Billgrantia hypersalina sp. nov. LNSP4103-1T: A Halotolerant Bioplastic-Producing Bacterium from Saline Agricultural Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Reactivation of Microbial Resources
2.2. Analysis of the 16S rRNA Gene Sequence
2.3. Genomic Analysis
2.4. Phenotypic Analysis
2.5. Chemotaxonomic Analysis
2.6. Statistical Analysis
3. Results
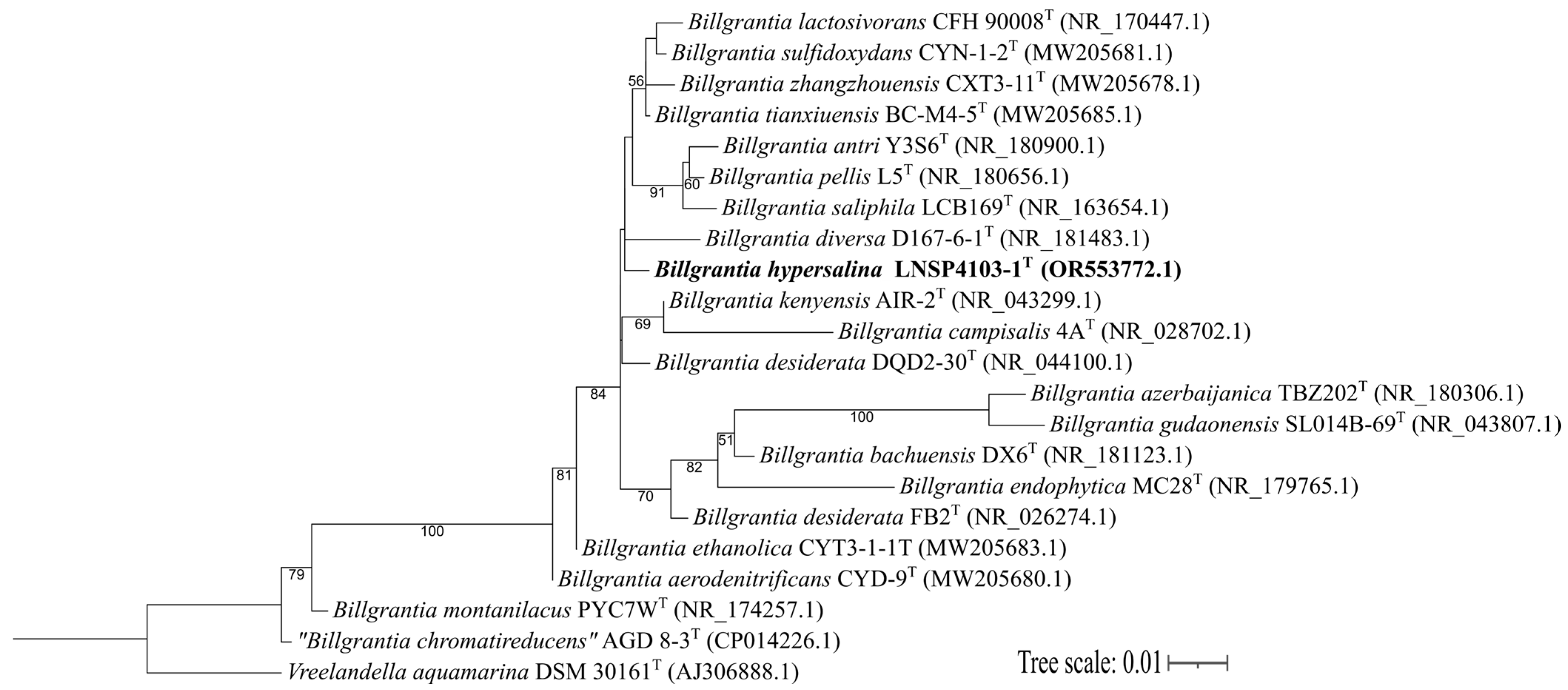
3.1. Analysis of the 16S rRNA Gene Sequence
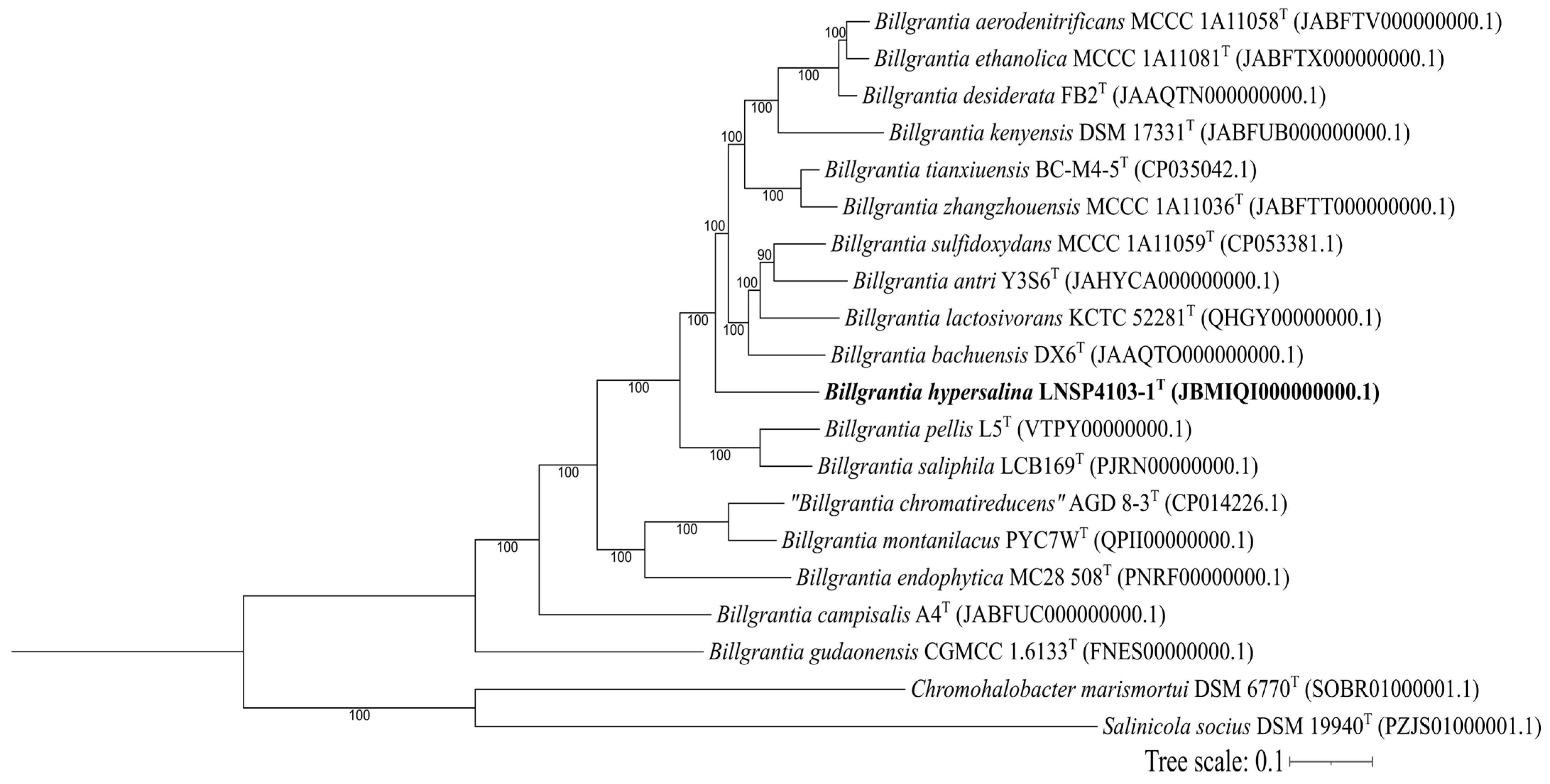
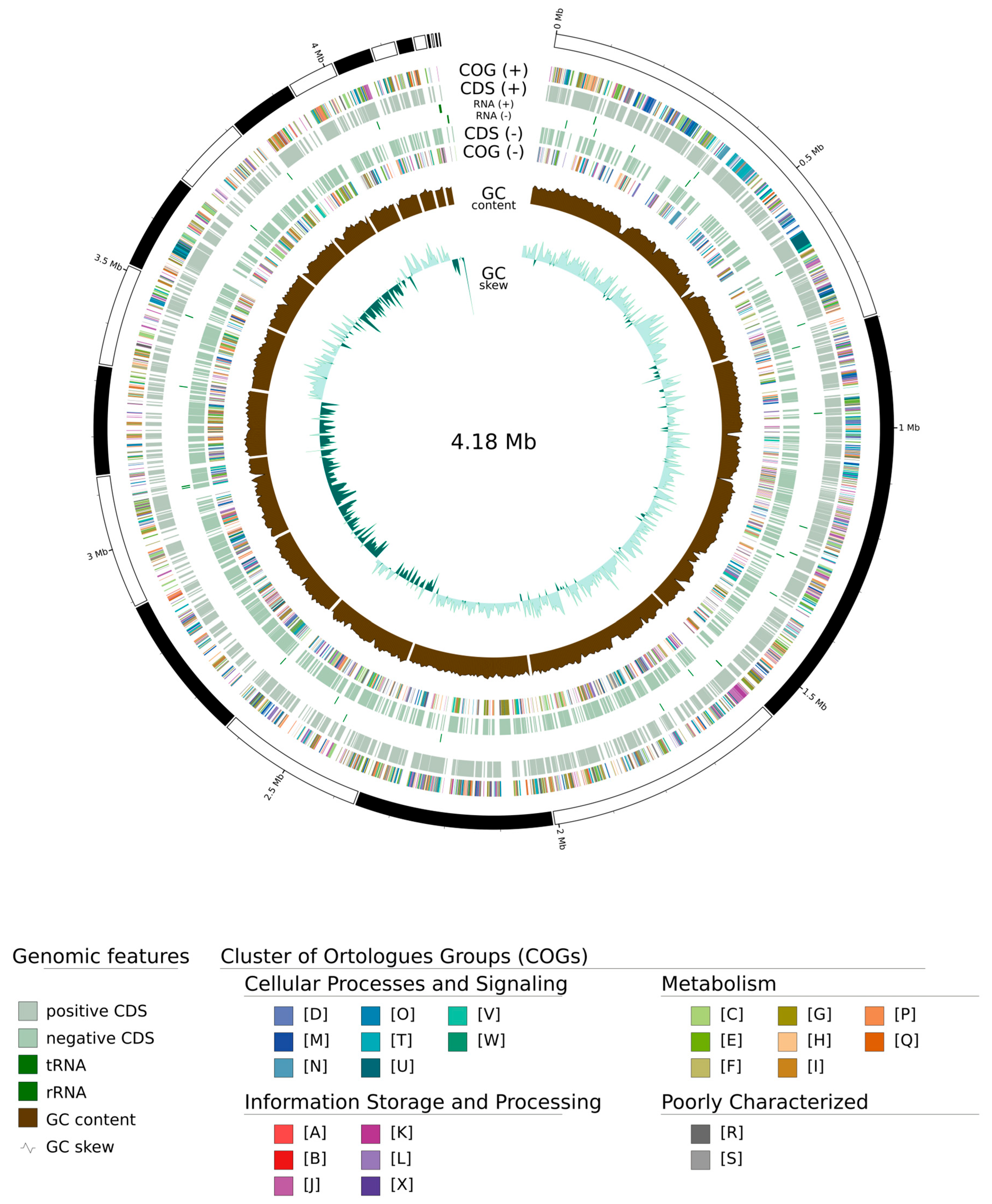
3.2. Genomic Analysis
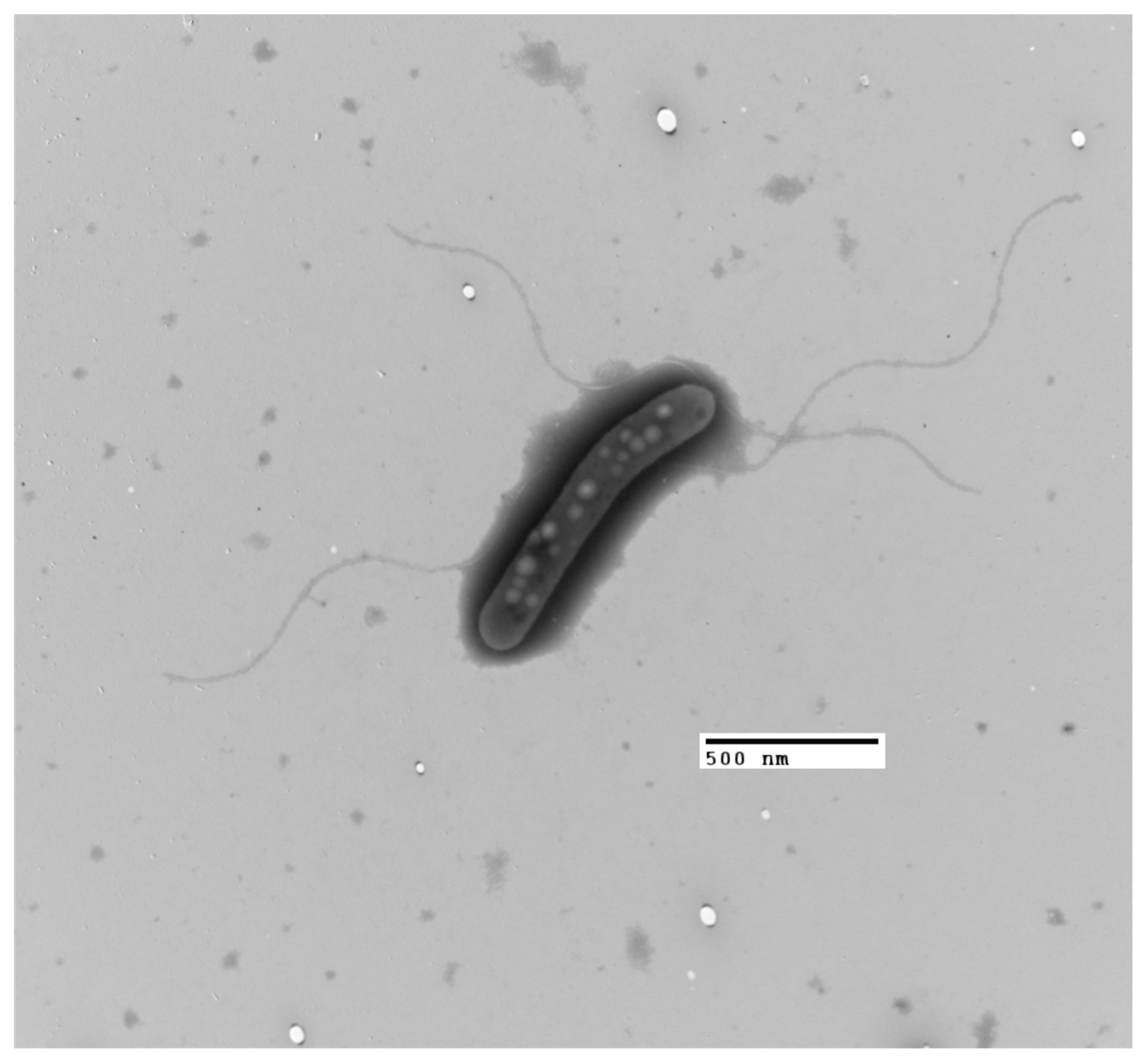
3.3. Phenotypic Analysis
3.4. Chemotaxonomic Analysis
4. Discussion
4.1. Proposal of Billgrantia hypersalina sp. nov.
4.2. Description of Billgrantia hypersalina sp. nov.
4.3. Metabolic Pathway, Production of Polyhydroxyalkanoates, and Osmoadaptation Mechanisms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PHA | Polyhydroxyalkanoates |
| DPG | Diphosphatidylglycerol |
| PE | Phosphatidylethanolamine |
| PG | Phosphatidylglycerol |
| ANI | Average Nucleotide Identity |
| TSB | Tryptic Soy Broth |
| PCR | Polymerase Chain Reaction |
| AAI | Average Aminoacidic Identity |
| GGDC | Genome-to-Genome Distance Calculator |
| BLAST | Basic Local Alignment Search Tool |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LB | Luria–Bertani |
| TSA | Tryptic Soy Agar |
| OD | Optical Density |
| MSM | Mineral Salt Broth |
| CLSI | Clinical & Laboratory Standards Institute |
| MAM | Modified Accumulation Medium |
| dDDH | Digital DNA-DNA Hybridization |
| TETRA | Tetranucleotide Signature Frequency Correlation Coefficient |
| GC | Guanine–Cytosine |
| COG | Clusters of Orthologous Genes |
| CDS | Coding DNA Sequence |
| ATCC | American Type Culture Collection |
| CAIM | Collection of Aquatic Important Microorganisms |
References
- Mazloom, R.; Pierce-Ward, N.T.; Sharma, P.; Pritchard, L.; Brown, C.T.; Vinatzer, B.A.; Heath, L.S. LINgroups as a Robust Principled Approach to Compare and Integrate Multiple Bacterial Taxonomies. IEEE/ACM Trans. Comput. Biol. Bioinform. 2024, 21, 2304–2314. [Google Scholar] [CrossRef]
- de la Haba, R.R.; Arahal, D.R.; Sánchez-Porro, C.; Chuvochina, M.; Wittouck, S.; Hugenholtz, P.; Ventosa, A. A long-awaited taxogenomic investigation of the family Halomonadaceae. Front. Microbiol. 2023, 14, 1293707, Erratum in Front. Microbiol. 2025, 16, 1549745. https://doi.org/10.3389/fmicb.2025.1549745. [Google Scholar] [CrossRef]
- Ventosa, A.; de la Haba, R.R.; Arahal, D.R.; Sánchez-Porro, C. Halomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; Wiley: Hoboken, NJ, USA. [CrossRef]
- Zhou, W. Microbial Synthesis and Degradation of Polyhydroxyalkanoates (PHAs). Ph.D. Thesis, Products and Processes for Biotechnology, Faculty of Science and Engineering, Engineering and Technology Institute Groningen, Groningen, The Netherlands, 2023. [Google Scholar] [CrossRef]
- Gautam, S.; Gautam, A.; Pawaday, J.; Kanzariya, R.K.; Yao, Z. Current Status and Challenges in the Commercial Production of Polyhydroxyalkanoate-Based Bioplastic: A Review. Processes 2024, 12, 1720. [Google Scholar] [CrossRef]
- Kucera, D.; Pernicová, I.; Kovalcik, A.; Koller, M.; Mullerova, L.; Sedlacek, P.; Mravec, F.; Nebesarova, J.; Kalina, M.; Marova, I.; et al. Characterization of the promising poly(3-hydroxybutyrate) producing halophilic bacterium Halomonas halophila. Bioresour. Technol. 2018, 256, 552–556. [Google Scholar] [CrossRef]
- Leandro, T.; Oliveira, M.C.; da Fonseca, M.M.R.; Cesário, M.T. Co-Production of Poly(3-hydroxybutyrate) and Gluconic Acid from Glucose by Halomonas elongata. Bioengineering 2023, 10, 643. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Sudesh, K.; Bazire, A.; Elain, A.; Tan, H.T.; Lim, H.; Bruzaud, S. PHA Production and PHA Synthases of the Halophilic Bacterium Halomonas sp. SF2003. Bioengineering 2020, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Mozejko-Ciesielska, J.; Moraczewski, K.; Czaplicki, S.; Singh, V. Production and characterization of polyhydroxyalkanoates by Halomonas alkaliantarctica utilizing dairy waste as feedstock. Sci. Rep. 2023, 13, 22289. [Google Scholar] [CrossRef]
- Pernicova, I.; Kucera, D.; Nebesarova, J.; Kalina, M.; Novackova, I.; Koller, M.; Obruca, S. Production of polyhydroxyalkanoates on waste frying oil employing selected Halomonas strains. Bioresour. Technol. 2019, 292, 122028. [Google Scholar] [CrossRef]
- Christensen, M.; Jablonski, P.; Altermark, B.; Irgum, K.; Hansen, H. High natural PHA production from acetate in Cobetia sp. MC34 and Cobetia marina DSM 4741T and in silico analyses of the genus specific PhaC2 polymerase variant. Microb. Cell Factories 2021, 20, 225. [Google Scholar] [CrossRef]
- Guevara-Luna, J.; Arroyo-Herrera, I.; Tapia-García, E.Y.; Santos, P.E.-D.L.; Ortega-Nava, A.J.; Vásquez-Murrieta, M.S. Diversity and Biotechnological Potential of Cultivable Halophilic and Halotolerant Bacteria from the “Los Negritos” Geothermal Area. Microorganisms 2024, 12, 482. [Google Scholar] [CrossRef] [PubMed]
- Caton, T.; Witte, L.; Ngyuen, H.; Buchheim, J.; Buchheim, M.; Schneegurt, M. Halotolerant aerobic heterotrophic bacteria from the Great Salt Plains of Oklahoma. Microb. Ecol. 2004, 48, 449–462. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Gouy, M.; Tannier, E.; Comte, N.; Parsons, D.P. Seaview Version 5: A Multiplatform Software for Multiple Sequence Alignment, Molecular Phylogenetic Analyses, and Tree Reconciliation. In Multiple Sequence Alignment; Methods in Molecular Biology; Humana: New York, NY, USA, 2021; Volume 2231, pp. 241–260. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 September 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Camargo, A.P.; Roux, S.; Schulz, F.; Babinski, M.; Xu, Y.; Hu, B.; Chain, P.S.G.; Nayfach, S.; Kyrpides, N.C. Identification of mobile genetic elements with geNomad. Nat. Biotechnol. 2024, 42, 1303–1312. [Google Scholar] [CrossRef]
- Chklovski, A.; Parks, D.H.; Woodcroft, B.J.; Tyson, G.W. CheckM2: A rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 2023, 20, 1203–1212, Erratum in Nat. Methods 2024, 21, 735. https://doi.org/10.1038/s41592-024-02248-z. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Cumsille, A.; Durán, R.E.; Rodríguez-Delherbe, A.; Saona-Urmeneta, V.; Cámara, B.; Seeger, M.; Araya, M.; Jara, N.; Buil-Aranda, C. GenoVi, an open-source automated circular genome visualizer for bacteria and archaea. PLoS Comput. Biol. 2023, 19, e1010998. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2015, 8, 12–24. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Kim, D.; Park, S.; Chun, J. Introducing EzAAI: A pipeline for high-throughput calculations of prokaryotic average amino acid identity. J. Microbiol. 2021, 59, 476–480. [Google Scholar] [CrossRef]
- Riesco, R.; Trujillo, M.E. Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2024, 74, 006300. [Google Scholar] [CrossRef] [PubMed]
- Viver, T.; Conrad, R.E.; Rodriguez-R, L.M.; Ramírez, A.S.; Venter, S.N.; Rocha-Cárdenas, J.; Llabrés, M.; Amann, R.; Konstantinidis, K.T.; Rossello-Mora, R. Towards estimating the number of strains that make up a natural bacterial population. Nat. Commun. 2024, 15, 544. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 50, D801–D807. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Shang, J.; Xu, L.; Yang, R.; Zhao, Y.; Tang, S.-K.; Sun, J.-Q. Halomonas alkalisoli sp. nov. a novel haloalkalophilic species from saline-alkaline soil, and reclassification of Halomonas daqingensis Wu et al. 2018 as a later heterotypic synonym of Halomonas desiderata Berendes et al. 1996. Syst. Appl. Microbiol. 2022, 45, 126351. [Google Scholar] [CrossRef]
- Rathi, D.-N.; Amir, H.; Abed, R.; Kosugi, A.; Arai, T.; Sulaiman, O.; Hashim, R.; Sudesh, K. Polyhydroxyalkanoate biosynthesis and simplified polymer recovery by a novel moderately halophilic bacterium isolated from hypersaline microbial mats. J. Appl. Microbiol. 2013, 114, 384–395. [Google Scholar] [CrossRef]
- Sav, A.R.; Mittal, A.K.; Thorat, A.A.; Dubey, S.; Banerjee, U.C. A Comparative study on the production of PHA by three different Pseudomonas sp. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 940–954. [Google Scholar]
- Cabrera-Santos, D.; Ordoñez-Salanueva, C.A.; Sampayo-Maldonado, S.; Campos, J.E.; Orozco-Segovia, A.; Flores-Ortiz, C.M. Chia (Salvia hispanica L.) Seed Soaking, Germination, and Fatty Acid Behavior at Different Temperatures. Agriculture 2021, 11, 498. [Google Scholar] [CrossRef]
- Abby, S.S.; Kazemzadeh, K.; Vragniau, C.; Pelosi, L.; Pierrel, F. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148259. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Baddiley, J.; Buchanan, J.G.; Handschumacher, R.E.; Prescott, J.F. 551. Chemical studies in the biosynthesis of purine nucleotides. Part, I. The preparation of n-glycylglycosylamines. J. Chem. Soc. 1956, 2818–2823. [Google Scholar] [CrossRef]
- Cronan, J.E. Unsaturated fatty acid synthesis in bacteria: Mechanisms and regulation of canonical and remarkably noncanonical pathways. Biochimie 2024, 218, 137–151. [Google Scholar] [CrossRef]
- Ernster, L.; Dallner, G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta 1995, 1271, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Na Rhie, M.; Kim, H.T.; Joo, J.C.; Cho, I.J.; Son, J.; Jo, S.Y.; Sohn, Y.J.; Baritugo, K.-A.; Pyo, J.; et al. Metabolic engineering for the synthesis of polyesters: A 100-year journey from polyhydroxyalkanoates to non-natural microbial polyesters. Metab. Eng. 2020, 58, 47–81. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.; Yin, J.; Chen, Q.G. 29—Production of—Polyhydroxyalkanoates. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Negi, S., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 655–692. [Google Scholar] [CrossRef]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.-W.; Zhang, H.-Y.; Lu, P.-F.; Peng, Y.-Z. Effects of carbon sources on the enrichment of halophilic polyhydroxyalkanoate-storing mixed microbial culture in an aerobic dynamic feeding process. Sci. Rep. 2016, 6, 30766. [Google Scholar] [CrossRef] [PubMed]
- Dutta, B.; Bandopadhyay, R. Biotechnological potentials of halophilic microorganisms and their impact on mankind. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 75. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Substrate | Organism | Type of PHA | Concentration | Reference |
|---|---|---|---|---|
| Cheese whey hydrolysate, corn stover. | Halomonas halophila | P(3HB) | 82% PHB (w/w to CDW). | [6] |
| Glucose | Halomonas elongata | P(3HB) | 21.2 g L−1 (P(3HB), 53.0% (w/w CDW). | [7] |
| Glucose | Halomonas sp. SF2003 | P(3HB)/P(3HB-co-3HV) | ~2.25 g L−1 | [8] |
| Cheese whey mother liquor | Halomonas alkaliantarctica | P(3HB)/P(3HB-co-3HV) | ~20.1% CDM at 72 h | [9] |
| Waste frying oil | Halomonas hydrothermalis | P(3HB) y P(3HB-co-3HV) | 0.42 g/L, 50.15 mol% 3 HV fraction | [10] |
| Acetate, glycerol, glucose | Cobetia marina | PHB/PHBV | ~2.5 g PHB/L, 61% with acetate. | [11] |
| Strain | dDDH a (%) | ANIb b (%) | ANIm c (%) | FastANI d (%) | TETRA e | AAI f (%) |
|---|---|---|---|---|---|---|
| B. lactosivorans KCTC 52281T | 28.3 | 84.60 | 86.79 | 85.87 | 0.9630 | 87.43 |
| B. sulfidoxydans MCCC 1A11059T | 28.2 | 84.70 | 86.81 | 85.77 | 0.9734 | 87.76 |
| B. bachuensis DX6T | 27.4 | 84.06 | 86.43 | 84.95 | 0.9957 | 87.83 |
| B. tianxiuensis BC-M4-5T | 27.3 | 83.80 | 86.25 | 84.80 | 0.9929 | 87.45 |
| B. desiderata FB2T | 26.8 | 83.50 | 86.27 | 84.68 | 0.9745 | 86.32 |
| B. antri Y3S6T | 26.8 | 83.54 | 86.22 | 84.69 | 0.9943 | 87.30 |
| Substrate | LNSP4103-1T | KCTC 52281T | MCCC 1A11059T | FB2T |
|---|---|---|---|---|
| Mobility | − | + | + | + |
| Pigmentation | Yellow | Yellow | Yellow | White |
| EPS | + | + | + | NA |
| PHA | + | + | + | + |
| NaCl range (optimal, %, p/v) | 0–30.0 (0–12.0) | 1.0–12.0 (4.0–10.0) | 0–18.0 (2.0–6.0) | 0.0–18.0 (8.0–9.0) |
| pH range (optimal) | 7.0–12.0 (7.0–8.0) | 6.0–9.0 (7.0–8.0) | 6.0–10.0 (7.0–8.0) | 7.0–11.0 (9.0–9.5) |
| Oxidase | + | + | + | + |
| Reduction of NO3 | + | + | + | + |
| Production of indole | − | − | − | − |
| Production of H2S | + | − | − | NA |
| Methyl red | − | − | w | NA |
| Voges–Proskauer | − | − | w | − |
| Urease | − | − | − | NA |
| Lysine decarboxylase | + | − | − | NA |
| Ornithine decarboxylase | + | − | − | NA |
| Phenylalanine deaminase | + | − | NA | + |
| Use of carbohydrates | ||||
| D-glucose | + | + | + | + |
| Glycerol | − | + | + | + |
| D-xylose | − | NA | NA | + |
| Succinic acid | − | NA | + | + |
| D-ribose | − | NA | NA | + |
| D-mannose | + | + | − | + |
| Malic acid | − | NA | + | NA |
| D-maltose | − | + | + | + |
| Starch | − | − | − | − |
| D-fructose | + | + | NA | NA |
| Mannitol | + | + | + | NA |
| Propionic acid | + | NA | + | NA |
| Acetate | + | NA | + | + |
| Sodium gluconate | + | NA | + | + |
| L-lactate | + | NA | + | + |
| L-arabinose | − | + | + | + |
| D-sorbitol | − | + | NA | NA |
| D-sucrose | + | + | + | + |
| α-lactose | − | + | NA | NA |
| Citrate | + | − | + | + |
| D-galactose | − | + | NA | + |
| %G + C | 63.75 | 66.7 | 66 | 66 |
| Environment | Saline-sodic soil | Salt lake | Coastal surface sediments | Municipal sewerage |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moran-Olvera, P.M.; Guevara-Luna, J.; Arroyo-Herrera, I.; Larios-Serrato, V.; Flores-Ortiz, C.M.; Hernández-Portilla, L.B.; López-Villegas, E.; Estrada-de los Santos, P.; Hernández-García, J.A.; Vásquez-Murrieta, M.S. Billgrantia hypersalina sp. nov. LNSP4103-1T: A Halotolerant Bioplastic-Producing Bacterium from Saline Agricultural Soil. Microorganisms 2025, 13, 2683. https://doi.org/10.3390/microorganisms13122683
Moran-Olvera PM, Guevara-Luna J, Arroyo-Herrera I, Larios-Serrato V, Flores-Ortiz CM, Hernández-Portilla LB, López-Villegas E, Estrada-de los Santos P, Hernández-García JA, Vásquez-Murrieta MS. Billgrantia hypersalina sp. nov. LNSP4103-1T: A Halotolerant Bioplastic-Producing Bacterium from Saline Agricultural Soil. Microorganisms. 2025; 13(12):2683. https://doi.org/10.3390/microorganisms13122683
Chicago/Turabian StyleMoran-Olvera, Pedro Mauricio, Joseph Guevara-Luna, Ivan Arroyo-Herrera, Violeta Larios-Serrato, César Mateo Flores-Ortiz, Luis Barbo Hernández-Portilla, Edgar López-Villegas, Paulina Estrada-de los Santos, Juan Alfredo Hernández-García, and María Soledad Vásquez-Murrieta. 2025. "Billgrantia hypersalina sp. nov. LNSP4103-1T: A Halotolerant Bioplastic-Producing Bacterium from Saline Agricultural Soil" Microorganisms 13, no. 12: 2683. https://doi.org/10.3390/microorganisms13122683
APA StyleMoran-Olvera, P. M., Guevara-Luna, J., Arroyo-Herrera, I., Larios-Serrato, V., Flores-Ortiz, C. M., Hernández-Portilla, L. B., López-Villegas, E., Estrada-de los Santos, P., Hernández-García, J. A., & Vásquez-Murrieta, M. S. (2025). Billgrantia hypersalina sp. nov. LNSP4103-1T: A Halotolerant Bioplastic-Producing Bacterium from Saline Agricultural Soil. Microorganisms, 13(12), 2683. https://doi.org/10.3390/microorganisms13122683








