Rhizosphere Microbiome and Nutrient Fluxes Reveal Subtle Biosafety Signals in Transgenic Cotton
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cultivation
2.2. Soil Sampling
2.3. Morphological Assessment and Biomass Determination
2.4. Soil Chemical Properties Analysis
2.5. Microbiome Sequencing
2.6. Bioinformatics Analysis
2.7. Taxonomic Annotation
2.8. Alpha Diversity
2.9. Beta Diversity
2.10. Redundancy Analysis
2.11. Correlation and Mantel Test
3. Results
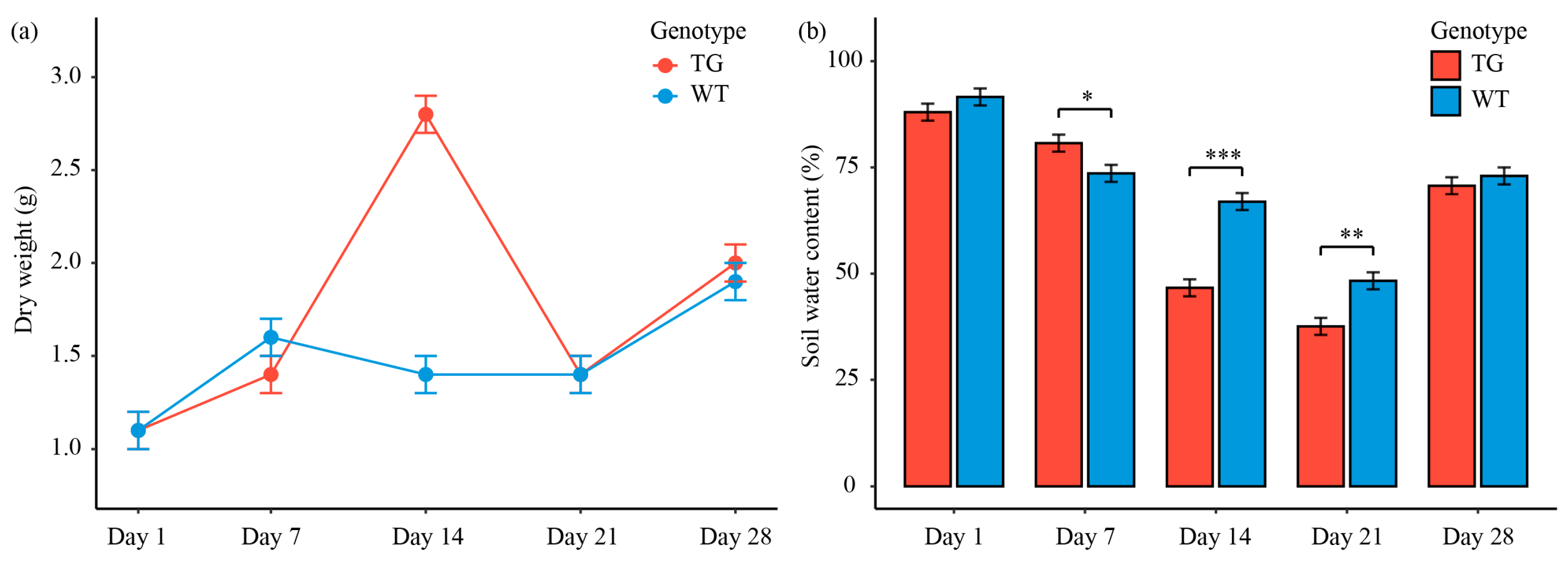
3.1. Transgenic Cotton Demonstrate Enhanced Above-Ground Biomass Accumulation with Comparable Soil Water Retention
3.2. Bacterial α- and β-Diversity Exhibit Temporal Variation with Minimal Genotype Effects
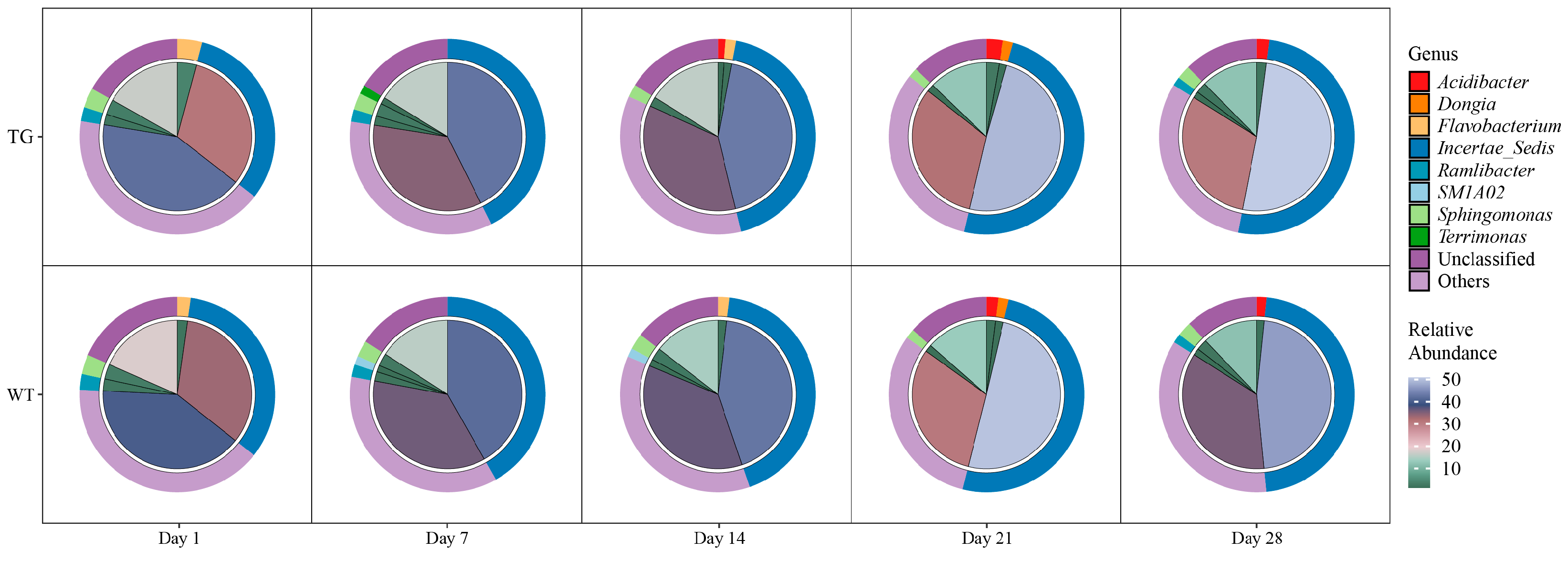
3.3. Genus-Level Composition Exhibits Temporal Dynamics with Minimal Genotype Effect
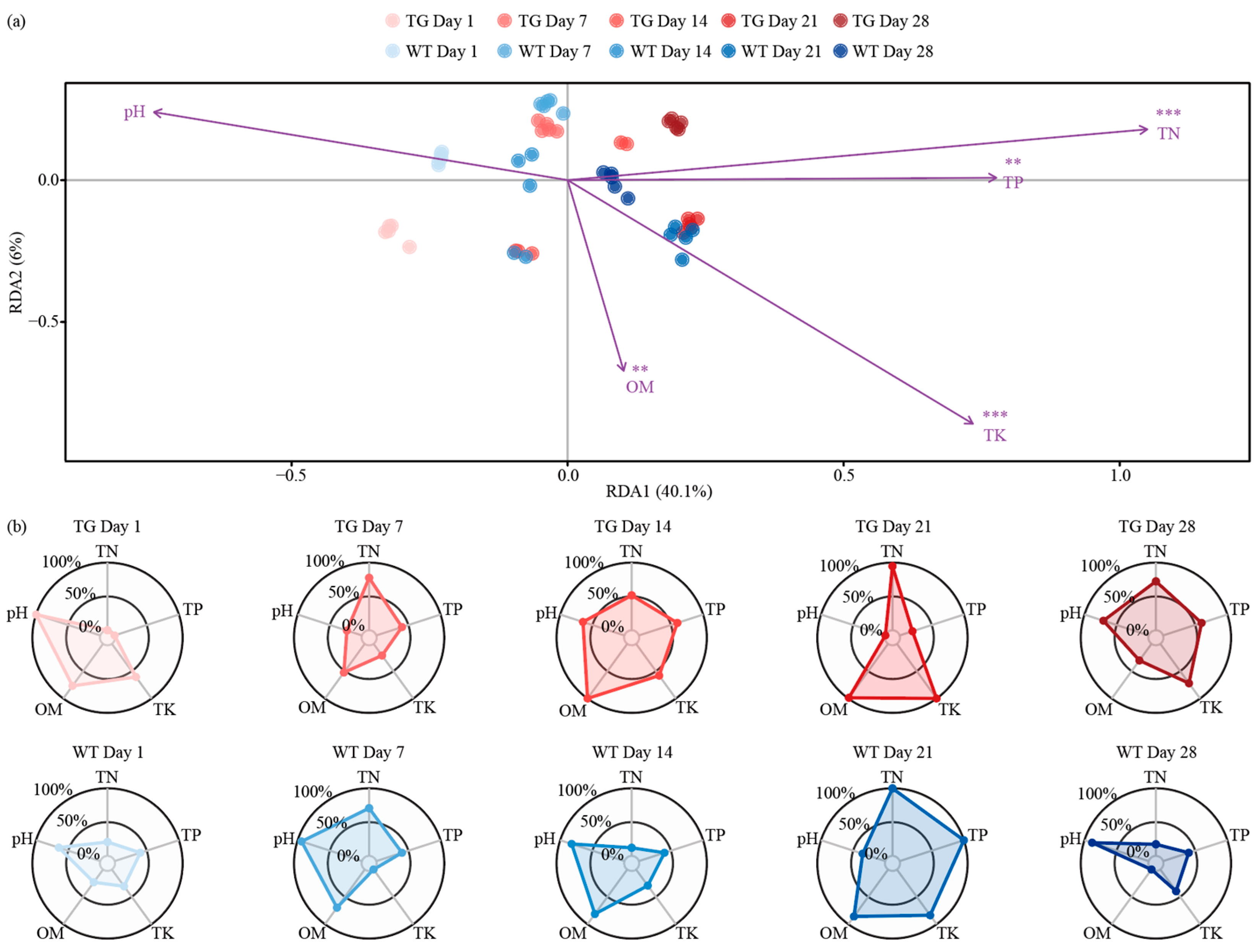
3.4. Temporal Dynamics of Genotype-Modulated Rhizosphere Nutrient Gradients
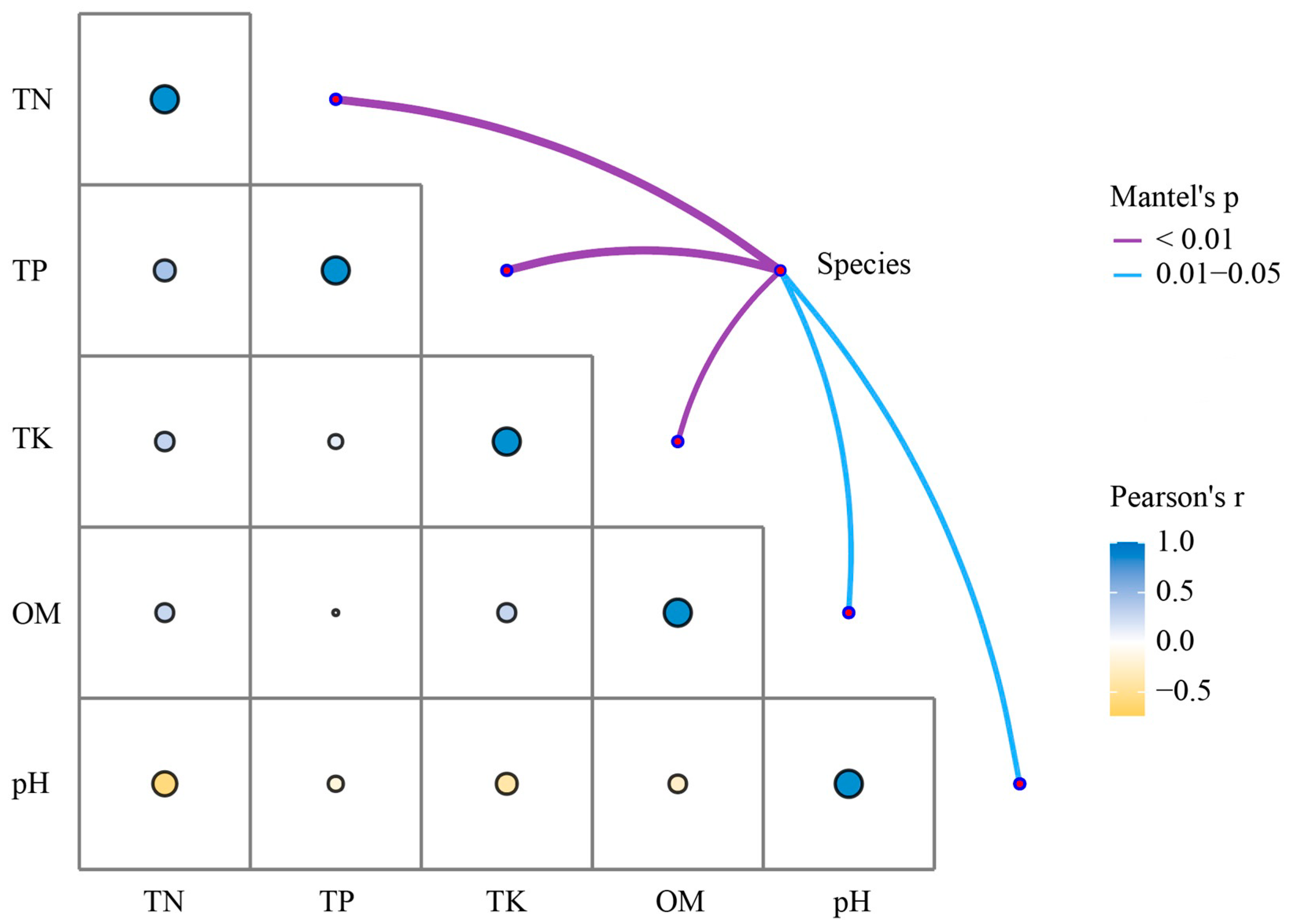
3.5. Microbiome Composition Correlates Moderately with Nutrient Profiles Diversity
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OTU | operational taxonomic unit |
| PCoA | principal coordinates analysis |
| PERMANOVA | permutational multivariate analysis of variance |
| RDA | redundancy analysis |
| OM | organic matter |
| TN | total nitrogen |
| TP | total phosphorus |
| TK | total potassium |
| TG | transgenic |
| WT | wild-type |
References
- Haichar, F.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef]
- Briefs, I. Global status of commercialized biotech/GM crops in 2017: Biotech crop adoption surges as economic benefits accumulate in 22 years. ISAAA Briefs 2017, 53, 25–26. [Google Scholar]
- Erkossa, T.; Itanna, F.; Stahr, K. Indexing soil quality: A new paradigm in soil science research. Soil Res. 2007, 45, 129–137. [Google Scholar] [CrossRef]
- Karthik, K.; Nandiganti, M.; Thangaraj, A.; Singh, S.; Mishra, P.; Rathinam, M.; Sharma, M.; Singh, N.K.; Dash, P.K.; Sreevathsa, R. Transgenic Cotton (Gossypium hirsutum L.) to Combat Weed Vagaries: Utility of an Apical Meristem-Targeted in planta Transformation Strategy to Introgress a Modified CP4-EPSPS Gene for Glyphosate Tolerance. Front. Plant. Sci. 2020, 11, 768. [Google Scholar] [CrossRef]
- Vulchi, R.; Bagavathiannan, M.; Nolte, S.A. History of Herbicide-Resistant Traits in Cotton in the U.S. and the Importance of Integrated Weed Management for Technology Stewardship. Plants 2022, 11, 1189. [Google Scholar] [CrossRef]
- Green, J.M. The benefits of herbicide-resistant crops. Pest Manag. Sci. 2012, 68, 1323–1331. [Google Scholar] [CrossRef]
- Wolfenbarger, L.L.; Phifer, P.R. The ecological risks and benefits of genetically engineered plants. Science 2000, 290, 2088–2093. [Google Scholar] [CrossRef]
- Liang, J.; Sun, S.; Ji, J.; Wu, H.; Meng, F.; Zhang, M.; Zheng, X.; Wu, C.; Zhang, Z. Comparison of the rhizosphere bacterial communities of Zigongdongdou soybean and a high-methionine transgenic line of this cultivar. PLoS ONE 2014, 9, e103343. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Xie, M.; Li, Q.; Zhang, X.-L.; Zhang, Z.-R. Monitoring changes in the actinobacterial field communities present in the rhizosphere soil of a transgenic cotton producing Cry1Ab/Ac proteins. Crop Prot. 2017, 91, 1–7. [Google Scholar] [CrossRef]
- Castaldini, M.; Turrini, A.; Sbrana, C.; Benedetti, A.; Marchionni, M.; Mocali, S.; Fabiani, A.; Landi, S.; Santomassimo, F.; Pietrangeli, B.; et al. Impact of Bt corn on rhizospheric and soil eubacterial communities and on beneficial mycorrhizal symbiosis in experimental microcosms. Appl. Environ. Microbiol. 2005, 71, 6719–6729. [Google Scholar] [CrossRef]
- Qiao, Q.; Wang, F.; Zhang, J.; Chen, Y.; Zhang, C.; Liu, G.; Zhang, H.; Ma, C.; Zhang, J. The variation in the rhizosphere microbiome of cotton with soil type, genotype and developmental stage. Sci. Rep. 2017, 7, 3940. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Schimel, J.P.; Bennett, J. Nitrogen mineralization: Challenges of a changing paradigm. Ecology 2004, 85, 591–602. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Crist, T.O.; Chase, J.M.; Vellend, M.; Inouye, B.D.; Freestone, A.L.; Sanders, N.J.; Cornell, H.V.; Comita, L.S.; Davies, K.F.; et al. Navigating the multiple meanings of β diversity: A roadmap for the practicing ecologist. Ecol. Lett. 2010, 14, 19–28, Erratum in Ecol. Lett. 2011, 14, 210. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2024, 42, 715–718, Erratum in Nat. Biotechnol. 2024, 42, 813. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Shahmoradi, Z.S.; Tohidfar, M.; Marashi, H.; Malekzadeh-Shafaroudi, S.; Karimi, E. Cultivation Effect of Chitinase-Transgenic Cotton on Functional Bacteria and Fungi in Rhizosphere and Bulk Soil. Iran. J. Biotechnol. 2019, 17, e1982. [Google Scholar] [CrossRef]
- Saxena, D.; Flores, S.; Stotzky, G. Bt toxin is released in root exudates from 12 transgenic corn hybrids representing three transformation events. Soil Biol. Biochem. 2002, 34, 133–137. [Google Scholar] [CrossRef]
- Turrini, A.; Sbrana, C.; Nuti, M.P.; Pietrangeli, B.M.; Giovannetti, M. Development of a model system to assess the impact of genetically modified corn and aubergine plants on arbuscular mycorrhizal fungi. Plant Soil 2005, 266, 69–75. [Google Scholar] [CrossRef]
- Cotta, S.R.; Dias, A.C.F.; Marriel, I.E.; Gomes, E.A.; van Elsas, J.D.; Seldin, L. Temporal dynamics of microbial communities in the rhizosphere of two genetically modified (GM) maize hybrids in tropical agrosystems. Antonie Leeuwenhoek 2013, 103, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, K.E.; Germida, J.J. Impact of genetically modified crops on soil- and plant-associated microbial communities. J. Environ. Qual. 2004, 33, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.D.; Zou, H.L.; Chu, L.M.; Liao, B.; Ye, C.M.; Lan, C.Y. Field Released Transgenic Papaya Affects Microbial Communities and Enzyme Activities in Soil. Plant Soil 2006, 285, 347–358. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Ge, L.; Hu, C.; Wu, G.; Sun, Y.; Song, L.; Wu, X.; Pan, A.; Xu, Q.; et al. Environmental Behaviors of Bacillus thuringiensis (Bt) Insecticidal Proteins and Their Effects on Microbial Ecology. Plants 2022, 11, 1212. [Google Scholar] [CrossRef]
- Lukow, T.; Dunfield, P.F.; Liesack, W. Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol. Ecol. 2000, 32, 241–247. [Google Scholar] [CrossRef]
- Baumgarte, S.; Tebbe, C.C. Field studies on the environmental fate of the Cry1Ab Bt-toxin produced by transgenic maize (MON810) and its effect on bacterial communities in the maize rhizosphere. Mol. Ecol. 2005, 14, 2539–2551. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Duan, Y.; Wei, R.; Yuan, Y.; Yan, H.; Tang, T.; Shang, H. Rhizosphere Microbiome and Nutrient Fluxes Reveal Subtle Biosafety Signals in Transgenic Cotton. Microorganisms 2025, 13, 2702. https://doi.org/10.3390/microorganisms13122702
Yang Z, Duan Y, Wei R, Yuan Y, Yan H, Tang T, Shang H. Rhizosphere Microbiome and Nutrient Fluxes Reveal Subtle Biosafety Signals in Transgenic Cotton. Microorganisms. 2025; 13(12):2702. https://doi.org/10.3390/microorganisms13122702
Chicago/Turabian StyleYang, Zheng, Yuhang Duan, Renhui Wei, Youlu Yuan, Haoliang Yan, Tong Tang, and Haihong Shang. 2025. "Rhizosphere Microbiome and Nutrient Fluxes Reveal Subtle Biosafety Signals in Transgenic Cotton" Microorganisms 13, no. 12: 2702. https://doi.org/10.3390/microorganisms13122702
APA StyleYang, Z., Duan, Y., Wei, R., Yuan, Y., Yan, H., Tang, T., & Shang, H. (2025). Rhizosphere Microbiome and Nutrient Fluxes Reveal Subtle Biosafety Signals in Transgenic Cotton. Microorganisms, 13(12), 2702. https://doi.org/10.3390/microorganisms13122702






