MACNeXt-Based Bacteria Species Detection
Abstract
1. Introduction
1.1. Literature Gaps
1.2. Motivation and Our Model
1.3. Novelties
1.4. Contributions
2. Materials and Methods
2.1. Materials
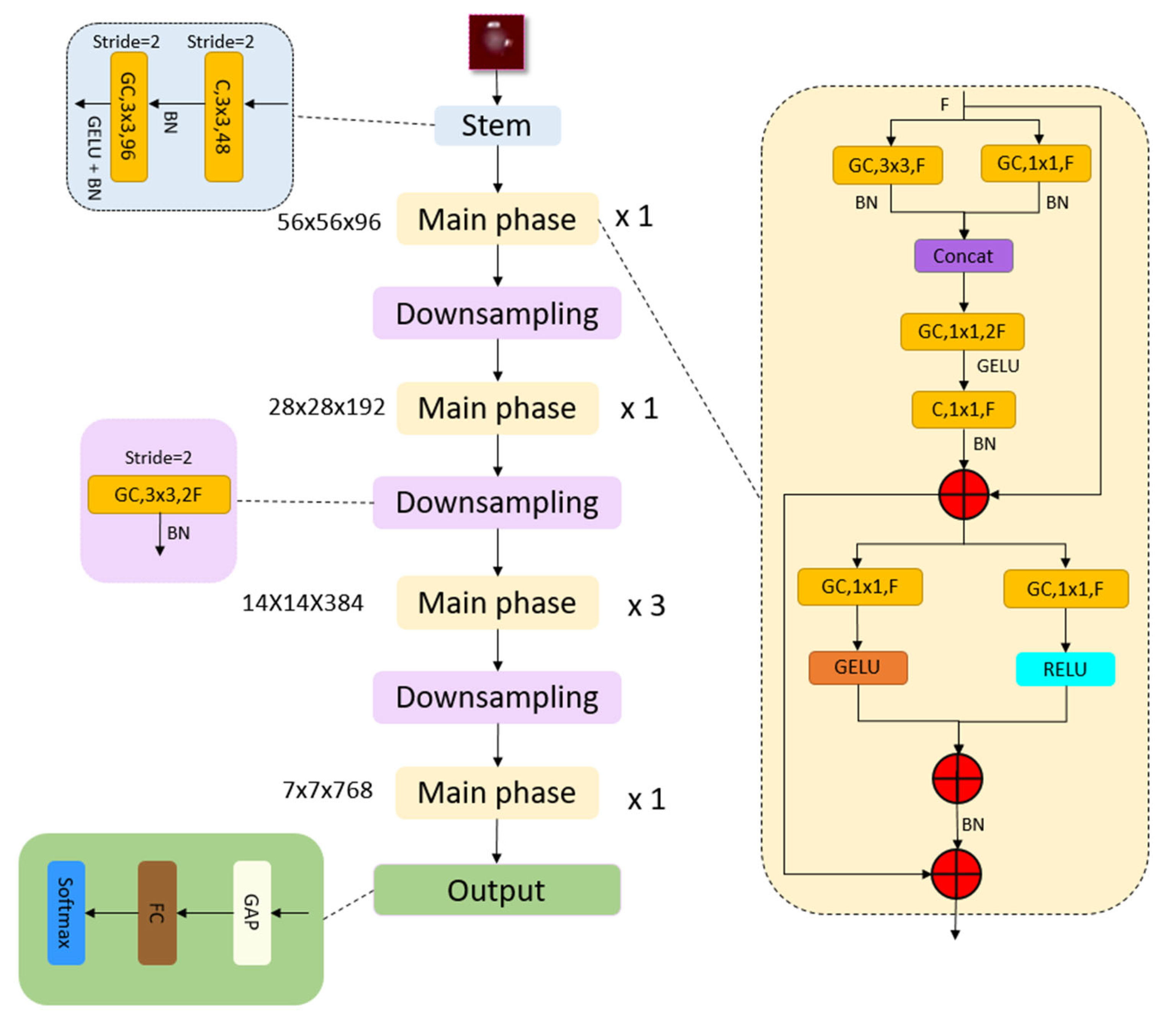
2.2. The Proposed MACNeXt
| Algorithm 1. Pseudocode of the introduced MACNeXt CNN architecture |
| Input: RGB image I of size 224 × 224 × 3 Output: Class probabilities y_hat |
| # ---------- Stem Block ---------- # Conv 3 × 3, 48 filters, stride 2 T1 = Conv 3 × 3(I, filters = 48, stride = 2) T1 = BatchNorm(T1) # Grouped Conv 3 × 3, 96 filters, stride 2 + GELU T1 = GroupConv 3 × 3(T1, filters = 96, stride = 2) T1 = GELU(T1) T1 = BatchNorm(T1) # T1: 56 × 56 × 96 (Tens1) # ---------- Stage configuration ---------- # Feature sizes per stage F = [96, 192, 384, 768] # Block repeat numbers per stage R = [1, 1, 3, 1] T = T1 # ---------- Main Stages + Downsampling ---------- for s in 1...4 do # stage index # ----- Repeat MACNeXt main block R[s] times ----- for r in 1..R[s] do T = MACNeXt_Block(T, F[s]) end for # ----- Apply downsampling after first 3 stages ----- if s < 4 then # Grouped Conv 3 × 3, stride 2, channels: F[s] → 2·F[s] T = GroupConv 3 × 3(T, filters = 2 × F[s], stride = 2) T = BatchNorm(T) # now channel size becomes F[s + 1] end if end for # ---------- Classification Head ---------- T_gap = GlobalAveragePooling(T) # 1 × 1 × 768 z = FullyConnected(T_gap, n) # logits, Herein, n is the number of outputs y_hat = Softmax(z) # probabilities return y_hat # ---------- Definition of MACNeXt main block ---------- # Input: tensor X with F channels # Output: tensor Y with F channels function MACNeXt_Block(X, F): # (1) Parallel 3 × 3 and 1 × 1 grouped convolutions + BN P1 = GroupConv 3 × 3(X, filters = F, stride = 1) P1 = BatchNorm(P1) P2 = GroupConv 1 × 1(X, filters = F) P2 = BatchNorm(P2) # (2) Concatenate and expand (inverted bottleneck) C = ConcatChannels(P1, P2) # 2F channels E = GroupConv 1 × 1(C, filters = 2 × F) # expansion # (3) GELU + squeeze to F channels + BN E = GELU(E) S = Conv 1 × 1(E, filters = F) S = BatchNorm(S) # (4) First residual connection R1 = X + S # (5) Dual activations on R1 G_branch = GELU(GroupConv 1 × 1(R1, filters = F)) R_branch = ReLU(GroupConv 1 × 1(R1, filters = F)) # (6) Fuse branches + BN Fused = G_branch + R_branch Fused = BatchNorm(Fused) # (7) Second residual connection Y = R1 + Fused return Y end function |
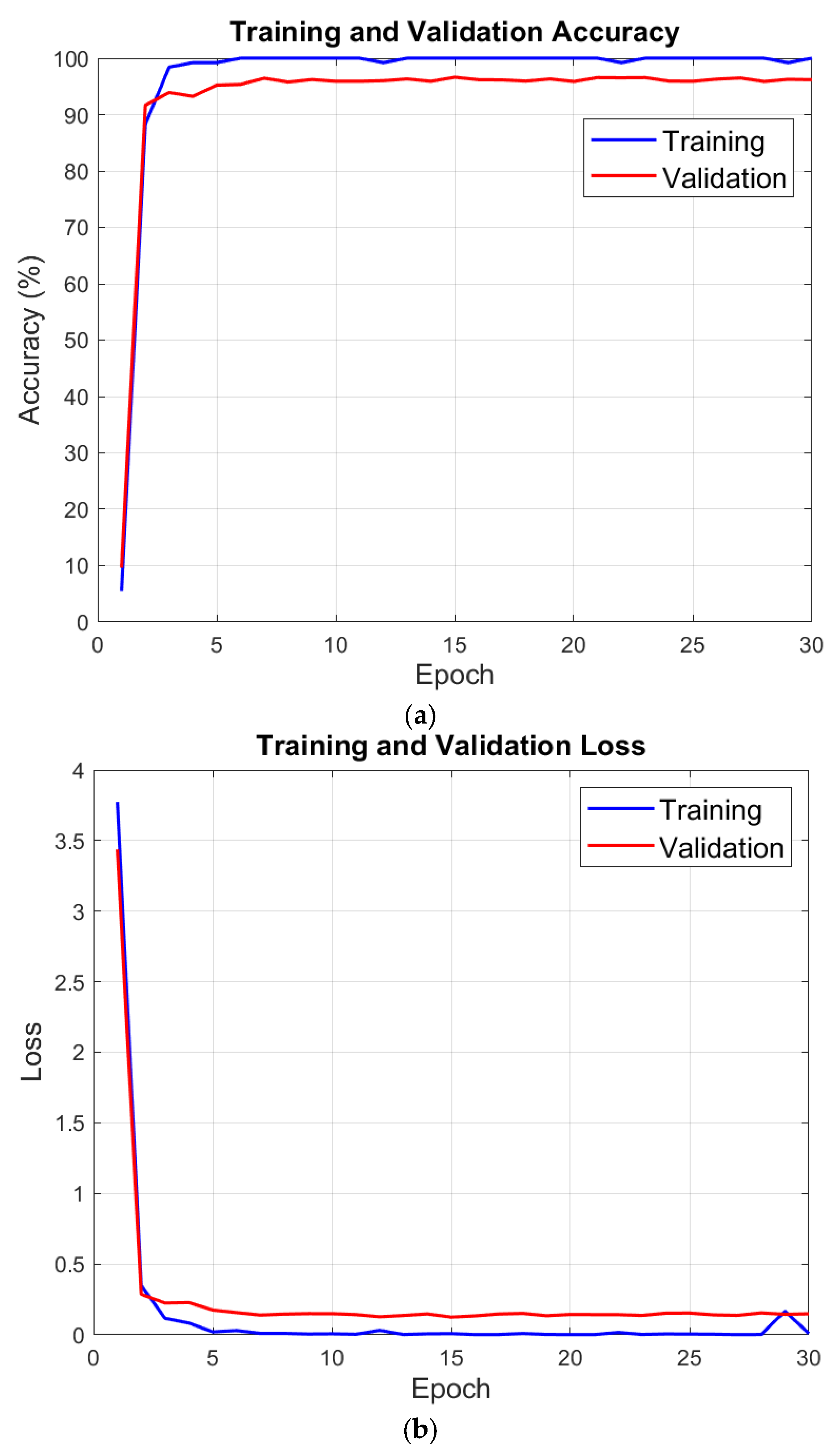
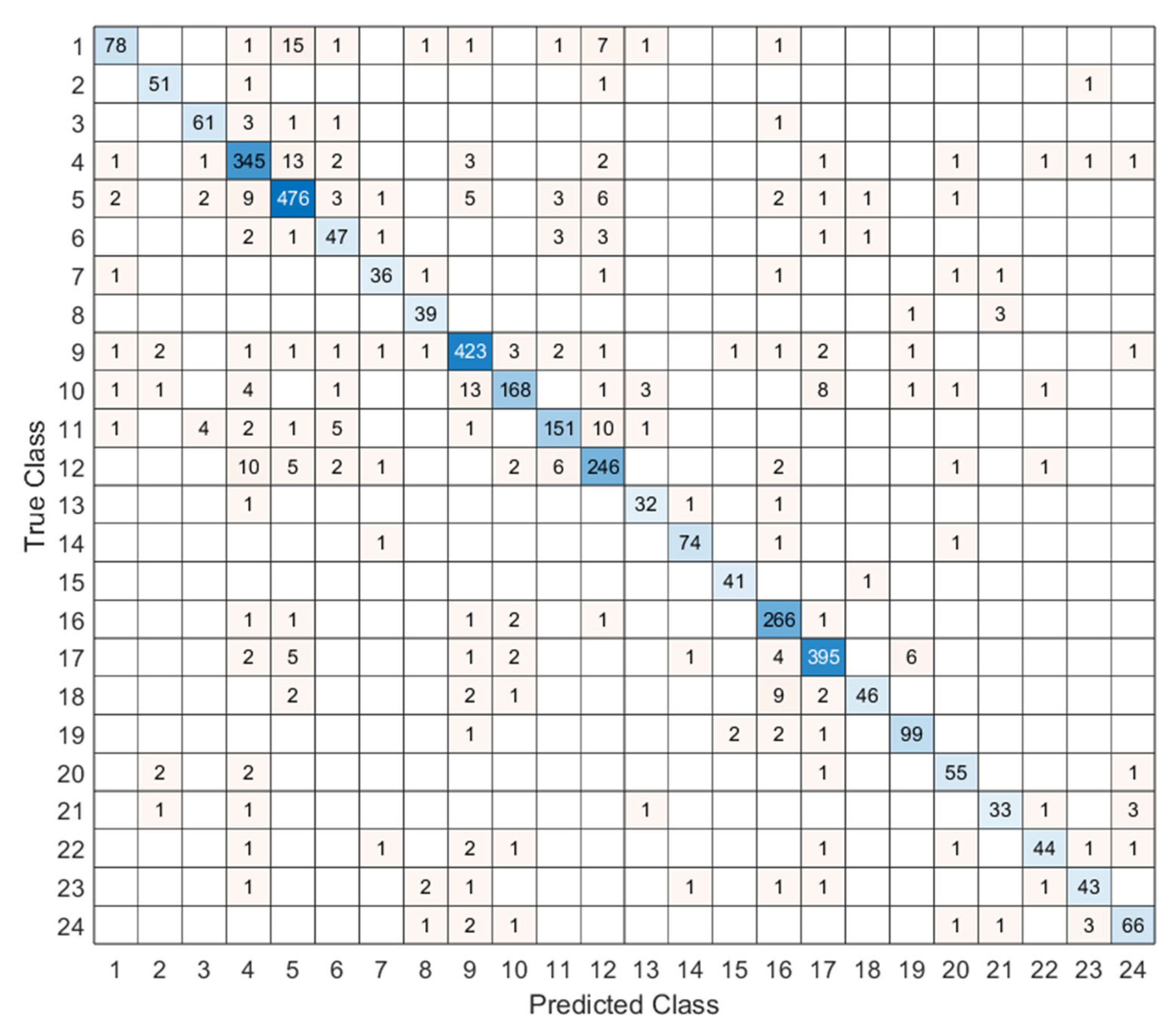
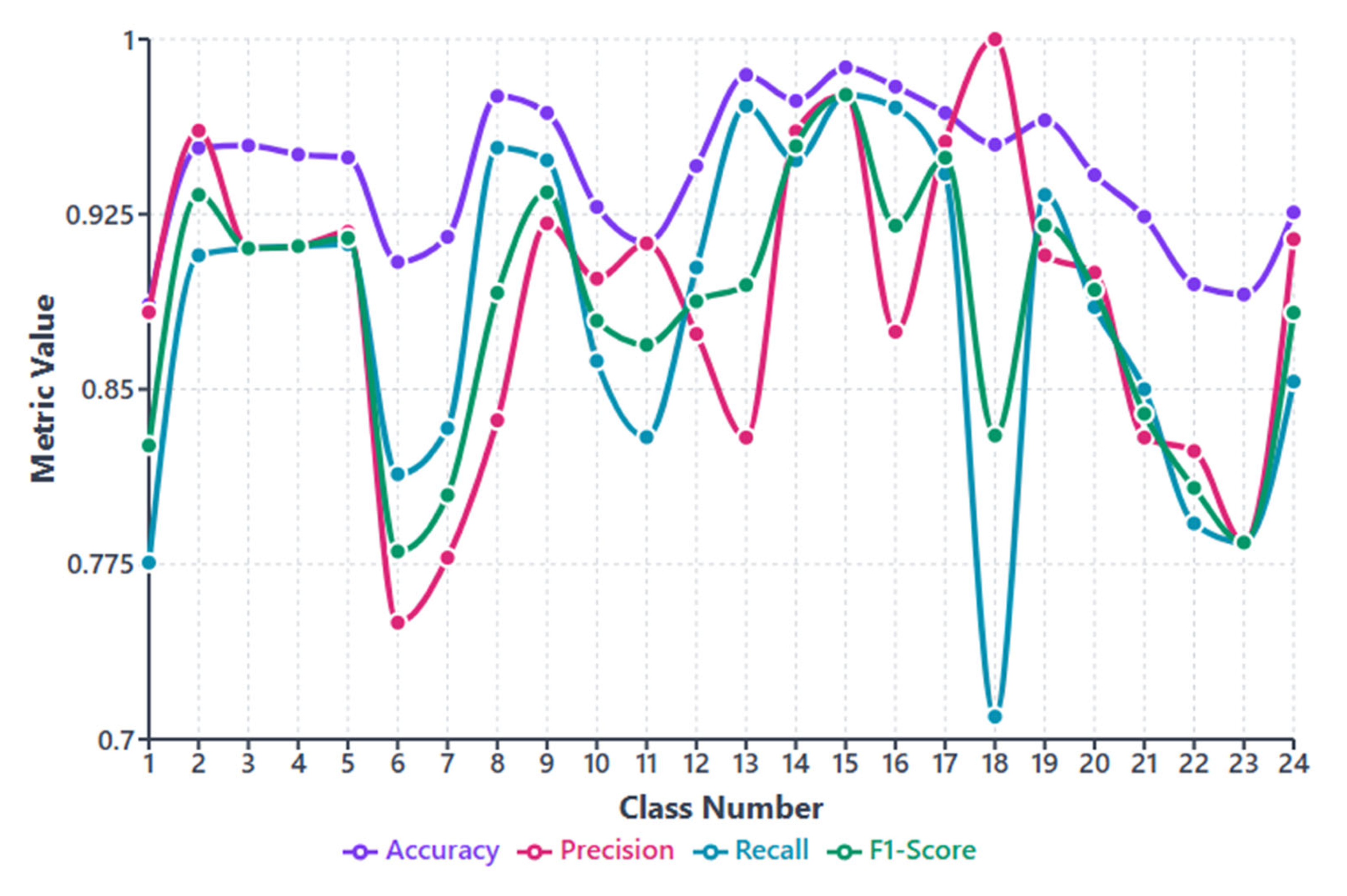
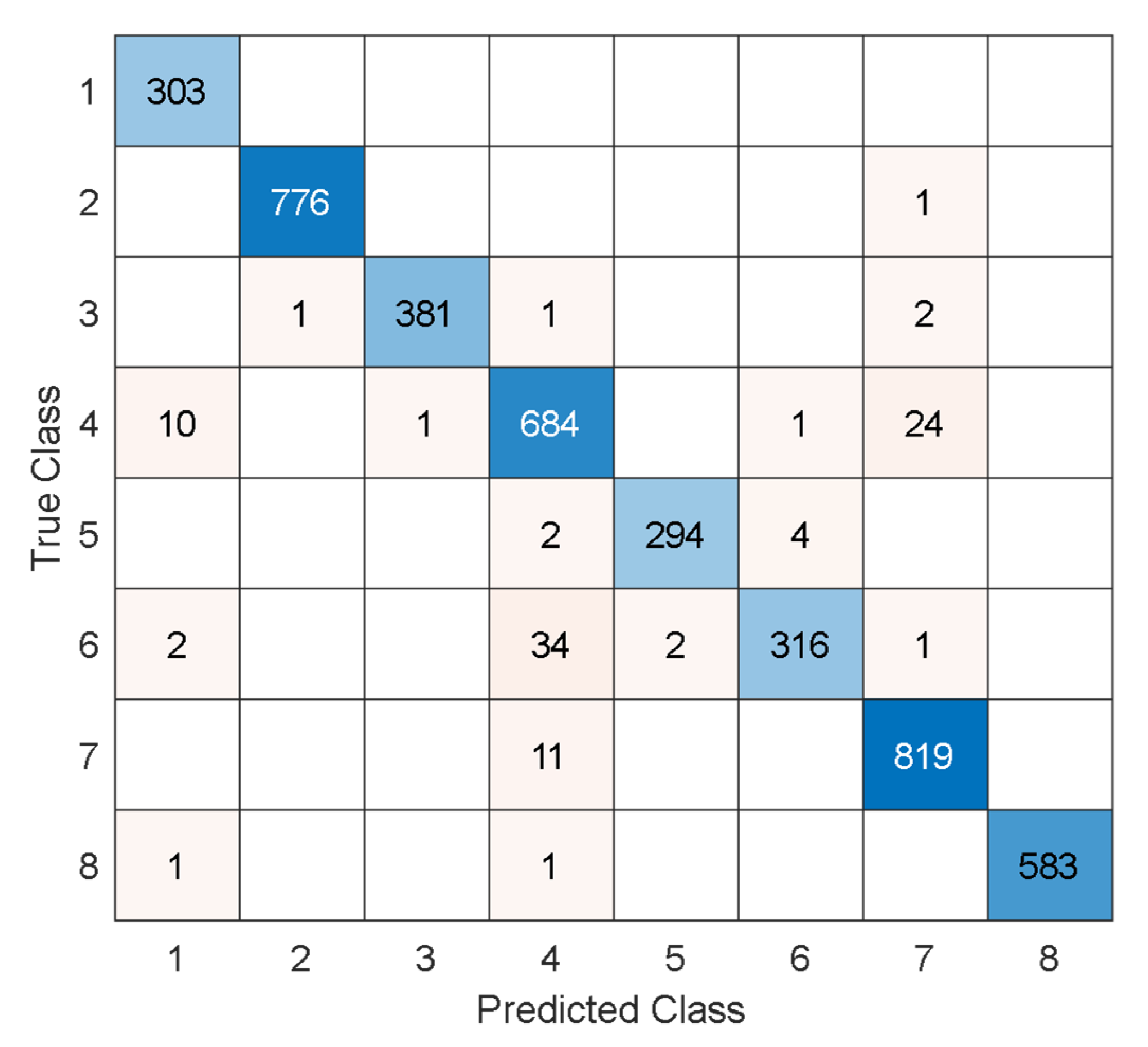
3. Performance Evaluation
4. Discussion
4.1. Comparative Results
4.2. Future Directions
- -
- Our proposed MACNeXt model will be evaluated on additional bacterial groups. These additional species are linked to the biocorrosion of medical implants.
- -
- These bacteria affect material degradation and can influence long-term implant performance.
- -
- The introduced MACNeXt can be adapted to these cases by training on corrosion-related colony images.
- -
- We plan to collect corrosion-focused datasets from laboratory and clinical environments.
- -
- The introduced MACNeXt architecture will be tested under both clinical and material-science settings to explore broader use cases.
- -
- A Transformer-based version of the model will also be investigated to compare feature extraction, attention behavior, and scalability under the same systematic workflow.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, P.; Skiba, D.; Sawicka, B. The Indispensable Role of Bacteria in Human Life and Environmental Health. J. Cell Tissue Res. 2024, 24, 7495–7507. [Google Scholar]
- Akoijam, N.; Kalita, D.; Joshi, S. Bacteria and their industrial importance. In Industrial Microbiology and Biotechnology; Springer: Singapore, Singapore, 2022; pp. 63–79. [Google Scholar]
- Habibur, M.; Ara, S.A.; Bulbul, A.; Moshiur, M.; Hasan, M.M.; Chandra, D.; Shahana, M.; Al, A. Artificial intelligence for improved diagnosis and treatment of bacterial infections. Microb. Bioact. 2024, 7, 521. [Google Scholar] [CrossRef]
- Deusenbery, C.; Wang, Y.; Shukla, A. Recent innovations in bacterial infection detection and treatment. ACS Infect. Dis. 2021, 7, 695–720. [Google Scholar] [CrossRef] [PubMed]
- Váradi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, C.; Rahaman, M.M.; Yao, Y.; Ma, P.; Zhang, J.; Zhao, X.; Jiang, T.; Grzegorzek, M. A comprehensive review of image analysis methods for microorganism counting: From classical image processing to deep learning approaches. Artif. Intell. Rev. 2022, 55, 2875–2944. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Yin, Y.; Zhang, J.; Grzegorzek, M. Applications of artificial neural networks in microorganism image analysis: A comprehensive review from conventional multilayer perceptron to popular convolutional neural network and potential visual transformer. Artif. Intell. Rev. 2023, 56, 1013–1070. [Google Scholar] [CrossRef]
- Li, Y.; Liu, T.; Koydemir, H.C.; Wang, H.; O’Riordan, K.; Bai, B.; Haga, Y.; Kobashi, J.; Tanaka, H.; Tamaru, T. Deep learning-enabled detection and classification of bacterial colonies using a thin-film transistor (TFT) image sensor. ACS Photonics 2022, 9, 2455–2466. [Google Scholar] [CrossRef]
- Yang, F.; Zhong, Y.; Yang, H.; Wan, Y.; Hu, Z.; Peng, S. Microbial colony detection based on deep learning. Appl. Sci. 2023, 13, 10568. [Google Scholar] [CrossRef]
- Wu, Y.; Gadsden, S.A. Machine learning algorithms in microbial classification: A comparative analysis. Front. Artif. Intell. 2023, 6, 1200994. [Google Scholar] [CrossRef]
- Makrai, L.; Fodróczy, B.; Nagy, S.Á.; Czeiszing, P.; Csabai, I.; Szita, G.; Solymosi, N. Annotated dataset for deep-learning-based bacterial colony detection. Sci. Data 2023, 10, 497. [Google Scholar] [CrossRef]
- Talo, M. An automated deep learning approach for bacterial image classification. arXiv 2019, arXiv:1912.08765. [Google Scholar] [CrossRef]
- Akbar, S.A.; Ghazali, K.H.; Hasan, H.; Mohamed, Z.; Aji, W.S.; Yudhana, A. Rapid bacterial colony classification using deep learning. Indones. J. Electr. Eng. Comput. Sci. 2022, 6, 352–361. [Google Scholar] [CrossRef]
- Gallardo Garcia, R.; Jarquin Rodriguez, S.; Beltran Martinez, B.; Hernandez Gracidas, C.; Martinez Torres, R. Efficient deep learning architectures for fast identification of bacterial strains in resource-constrained devices. Multimed. Tools Appl. 2022, 81, 39915–39944. [Google Scholar] [CrossRef]
- Mai, D.-T.; Ishibashi, K. Small-scale depthwise separable convolutional neural networks for bacteria classification. Electronics 2021, 10, 3005. [Google Scholar] [CrossRef]
- Zieliński, B.; Sroka-Oleksiak, A.; Rymarczyk, D.; Piekarczyk, A.; Brzychczy-Włoch, M. Deep learning approach to describe and classify fungi microscopic images. PLoS ONE 2020, 15, e0234806. [Google Scholar] [CrossRef] [PubMed]
- Hallström, E.; Kandavalli, V.; Wählby, C.; Hast, A. Rapid label-free identification of seven bacterial species using microfluidics, single-cell time-lapse phase-contrast microscopy, and deep learning-based image and video classification. PLoS ONE 2025, 20, e0330265. [Google Scholar] [CrossRef]
- Kang, R.; Park, B.; Eady, M.; Ouyang, Q.; Chen, K. Classification of foodborne bacteria using hyperspectral microscope imaging technology coupled with convolutional neural networks. Appl. Microbiol. Biotechnol. 2020, 104, 3157–3166. [Google Scholar] [CrossRef]
- Abd Elaziz, M.; Sarkar, U.; Nag, S.; Hinojosa, S.; Oliva, D. Improving image thresholding by the type II fuzzy entropy and a hybrid optimization algorithm. Soft Comput. 2020, 24, 14885–14905. [Google Scholar] [CrossRef]
- Giuliano, C.; Patel, C.R.; Kale-Pradhan, P.B. A guide to bacterial culture identification and results interpretation. Pharm. Ther. 2019, 44, 192. [Google Scholar]
- Parasuraman, P.; Busi, S.; Lee, J.-K. Standard microbiological techniques (staining, morphological and cultural characteristics, biochemical properties, and serotyping) in the detection of ESKAPE pathogens. In ESKAPE Pathogens: Detection, Mechanisms and Treatment Strategies; Springer: Singapore, Singapore, 2024; pp. 119–155. [Google Scholar]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131.e1129. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Xu, J.; Xue, K.; Zhang, K. Current status and future trends of clinical diagnoses via image-based deep learning. Theranostics 2019, 9, 7556. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Szegedy, C.; Zaremba, W.; Sutskever, I.; Bruna, J.; Erhan, D.; Goodfellow, I.; Fergus, R. Intriguing properties of neural networks. arXiv 2013, arXiv:1312.6199. [Google Scholar]
- Acevedo, A.; Alférez, S.; Merino, A.; Puigví, L.; Rodellar, J. Recognition of peripheral blood cell images using convolutional neural networks. Comput. Methods Programs Biomed. 2019, 180, 105020. [Google Scholar] [CrossRef]
- Acevedo, A.; Merino, A.; Alférez, S.; Molina, Á.; Boldú, L.; Rodellar, J. A dataset of microscopic peripheral blood cell images for development of automatic recognition systems. Data Brief 2020, 30, 105474. [Google Scholar] [CrossRef]
- Dwivedi, K.; Dutta, M.K. Microcell-Net: A deep neural network for multi-class classification of microscopic blood cell images. Expert Syst. 2023, 40, e13295. [Google Scholar] [CrossRef]
- Bilal, O.; Asif, S.; Zhao, M.; Khan, S.U.R.; Li, Y. An amalgamation of deep neural networks optimized with Salp swarm algorithm for cervical cancer detection. Comput. Electr. Eng. 2025, 123, 110106. [Google Scholar] [CrossRef]
- Khan, S.; Sajjad, M.; Abbas, N.; Escorcia-Gutierrez, J.; Gamarra, M.; Muhammad, K. Efficient leukocytes detection and classification in microscopic blood images using convolutional neural network coupled with a dual attention network. Comput. Biol. Med. 2024, 174, 108146. [Google Scholar] [CrossRef]
- Fırat, H. Classification of microscopic peripheral blood cell images using multibranch lightweight CNN-based model. Neural Comput. Appl. 2024, 36, 1599–1620. [Google Scholar] [CrossRef]
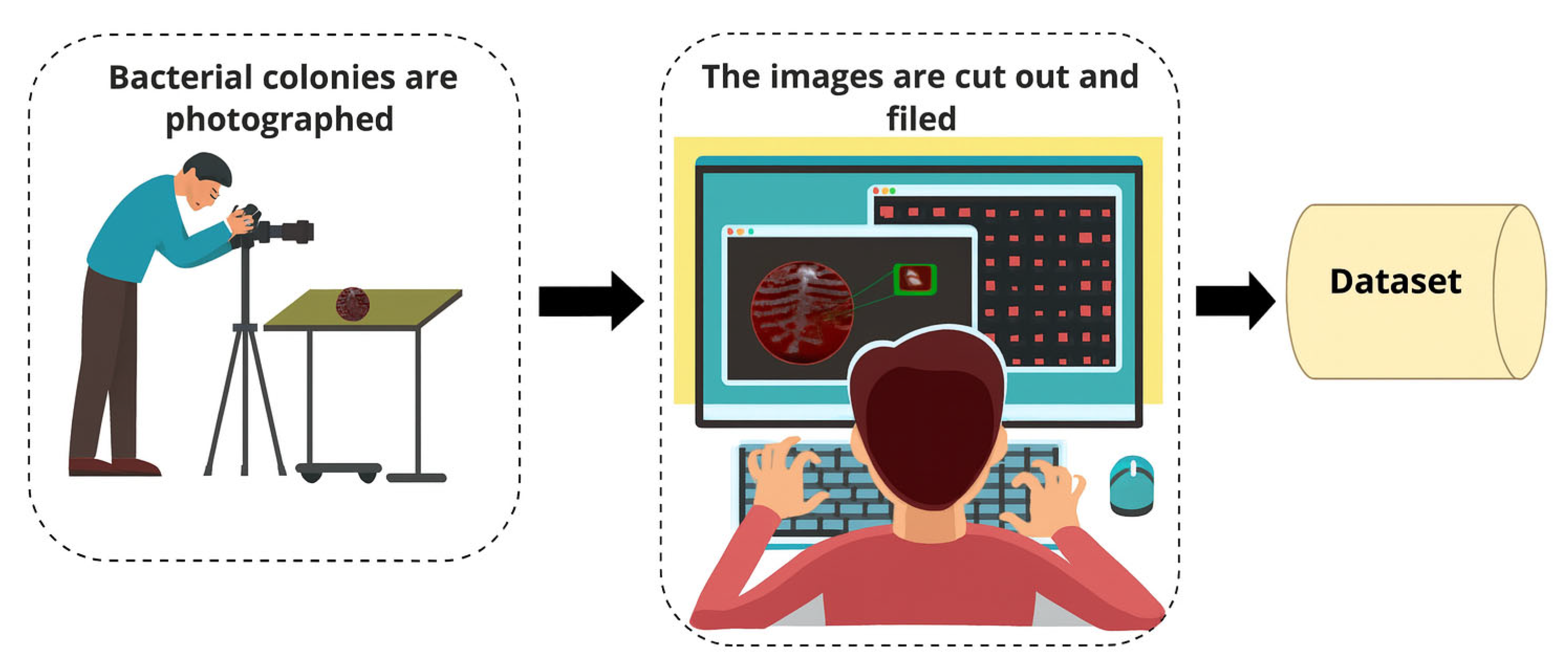

| Class No. | Bacterial Species | Number of Samples | Representative Image | Class No. | Bacterial Species | Number of Samples | Representative Image |
|---|---|---|---|---|---|---|---|
| 1 | Acinetobacter baumannii | 535 |  | 13 | Morganella morganii | 177 |  |
| 2 | Burkholderia cepacia | 272 |  | 14 | Proteus mirabilis | 387 |  |
| 3 | Citrobacter freundii | 337 |  | 15 | Staphylococcus saprophyticus | 209 |  |
| 4 | Hemolytic Escherichia coli | 1860 |  | 16 | Staphylococcus aureus | 1364 |  |
| 5 | Non-hemolytic Escherichia coli | 2562 |  | 17 | Staphylococcus epidermidis | 2078 |  |
| 6 | Enterobacter aerogenes | 292 |  | 18 | Staphylococcus haemolyticus | 311 |  |
| 7 | Enterobacter cloacae | 208 |  | 19 | Staphylococcus hominis | 527 |  |
| 8 | Enterococcus gallinarum | 215 |  | 20 | Stenotrophomonas maltophilia | 306 |  |
| 9 | Enterococcus faecalis | 2214 |  | 21 | Streptococcus pyogenes | 199 |  |
| 10 | Enterococcus faecium | 1013 |  | 22 | Streptococcus agalactiae | 265 |  |
| 11 | Klebsiella oxytoca | 882 |  | 23 | Streptococcus anginosus | 256 |  |
| 12 | Klebsiella pneumoniae | 1378 |  | 24 | Streptococcus mitis | 374 |  |
| Layer | Operation | Input | Output |
|---|---|---|---|
| Stem | 224 × 224 × 3 | 56 × 56 × 96 | |
| Main Stage 1 | 56 × 56 × 96 | 56 × 56 × 96 | |
| Downsampling 1 | 56 × 56 × 96 | 28 × 28 × 192 | |
| Main Stage 2 | 28 × 28 × 192 | 28 × 28 × 192 | |
| Downsampling 2 | 28 × 28 × 192 | 14 × 14 × 384 | |
| Main Stage 3 | 14 × 14 × 384 | 14 × 14 × 384 | |
| Downsampling 3 | 14 × 14 × 384 | 7 × 7 × 768 | |
| Main Stage 4 | 7 × 7 × 768 | 7 × 7 × 768 | |
| Output | GAP Layer, Fully Connected Layer, Softmax Layer | 7 × 7 × 768 | Number of classes |
| Total Learnable Parameters | 4.4 Million | ||
| Parameters | Value |
|---|---|
| Split Ratio | Training: 64%, Validation: 16%, Test: 20% |
| Solver | SGDM |
| Epoch | 30 |
| Mini Batch Size | 128 |
| Initial Learning rate | 0.01 |
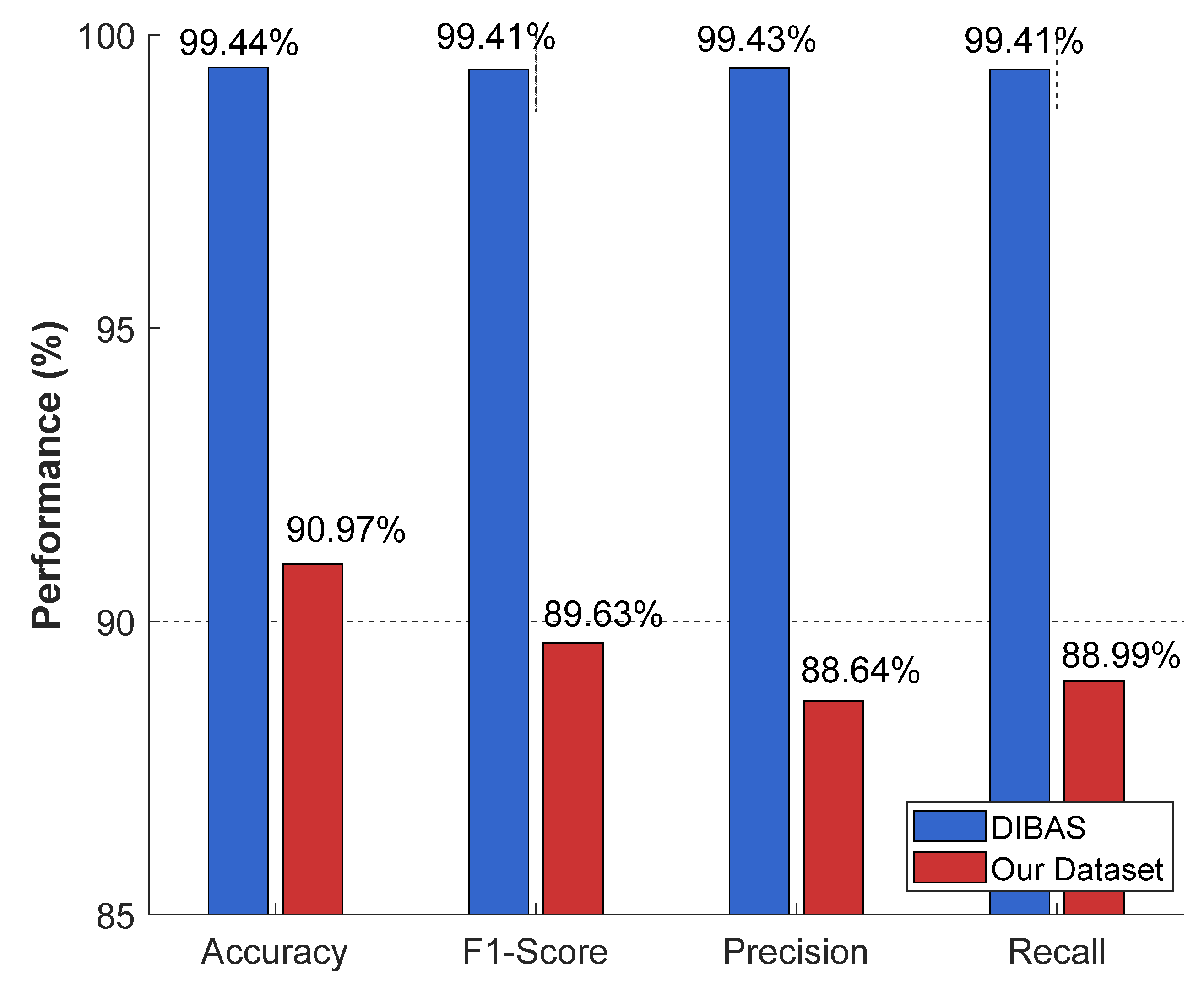
| Metric | Results (%) |
|---|---|
| Accuracy | 90.97 |
| Precision | 89.63 |
| Recall | 88.64 |
| F1-Score | 88.99 |
| Author(s) | Aim/Number of Classes | Method | Performance (%) |
|---|---|---|---|
| Li et al. [6] | 3-class bacterial colony detection (E. coli, Citrobacter, K. pneumoniae) | TFT-based lens-free imaging + ResNet | Accuracy = 97.3 |
| Yang et al. [7] | Bacterial colony detection | Data augmentation via style transfer + Cascade Mask R-CNN/YOLOv8x | YOLOv8x = 76.7 mAP |
| Wu ve Gadsden [8] | 33-class bacterial classification | DenseNet-121, ResNet-50, VGG16 | Accuracy = 99.08 |
| Makrai et al. [12] | Creation of a large-scale colony dataset containing 24 species | Bounding-box-based colony annotation | Dataset presentation |
| Talo [13] | 33-class bacterial classification (DIBaS dataset) | Transfer learning + ResNet-50 | Accuracy = 99.2 |
| Akbar et al. [14] | Bacterial classification | Hybrid: ResNet-101 + SVM | Accuracy = 99.61 |
| Gallardo-Garcia et al. [15] | Evaluation of mobile-friendly models (DIBaS dataset) | EfficientNet-Lite0, MobileNetV2 | Accuracy = 97.38 |
| Mai ve Ishibashi [16] | Development of a lightweight, low-parameter model (DIBaS dataset) | Depthwise Separable CNN | High accuracy (value not specified) |
| Zieliński et al. [17] | 33-class bacterial classification (DIBaS dataset) | Convolutional Neural Network (CNN) | Accuracy = 97.24 |
| Hallström et al. [18] | 4-class bacterial classification (time-series images) | Video-based ResNet | Accuracy = 99.55 ± 0.25 |
| Kang et al. [19] | Single-cell-level bacterial classification | Hyperspectral microscopy (HMI) + 1D-CNN | Accuracy = 90.0 |
| Abd Elaziz et al. [20] | 33-class bacterial classification (DIBaS dataset) | Hybrid: Fractional-order orthogonal descriptors + semantic features | Accuracy= 98.68 |
| Our Method | 33-class bacterial classification (DIBaS dataset) | Proposed MACNeXt model | Accuracy= 99.44 Precision= 99.43 Recall= 99.41 F1-Score= 99.41 |
| Our Method | 24-class clinical bacterial classification (Our collected dataset) | Proposed MACNeXt model | Accuracy= 90.97 Precision= 89.63 Recall= 88.64 F1-Score= 88.99 |
| Metric | Results (%) |
|---|---|
| Training Accuracy | 100% |
| Validation Accuracy | 97.82% |
| Training Loss | 9.71 × 10−5 |
| Validation Loss | 0.0804 |
| Metric | Results (%) |
|---|---|
| Accuracy | 97.67 |
| Precision | 97.91 |
| Recall | 97.40 |
| F1-Score | 97.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aytac, O.; Senol, F.F.; Kivrak, T.; Toraman, Z.A.; Gun, M.V.; Goktas, O.F.; Dogan, S.; Tuncer, T. MACNeXt-Based Bacteria Species Detection. Microorganisms 2025, 13, 2689. https://doi.org/10.3390/microorganisms13122689
Aytac O, Senol FF, Kivrak T, Toraman ZA, Gun MV, Goktas OF, Dogan S, Tuncer T. MACNeXt-Based Bacteria Species Detection. Microorganisms. 2025; 13(12):2689. https://doi.org/10.3390/microorganisms13122689
Chicago/Turabian StyleAytac, Ozlem, Feray Ferda Senol, Tarik Kivrak, Zulal Asci Toraman, Mehmet Veysel Gun, Omer Faruk Goktas, Sengul Dogan, and Turker Tuncer. 2025. "MACNeXt-Based Bacteria Species Detection" Microorganisms 13, no. 12: 2689. https://doi.org/10.3390/microorganisms13122689
APA StyleAytac, O., Senol, F. F., Kivrak, T., Toraman, Z. A., Gun, M. V., Goktas, O. F., Dogan, S., & Tuncer, T. (2025). MACNeXt-Based Bacteria Species Detection. Microorganisms, 13(12), 2689. https://doi.org/10.3390/microorganisms13122689








