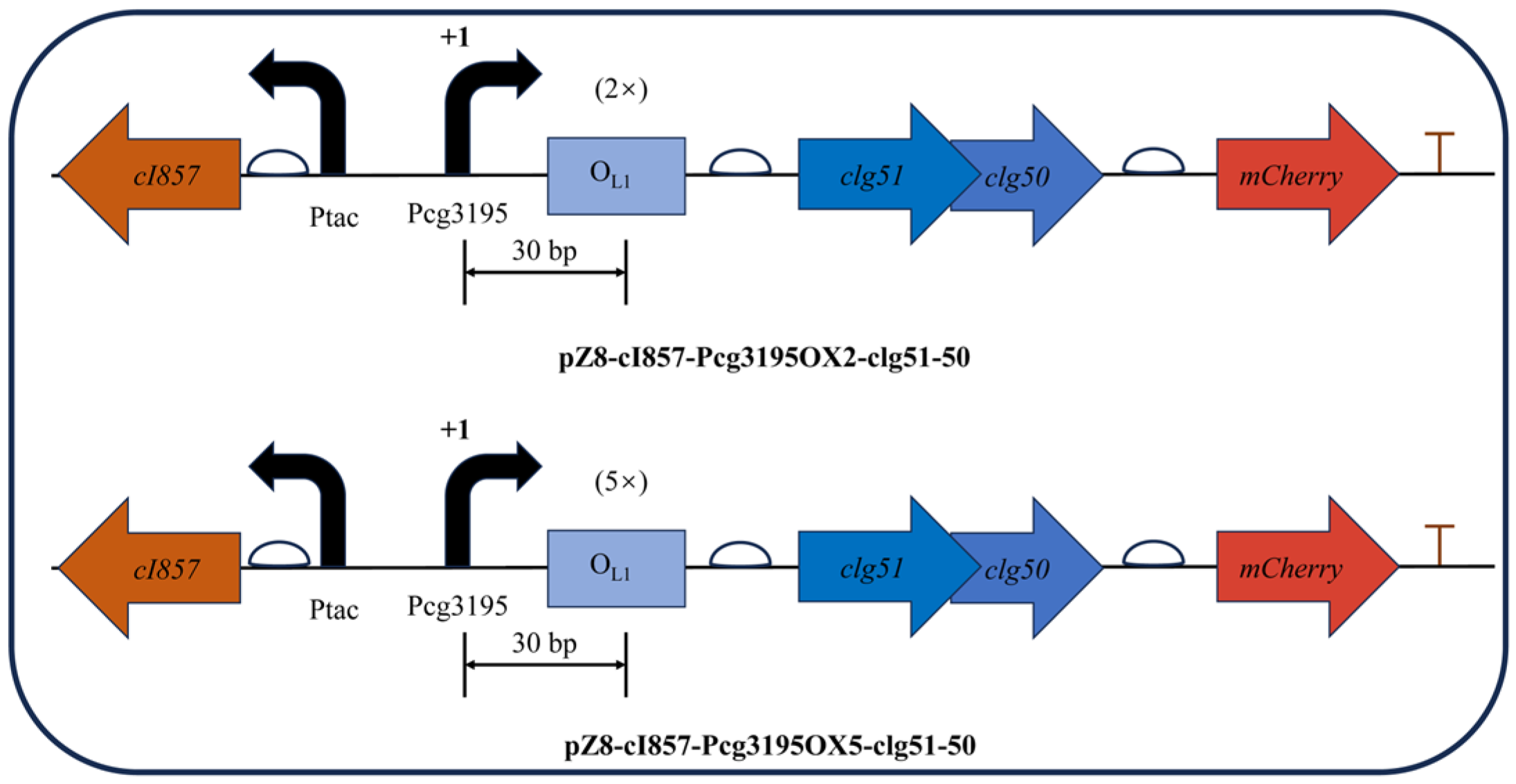
3.5. Dual-Cascade Regulatory Lysis System: Construction, Validation, and Temporal Dynamics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| C. glutamicum | Corynebacterium glutamicum |
| E. coil | Escherichia coli |
| SEM | Scanning electron microscopy |
| RFU | Relative fluorescence units |
References
- Zhao, N.; Qian, L.; Luo, G.; Zheng, S. Synthetic Biology Approaches to Access Renewable Carbon Source Utilization in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 9517–9529. [Google Scholar] [CrossRef] [PubMed]
- Plassmeier, J.; Li, Y.; Rueckert, C.; Sinskey, A.J. Metabolic Engineering Corynebacterium glutamicum to Produce Triacylglycerols. Metab. Eng. 2016, 33, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Burgardt, A.; Prell, C.; Wendisch, V.F. Utilization of a Wheat Sidestream for 5-Aminovalerate Production in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2021, 9, 732271. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Yoon, J.; Lee, S.-M.; Um, Y.; Han, S.O.; Woo, H.M. Modular Pathway Engineering of Corynebacterium glutamicum to Improve Xylose Utilization and Succinate Production. J. Biotechnol. 2017, 258, 69–78. [Google Scholar] [CrossRef]
- Su, H.; Lin, J.; Wang, G. Metabolic Engineering of Corynebacterium Crenatium for Enhancing Production of Higher Alcohols. Sci. Rep. 2016, 6, 39543. [Google Scholar] [CrossRef]
- Ikeda, M.; Takahashi, K.; Ohtake, T.; Imoto, R.; Kawakami, H.; Hayashi, M.; Takeno, S. A Futile Metabolic Cycle of Fatty Acyl Coenzyme A (Acyl-CoA) Hydrolysis and Resynthesis in Corynebacterium glutamicum and Its Disruption Leading to Fatty Acid Production. Appl. Environ. Microbiol. 2021, 87, e02469-20. [Google Scholar] [CrossRef]
- Walther, C.; Kellner, M.; Berkemeyer, M.; Brocard, C.; Dürauer, A. A Microscale Bacterial Cell Disruption Technique as First Step for Automated and Miniaturized Process Development. Process Biochem. 2017, 59, 207–215. [Google Scholar] [CrossRef]
- Rahman, M.M.; Hosano, N.; Hosano, H. Recovering Microalgal Bioresources: A Review of Cell Disruption Methods and Extraction Technologies. Molecules 2022, 27, 2786. [Google Scholar] [CrossRef]
- Danaeifar, M. New Horizons in Developing Cell Lysis Methods: A Review. Biotech. Bioeng. 2022, 119, 3007–3021. [Google Scholar] [CrossRef]
- Gao, Y.; Feng, X.; Xian, M.; Wang, Q.; Zhao, G. Inducible Cell Lysis Systems in Microbial Production of Bio-Based Chemicals. Appl. Microbiol. Biotechnol. 2013, 97, 7121–7129. [Google Scholar] [CrossRef]
- Liu, X.; Curtiss, R. Nickel-Inducible Lysis System in Synechocystis Sp. PCC 6803. Proc. Natl. Acad. Sci. USA 2009, 106, 21550–21554. [Google Scholar] [CrossRef]
- Wang, G.; Lu, X.; Zhu, Y.; Zhang, W.; Liu, J.; Wu, Y.; Yu, L.; Sun, D.; Cheng, F. A Light-Controlled Cell Lysis System in Bacteria. J. Ind. Microbiol. Biotechnol. 2018, 45, 429–432. [Google Scholar] [CrossRef]
- Liebl, W.; Klamer, R.; Schleifer, K.-H. Requirement of Chelating Compounds for the Growth of Corynebacterium glutamicum in Synthetic Media. Appl. Microbiol. Biotechnol. 1989, 32, 205–210. [Google Scholar] [CrossRef]
- Hünnefeld, M.; Viets, U.; Sharma, V.; Wirtz, A.; Hardy, A.; Frunzke, J. Genome Sequence of the Bacteriophage CL31 and Interaction with the Host Strain Corynebacterium glutamicum ATCC 13032. Viruses 2021, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.; Baumgart, M.; Bott, M. Development of a Single-Cell GlxR-Based cAMP Biosensor for Corynebacterium glutamicum. J. Biotechnol. 2017, 258, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Kim, T.-H.; Kim, Y.; Lee, H.-S. Identification and Characterization of glxR, a Gene Involved in Regulation of Glyoxylate Bypass in Corynebacterium glutamicum. J. Bacteriol. 2004, 186, 3453–3460. [Google Scholar] [CrossRef]
- Toyoda, K.; Inui, M. Regulons of Global Transcription Factors in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2016, 100, 45–60. [Google Scholar] [CrossRef]
- Jungwirth, B.; Sala, C.; Kohl, T.A.; Uplekar, S.; Baumbach, J.; Cole, S.T.; Pühler, A.; Tauch, A. High-Resolution Detection of DNA Binding Sites of the Global Transcriptional Regulator GlxR in Corynebacterium glutamicum. Microbiology 2013, 159, 12–22. [Google Scholar] [CrossRef]
- Hong, E.-J.; Park, J.-S.; Kim, Y.; Lee, H.-S. Role of Corynebacterium glutamicum sprA Encoding a Serine Protease in glxR-Mediated Global Gene Regulation. PLoS ONE 2014, 9, e93587. [Google Scholar] [CrossRef]
- Toyoda, K.; Teramoto, H.; Inui, M.; Yukawa, H. Genome-Wide Identification of In Vivo Binding Sites of GlxR, a Cyclic AMP Receptor Protein-Type Regulator in Corynebacterium glutamicum. J. Bacteriol. 2011, 193, 4123–4133. [Google Scholar] [CrossRef]
- Frunzke, J.; Bramkamp, M.; Schweitzer, J.-E.; Bott, M. Population Heterogeneity in Corynebacterium glutamicum ATCC 13032 Caused by Prophage CGP3. J. Bacteriol. 2008, 190, 5111–5119. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-U.; Jo, J.-H.; Kim, Y.-J.; Chung, S.-S.; Lee, J.-H.; Lee, H.H. Construction of Heat-Inducible Expression Vector of Corynebacterium glutamicum and C. ammoniagenes: Fusion of λ Operator with Promoters Isolated from C. ammoniagenes. J. Microbiol. Biotechnol. 2008, 18, 639–647. [Google Scholar] [PubMed]
- Hao, N.; Krishna, S.; Ahlgren-Berg, A.; Cutts, E.E.; Shearwin, K.E.; Dodd, I.B. Road Rules for Traffic on DNA—Systematic Analysis of Transcriptional Roadblocking in Vivo. Nucleic Acids Res. 2014, 42, 8861–8872. [Google Scholar] [CrossRef] [PubMed]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; São-José, C.; Pimentel, M. Diversity in Bacterial Lysis Systems: Bacteriophages Show the Way. FEMS Microbiol. Rev. 2013, 37, 554–571. [Google Scholar] [CrossRef]
- Jain, V.; Pohane, A.A. Insights into the Regulation of Bacteriophage Endolysin: Multiple Means to the Same End. Microbiology 2015, 161, 2269–2276. [Google Scholar] [CrossRef]
- São-José, C.; Parreira, R.; Vieira, G.; Santos, M.A. The N-Terminal Region of the Oenococcus oeni Bacteriophage fOg44 Lysin Behaves as a Bona Fide Signal Peptide in Escherichia coli and as a Cis -Inhibitory Element, Preventing Lytic Activity on Oenococcal Cells. J. Bacteriol. 2000, 182, 5823–5831. [Google Scholar] [CrossRef]
- Frias, M.J.; Melo-Cristino, J.; Ramirez, M. Export of the Pneumococcal Phage SV1 Lysin Requires Choline-containing Teichoic Acids and Is Holin-independent. Mol. Microbiol. 2013, 87, 430–445. [Google Scholar] [CrossRef]
- Wang, I.-N.; Smith, D.L.; Young, R. Holins: The Protein Clocks of Bacteriophage Infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage Endolysins as Novel Antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef]








| Strain or Plasmid | Description | Source |
|---|---|---|
| Strain | ||
| E. coli TOP 10 | F-mcrAΔ(mrr-hsdRMS-mcrBC) φ80lacZΔM15ΔlacX74recA1 araΔ139Δ(ara-leu)7697 galU galK rpsL (StrR)endA1nupG | Laboratory |
| C. glutamicum ATCC 13032 | Wild-type | ATCC |
| CGY1 | C. glutamicum ATCC 13032 harboring the pZ8-Pcg3195-mCherry | This work |
| CGY2 | C. glutamicum ATCC 13032 harboring the pZ8-Ptac-cg1974-mCherry | This work |
| CGY3 | C. glutamicum ATCC 13032 harboring the pZ8-Ptac-clg51-50-mCherry | This work |
| CGY4 | C. glutamicum ATCC 13032 harboring the pZ8-Pcg3195-clg51-50-mCherry | This work |
| CGY5 | C. glutamicum ATCC 13032 harboring the pZ8-cI857-CJ1OX2 | This work |
| CGY6 | C. glutamicum ATCC 13032 harboring the pZ8-cI857- Pcg3195OX2-clg51-50 | This work |
| CGY7 | C. glutamicum ATCC 13032 harboring the pZ8-cI857- Pcg3195OX5-clg51-50 | This work |
| Plasmid | ||
| pZ8-Ptac | KmR, E. coli–C.glutamicum shuttle expression vector carrying the tac promotor | Laboratory |
| pZ8-Pcg3195-mCherry | Derived from pZ8-Ptac by replacing the tac promotor with the Pcg3195, carrying the gene mCherry | This work |
| pZ8-Ptac-cg1974-mCherry | pZ8-Ptac carrying the genes cg1974, mCherry | This work |
| pZ8-Ptac-clg51-50-mCherry | pZ8-Ptac carrying the genes clg51, clg50, mCherry | This work |
| pZ8-Pcg3195-clg51-50-mCherry | pZ8-Pcg3195-mCherry carrying the genes clg51, clg50 | This work |
| pZ8-cI857-CJ1OX2 | Derived from pZ8-Ptac-clg51-50-mCherry by inserting the cI857 expression cassette and swapping the Pcg3195 for the CJ1OX2 to control the genes clg51, clg50, mCherry | This work |
| pZ8-cI857-Pcg3195OX2-clg51-50 | Derived from pZ8-cI857-CJ1OX2 by replacing the CJ1 promotor with the Pcg3195 and inserting two OL1 operator sites downstream of the Pcg3195 | This work |
| pZ8-cI857-Pcg3195OX5-clg51-50 | Derived from pZ8-cI857-CJ1OX2 by replacing the CJ1 promotor with the Pcg3195 and inserting five OL1 operator sites downstream of the Pcg3195 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Wang, S.; Wang, Q.; Ouyang, L.; Li, Y.; Zhang, L. Engineering Dual-Input Glucose- and Temperature-Sensitive Lysis Circuits in Corynebacterium glutamicum for Efficient Intracellular Product Recovery. Microorganisms 2025, 13, 2758. https://doi.org/10.3390/microorganisms13122758
Ye Z, Wang S, Wang Q, Ouyang L, Li Y, Zhang L. Engineering Dual-Input Glucose- and Temperature-Sensitive Lysis Circuits in Corynebacterium glutamicum for Efficient Intracellular Product Recovery. Microorganisms. 2025; 13(12):2758. https://doi.org/10.3390/microorganisms13122758
Chicago/Turabian StyleYe, Ziyu, Shihui Wang, Qiyue Wang, Liming Ouyang, Youyuan Li, and Lixin Zhang. 2025. "Engineering Dual-Input Glucose- and Temperature-Sensitive Lysis Circuits in Corynebacterium glutamicum for Efficient Intracellular Product Recovery" Microorganisms 13, no. 12: 2758. https://doi.org/10.3390/microorganisms13122758
APA StyleYe, Z., Wang, S., Wang, Q., Ouyang, L., Li, Y., & Zhang, L. (2025). Engineering Dual-Input Glucose- and Temperature-Sensitive Lysis Circuits in Corynebacterium glutamicum for Efficient Intracellular Product Recovery. Microorganisms, 13(12), 2758. https://doi.org/10.3390/microorganisms13122758








