HtrA Contributes to Biofilm Formation in Mycobacterium smegmatis by Downregulating the Cell Wall Amidase Ami3
Abstract
1. Introduction
2. Results
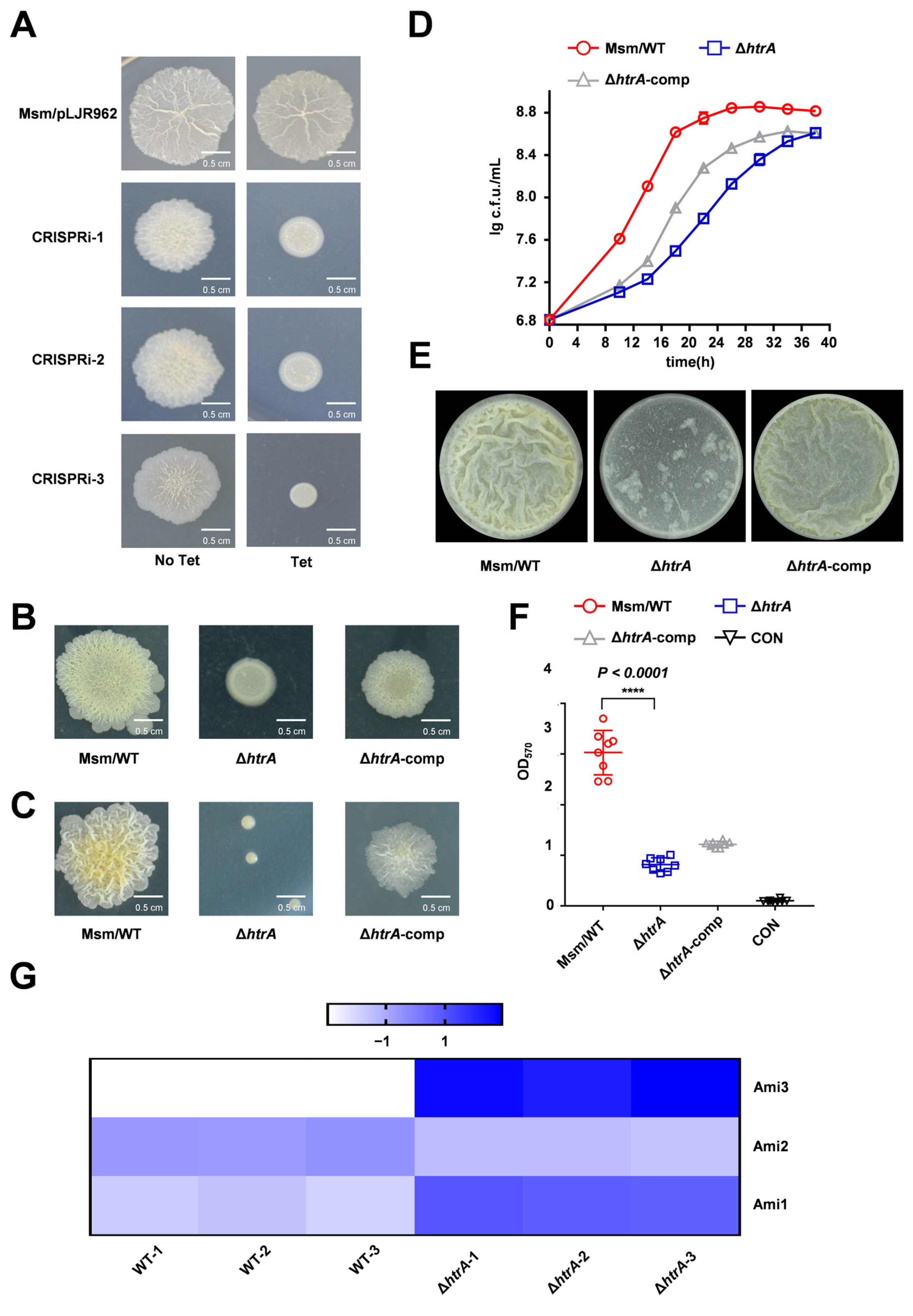
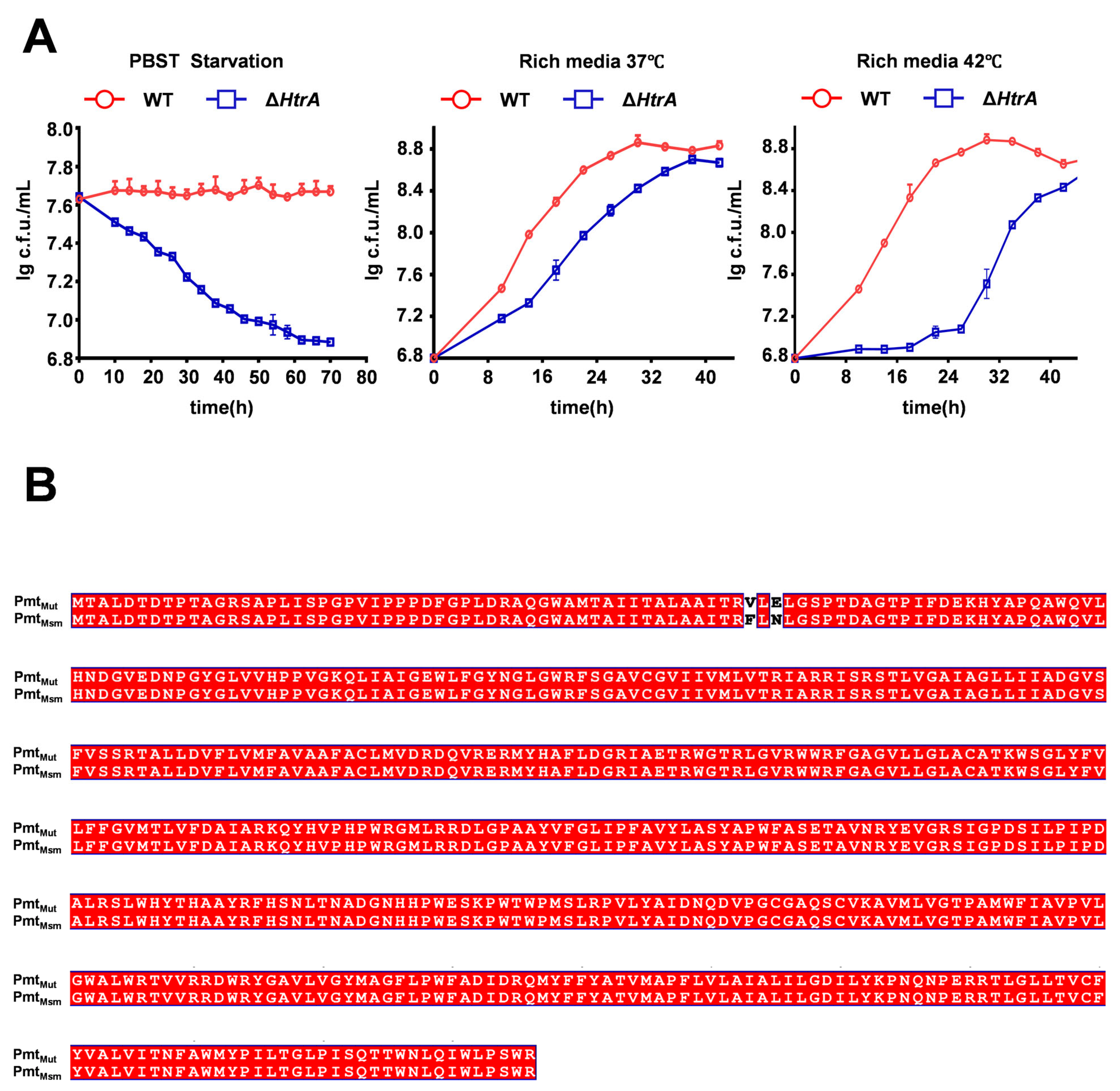
2.1. HtrA Is Essential for Growth and Biofilm Formation in M. smegmatis
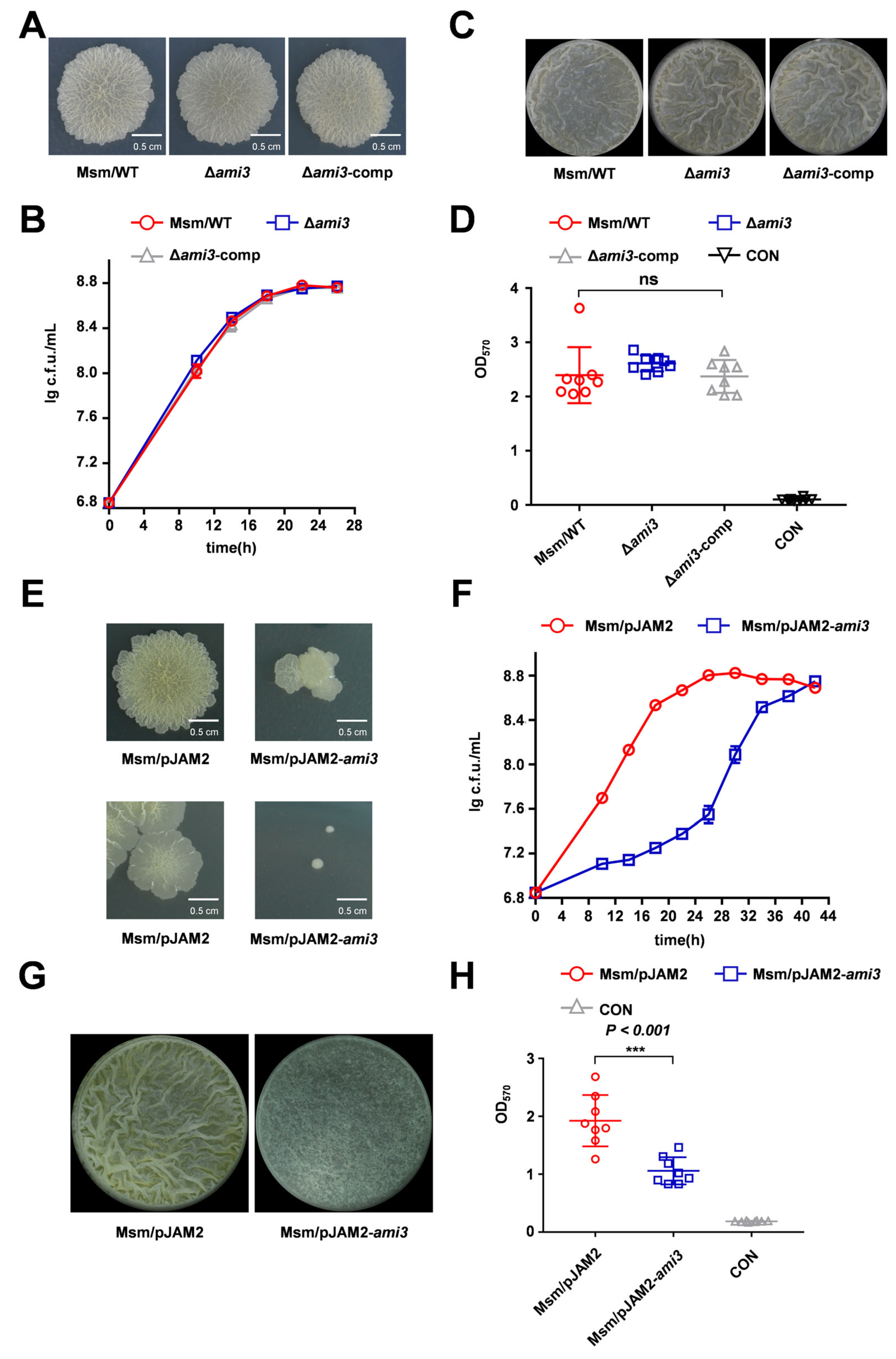
2.2. HtrA Modulates Mycobacterial Biofilm Formation Via Ami3
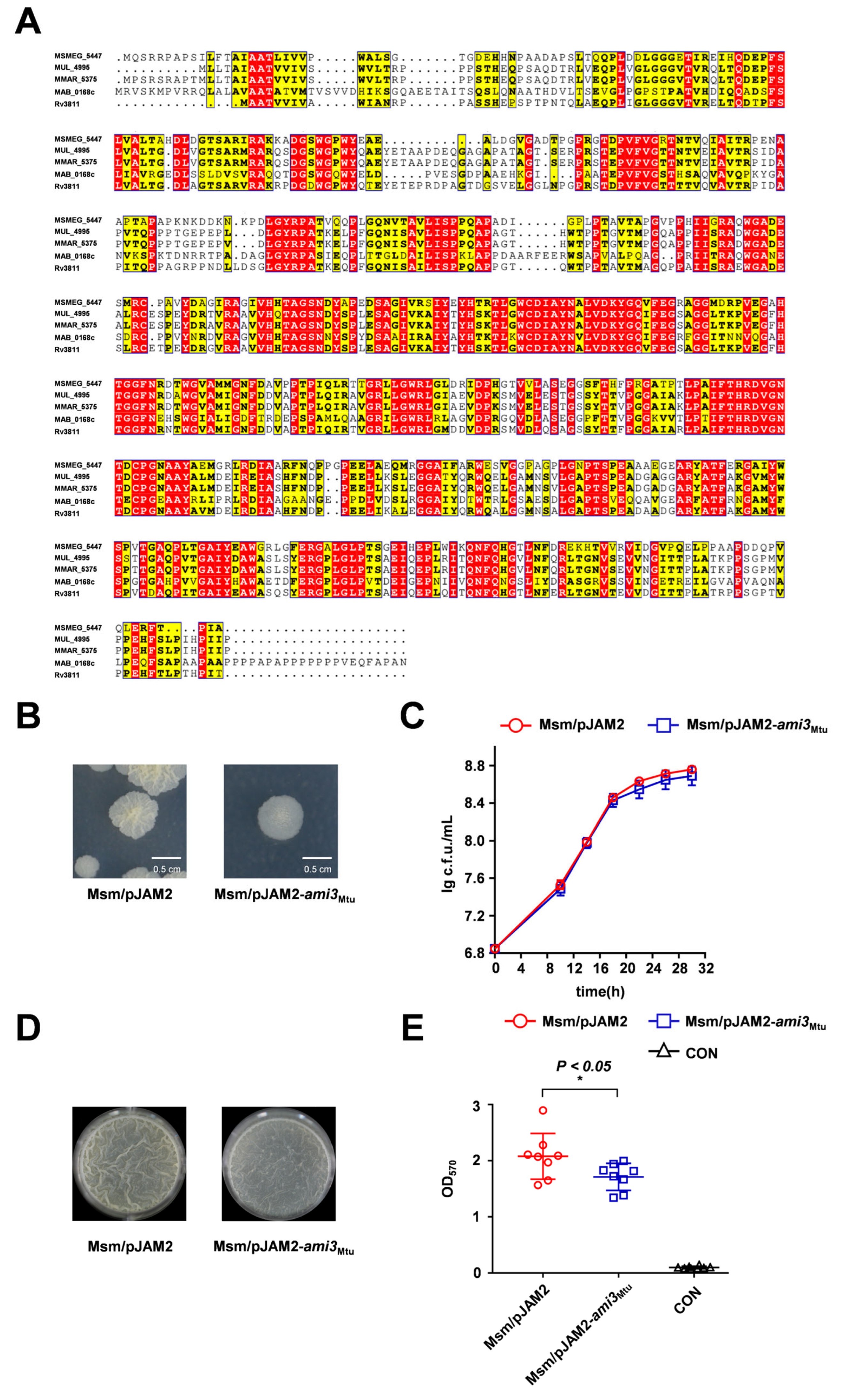
2.3. Ami3 Is Remarkably Conserved Across Mycobacteria
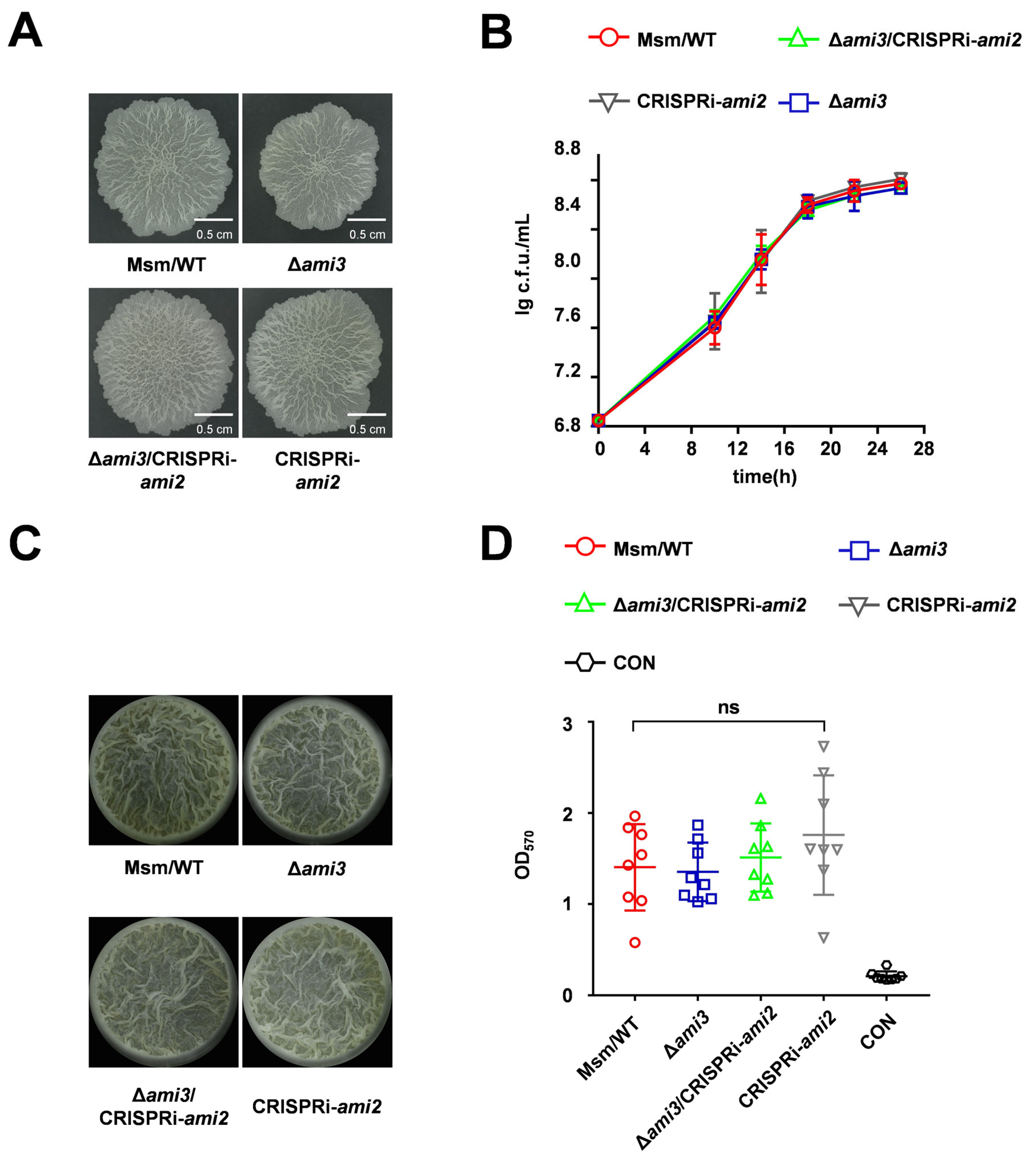
2.4. Ami3, but Not Ami2, Is a Key Regulator of Biofilm Formation in M. smegmatis
2.5. Identification of Novel Suppressor Mutations in Pmt
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Growth Conditions
4.2. Construction of the Deletion Mutants and Complementation Mycobacteria
4.3. Colony Morphology Assay
4.4. Biofilm Phenotyping Experiments
4.5. Growth Curve Measurement
4.6. Proteomic Assays
4.7. AlphaFold3 Analyses
4.8. CRISPRi Technology
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sambandan, D.; Dao, D.N.; Weinrick, B.C.; Vilchèze, C.; Gurcha, S.S.; Ojha, A.; Kremer, L.; Besra, G.S.; Hatfull, G.F.; Jacobs, W.R., Jr. Keto-mycolic acid-dependent pellicle formation confers tolerance to drug-sensitive Mycobacterium tuberculosis. mBio 2013, 4, e00222-13. [Google Scholar] [CrossRef]
- Lee, J.J.; Lee, S.K.; Song, N.; Nathan, T.O.; Swarts, B.M.; Eum, S.Y.; Ehrt, S.; Cho, S.N.; Eoh, H. Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis. Nat. Commun. 2019, 10, 2928. [Google Scholar] [CrossRef]
- Wang, F.; Sambandan, D.; Halder, R.; Wang, J.; Batt, S.M.; Weinrick, B.; Ahmad, I.; Yang, P.; Zhang, Y.; Kim, J.; et al. Identification of a small molecule with activity against drug-resistant and persistent tuberculosis. Proc. Natl. Acad. Sci. USA 2013, 110, E2510–E2517. [Google Scholar] [CrossRef]
- Prigozhin, D.M.; Mavrici, D.; Huizar, J.P.; Vansell, H.J.; Alber, T. Structural and biochemical analyses of Mycobacterium tuberculosis N-acetylmuramyl-L-alanine amidase Rv3717 point to a role in peptidoglycan fragment recycling. J. Biol. Chem. 2013, 288, 31549–31555. [Google Scholar] [CrossRef]
- Turapov, O.; Forti, F.; Kadhim, B.; Ghisotti, D.; Sassine, J.; Straatman-Iwanowska, A.; Bottrill, A.R.; Moynihan, P.J.; Wallis, R.; Barthe, P.; et al. Two Faces of CwlM, an Essential PknB Substrate, in Mycobacterium tuberculosis. Cell Rep. 2018, 25, 57–67.e55. [Google Scholar] [CrossRef] [PubMed]
- Ojha, A.K.; Baughn, A.D.; Sambandan, D.; Hsu, T.; Trivelli, X.; Guerardel, Y.; Alahari, A.; Kremer, L.; Jacobs, W.R., Jr.; Hatfull, G.F. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 2008, 69, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Dhar, N.; McKinney, J.D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007, 10, 30–38. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef]
- Spoering, A.L.; Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2017, 8, 2651. [Google Scholar] [CrossRef]
- Recht, J.; Martínez, A.; Torello, S.; Kolter, R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 2000, 182, 4348–4351. [Google Scholar] [CrossRef]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.R., Jr.; Hatfull, G.F. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 2005, 123, 861–873. [Google Scholar] [CrossRef]
- Lipinska, B.; Fayet, O.; Baird, L.; Georgopoulos, C. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 1989, 171, 1574–1584. [Google Scholar] [CrossRef]
- Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 2011, 12, 152–162. [Google Scholar] [CrossRef]
- Pallen, M.J.; Wren, B.W. The HtrA family of serine proteases. Mol. Microbiol. 1997, 26, 209–221. [Google Scholar] [CrossRef]
- Alba, B.M.; Leeds, J.A.; Onufryk, C.; Lu, C.Z.; Gross, C.A. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002, 16, 2156–2168. [Google Scholar] [CrossRef] [PubMed]
- Betton, J.M.; Sassoon, N.; Hofnung, M.; Laurent, M. Degradation versus aggregation of misfolded maltose-binding protein in the periplasm of Escherichia coli. J. Biol. Chem. 1998, 273, 8897–8902. [Google Scholar] [CrossRef] [PubMed]
- Krojer, T.; Sawa, J.; Schäfer, E.; Saibil, H.R.; Ehrmann, M.; Clausen, T. Structural basis for the regulated protease and chaperone function of DegP. Nature 2008, 453, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Iwanczyk, J.; Damjanovic, D.; Kooistra, J.; Leong, V.; Jomaa, A.; Ghirlando, R.; Ortega, J. Role of the PDZ domains in Escherichia coli DegP protein. J. Bacteriol. 2007, 189, 3176–3186. [Google Scholar] [CrossRef] [PubMed]
- Spiess, C.; Beil, A.; Ehrmann, M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 1999, 97, 339–347. [Google Scholar] [CrossRef]
- Baud, C.; Hodak, H.; Willery, E.; Drobecq, H.; Locht, C.; Jamin, M.; Jacob-Dubuisson, F. Role of DegP for two-partner secretion in Bordetella. Mol. Microbiol. 2009, 74, 315–329. [Google Scholar] [CrossRef]
- Biswas, S.; Biswas, I. Role of HtrA in surface protein expression and biofilm formation by Streptococcus mutans. Infect. Immun. 2005, 73, 6923–6934. [Google Scholar] [CrossRef]
- Hoy, B.; Löwer, M.; Weydig, C.; Carra, G.; Tegtmeyer, N.; Geppert, T.; Schröder, P.; Sewald, N.; Backert, S.; Schneider, G.; et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010, 11, 798–804. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Wessler, S.; Necchi, V.; Rohde, M.; Harrer, A.; Rau, T.T.; Asche, C.I.; Boehm, M.; Loessner, H.; Figueiredo, C.; et al. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell Host Microbe 2017, 22, 552–560.e555. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Matsukawa, M.; Horinishi, Y.; Nakai, K.; Shoji, A.; Yoneko, Y.; Yoshida, N.; Minagawa, S.; Gotoh, N. Interplay of flagellar motility and mucin degradation stimulates the association of Pseudomonas aeruginosa with human epithelial colorectal adenocarcinoma (Caco-2) cells. J. Infect. Chemother. 2013, 19, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.O.O. E-cadherin in gastric cancer. World J. Gastroenterol. 2006, 12, 199–203. [Google Scholar] [CrossRef] [PubMed]
- Noë, V.; Fingleton, B.; Jacobs, K.; Crawford, H.C.; Vermeulen, S.; Steelant, W.; Bruyneel, E.; Matrisian, L.M.; Mareel, M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J. Cell Sci. 2001, 114, 111–118. [Google Scholar] [CrossRef]
- Maretzky, T.; Reiss, K.; Ludwig, A.; Buchholz, J.; Scholz, F.; Proksch, E.; de Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc. Natl. Acad. Sci. USA 2005, 102, 9182–9187. [Google Scholar] [CrossRef]
- Altindis, E.; Fu, Y.; Mekalanos, J.J. Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. USA 2014, 111, E1548–E1556. [Google Scholar] [CrossRef]
- Xue, R.Y.; Liu, C.; Xiao, Q.T.; Sun, S.; Zou, Q.M.; Li, H.B. HtrA family proteases of bacterial pathogens: Pros and cons for their therapeutic use. Clin. Microbiol. Infect. 2021, 27, 559–564. [Google Scholar] [CrossRef]
- Gupta, A.K.; Behera, D.; Gopal, B. The crystal structure of Mycobacterium tuberculosis high-temperature requirement A protein reveals an autoregulatory mechanism. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 803–809. [Google Scholar] [CrossRef]
- Wu, K.J.; Boutte, C.C.; Ioerger, T.R.; Rubin, E.J. Mycobacterium smegmatis HtrA Blocks the Toxic Activity of a Putative Cell Wall Amidase. Cell Rep. 2019, 27, 2468–2479.e2463. [Google Scholar] [CrossRef]
- Park, H.; Kim, Y.T.; Choi, C.; Kim, S. Tripodal Lipoprotein Variants with C-Terminal Hydrophobic Residues Allosterically Modulate Activity of the DegP Protease. J. Mol. Biol. 2017, 429, 3090–3101. [Google Scholar] [CrossRef] [PubMed]
- Typas, A.; Banzhaf, M.; van den Berg van Saparoea, B.; Verheul, J.; Biboy, J.; Nichols, R.J.; Zietek, M.; Beilharz, K.; Kannenberg, K.; von Rechenberg, M.; et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 2010, 143, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Tielker, D.; Ernst, J.F. Protein-O-mannosyltransferases in virulence and development. Cell Mol. Life Sci. 2008, 65, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Machowski, E.E.; Senzani, S.; Ealand, C.; Kana, B.D. Comparative genomics for mycobacterial peptidoglycan remodelling enzymes reveals extensive genetic multiplicity. BMC Microbiol. 2014, 14, 75. [Google Scholar] [CrossRef]
- Boutte, C.C.; E Baer, C.; Papavinasasundaram, K.; Liu, W.; Chase, M.R.; Meniche, X.; Fortune, S.M.; Sassetti, C.M.; Ioerger, T.R.; Rubin, E.J. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. eLife 2016, 5, e14590. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, S.; Kumar, D.; Mishra, A.; Dewangan, R.P.; Shrivastava, P.; Ramachandran, S.; Taneja, B. The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2543–2554. [Google Scholar] [CrossRef]
- VanderVen, B.C.; Harder, J.D.; Crick, D.C.; Belisle, J.T. Export-Mediated Assembly of Mycobacterial Glycoproteins Parallels Eukaryotic Pathways. Science 2005, 309, 941–943. [Google Scholar] [CrossRef]
- Recht, J.; Kolter, R. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 2001, 183, 5718–5724. [Google Scholar] [CrossRef]
- Stack, H.M.; Sleator, R.D.; Bowers, M.; Hill, C.; Gahan, C.G. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 2005, 71, 4241–4247. [Google Scholar] [CrossRef]
- Zarzecka, U.; Modrak-Wójcik, A.; Figaj, D.; Apanowicz, M.; Lesner, A.; Bzowska, A.; Lipinska, B.; Zawilak-Pawlik, A.; Backert, S.; Skorko-Glonek, J. Properties of the HtrA Protease From Bacterium Helicobacter pylori Whose Activity Is Indispensable for Growth Under Stress Conditions. Front. Microbiol. 2019, 10, 961. [Google Scholar] [CrossRef]
- Abfalter, C.M.; Schubert, M.; Götz, C.; Schmidt, T.P.; Posselt, G.; Wessler, S. HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal 2016, 14, 30. [Google Scholar] [CrossRef]
- Backert, S.; Bernegger, S.; Skórko-Glonek, J.; Wessler, S. Extracellular HtrA serine proteases: An emerging new strategy in bacterial pathogenesis. Cell Microbiol. 2018, 20, e12845. [Google Scholar] [CrossRef]
- Larson, M.H.; A Gilbert, L.; Wang, X.; A Lim, W.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Li, Y.; Wei, Y.; Li, K.; Lu, J.; Liu, X.; Li, W. HtrA Contributes to Biofilm Formation in Mycobacterium smegmatis by Downregulating the Cell Wall Amidase Ami3. Microorganisms 2025, 13, 2688. https://doi.org/10.3390/microorganisms13122688
Zheng J, Li Y, Wei Y, Li K, Lu J, Liu X, Li W. HtrA Contributes to Biofilm Formation in Mycobacterium smegmatis by Downregulating the Cell Wall Amidase Ami3. Microorganisms. 2025; 13(12):2688. https://doi.org/10.3390/microorganisms13122688
Chicago/Turabian StyleZheng, Jiachen, Yueqi Li, Yizhang Wei, Kang Li, Jie Lu, Xiaolin Liu, and Weihui Li. 2025. "HtrA Contributes to Biofilm Formation in Mycobacterium smegmatis by Downregulating the Cell Wall Amidase Ami3" Microorganisms 13, no. 12: 2688. https://doi.org/10.3390/microorganisms13122688
APA StyleZheng, J., Li, Y., Wei, Y., Li, K., Lu, J., Liu, X., & Li, W. (2025). HtrA Contributes to Biofilm Formation in Mycobacterium smegmatis by Downregulating the Cell Wall Amidase Ami3. Microorganisms, 13(12), 2688. https://doi.org/10.3390/microorganisms13122688




