The Genomic Characteristics of a Novel Partitivirus Infecting Industrial Hemp in Yunnan, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Next-Generation Sequencing
2.3. RT-PCR and RACE
2.4. Sequence Analysis
2.5. RT-PCR Detection in Seeds
3. Results
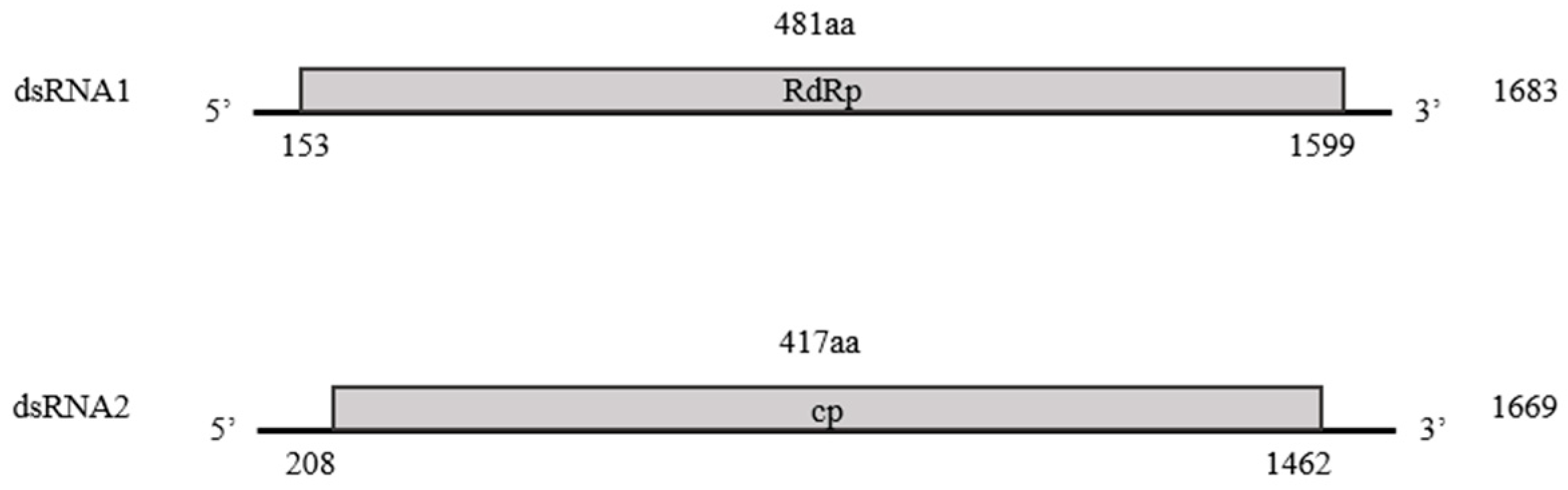
3.1. Viral Sequence Assembly
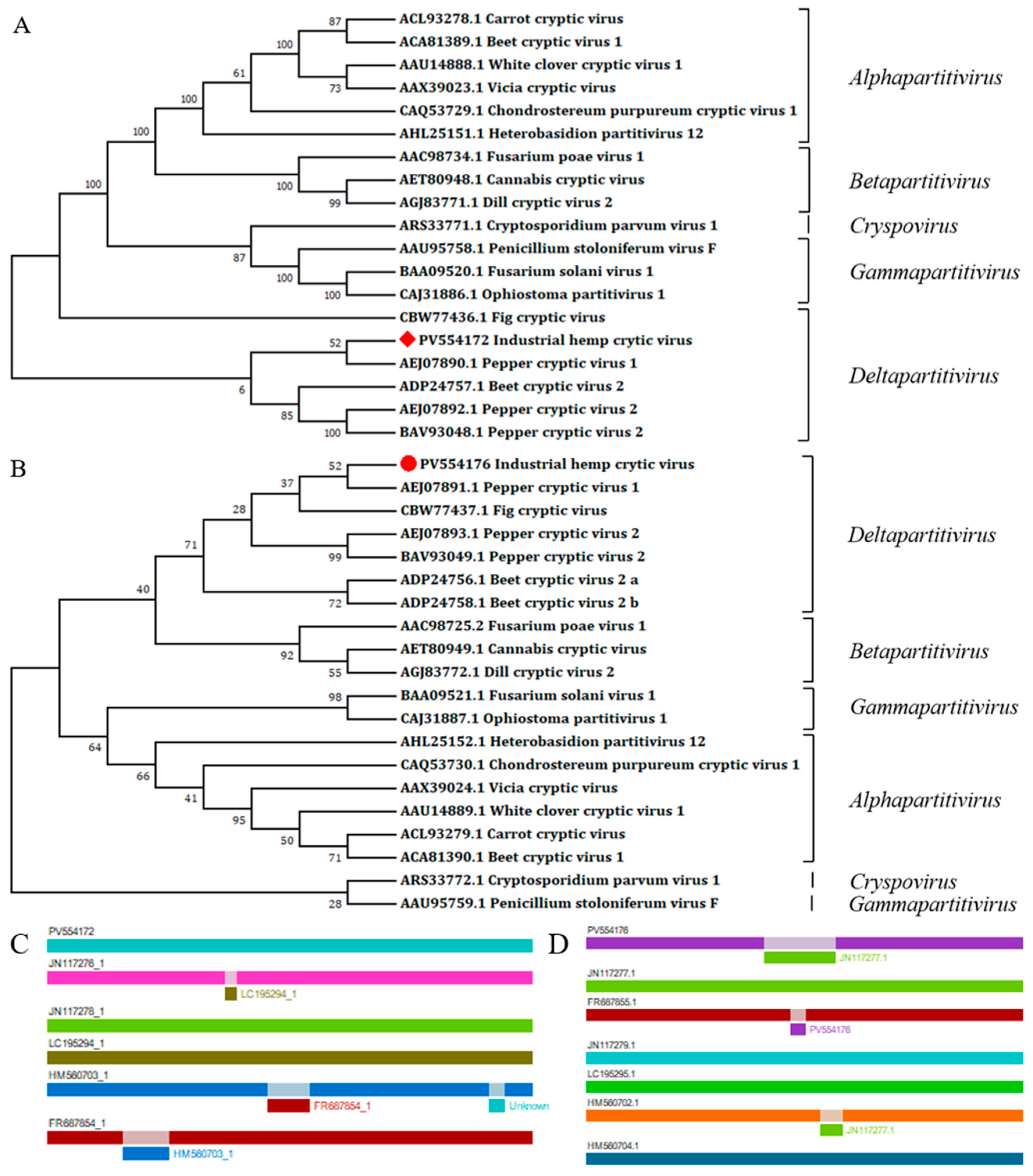
3.2. Viral Sequence Analysis
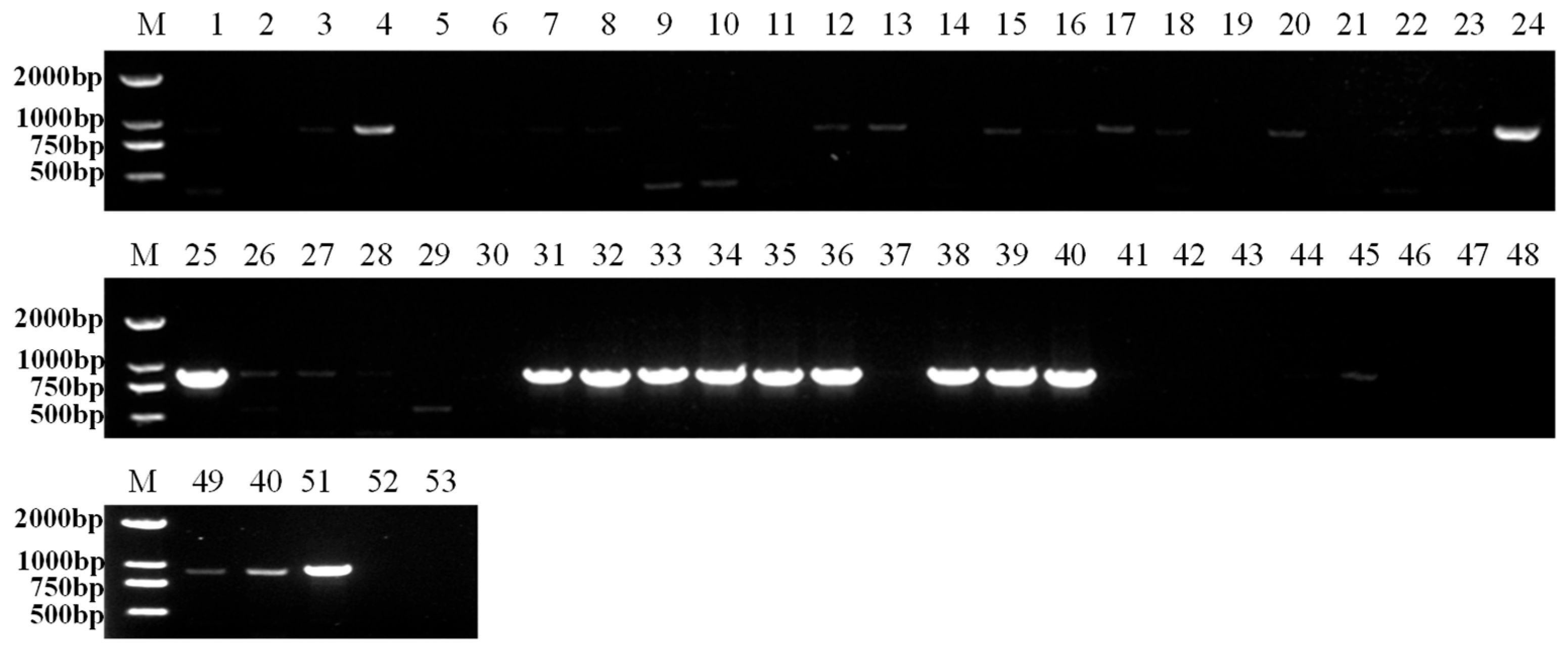
3.3. IHCV Detection in Seeds
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rupasinghe, H.P.V.; Davis, A.; Kumar, S.K.; Murray, B.; Zheljazkov, V.D. Industrial Hemp (Cannabis sativa subsp. sativa) as an Emerging Source for Value-Added Functional Food Ingredients and Nutraceuticals. Molecules 2020, 25, 4078. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Challenges towards Revitalizing Hemp: A Multifaceted Crop. Trends Plant Sci. 2017, 22, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Masson, R.; Dickey, L. First Report of Beet Curly Top Virus Infecting Industrial Hemp (Cannabis sativa) in Arizona. Plant Dis. 2021, 105, 1233. [Google Scholar] [CrossRef]
- Bektas, A.; Hardwick, K.M.; Waterman, K.; Kristof, J. The Occurrence of Hop Latent Viroid in Cannabis sativa with symptoms of Cannabis Stunting Disease in California. Plant Dis. 2019, 103, 2699–2700. [Google Scholar] [CrossRef]
- Jarugula, S.; Wagstaff, C.; Mitra, A.; Crowder, D.W.; Gang, D.R.; Naidu, R.A. First Reports of Beet Curly Top Virus, Citrus Yellow Vein-Associated Virus, and Hop Latent Viroid in Industrial Hemp (Cannabis sativa) in Washington State. Plant Dis. 2023, 107, 2897. [Google Scholar] [CrossRef]
- Miotti, N.; Passera, A.; Ratti, C.; Dall’ara, M.; Casati, P. A Guide to Cannabis Virology: From the Virome Investigation to the Development of Viral Biotechnological Tools. Viruses 2023, 15, 1532. [Google Scholar] [CrossRef]
- Pépin, N.; Hebert, F.O.; Joly, D.L. Genome-Wide Characterization of the MLO Gene Family in Cannabis sativa Reveals Two Genes as Strong Candidates for Powdery Mildew Susceptibility. Front. Plant Sci. 2021, 12, 729261. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Karl, E. Ein Beitrag Zur Analyse Der Virosen Des Hanfes Unter Beriicksichtigung Der Hanfplattlaus Als Virusvektor. Zentralblatt Bakteriol. Parasitenkd. Infekt. Hyg. 1970, 2, 16–22. [Google Scholar]
- Pitt, W.J.; Kairy, L.; Villa, E.; Nalam, V.J.; Nachappa, P. Virus Infection and Host Plant Suitability Affect Feeding Behaviors of Cannabis Aphid (Hemiptera: Aphididae), a Newly Described Vector of Potato Virus Y. Environ. Entomol. 2022, 51, 322–331. [Google Scholar] [CrossRef]
- Greathead, D.J. Hemp diseases and pests. Management and biological control. An advanced treatise J.M. McPartland, R.C. Clarke and D.P. Watson; CABI Publishing Wallingford. Crop Prot. 2001, 20, 351–352. [Google Scholar] [CrossRef]
- Righetti, L.; Paris, R.; Ratti, C.; Calassanzio, M.; Onofri, C.; Calzolari, D.; Menzel, W.; Knierim, D.; Magagnini, G.; Pacifico, D.; et al. Not the One, but the Only One: About Cannabis Cryptic Virus in Plants Showing ‘Hemp Streak’ Disease Symptoms. Eur. J. Plant Pathol. 2018, 150, 575–588. [Google Scholar] [CrossRef]
- Nibert, M.L.; Ghabrial, S.A.; Maiss, E.; Lesker, T.; Vainio, E.J.; Jiang, D.; Suzuki, N. Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res. 2014, 188, 128–141. [Google Scholar] [CrossRef]
- Ghabrial, S.; Ochoa, W.; Baker, T.; Nibert, M.L. Partitiviruses: General Features. In Encyclopedia of Virology; Academic Press: Cambridge, MA, USA, 2008; pp. 68–75. [Google Scholar] [CrossRef]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, W.F.; Havens, W.M.; Sinkovits, R.S.; Nibert, M.L.; Ghabrial, S.A.; Baker, T.S. Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure 2008, 16, 776–786. [Google Scholar] [CrossRef]
- Nibert, M.L.; Tang, J.; Xie, J.; Collier, A.M.; Ghabrial, S.A.; Baker, T.S.; Tao, Y.J. Chapter Three—3D Structures of Fungal Partitiviruses. In Advances in Virus Research; Ghabrial, S.A., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 59–85. [Google Scholar]
- Boccardo, G.; Lisa, V.; Luisoni, E.; Milne, R.G. Cryptic Plant Viruses. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA, 1987; pp. 171–214. [Google Scholar]
- Xiao, X.; Cheng, J.; Tang, J.; Fu, Y.; Jiang, D.; Baker, T.S.; Ghabrial, S.A.; Xie, J. A Novel Partitivirus That Confers Hypovirulence on Plant Pathogenic Fungi. J. Virol. 2014, 88, 10120–10133. [Google Scholar] [CrossRef]
- Rose, H.; Maiss, E. Plant and Protozoal Partitiviruses (Partitiviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 632–641. [Google Scholar]
- Szegő, A. Detection of high molecular weight dsRNA persisting in Dianthus species. Acta Biol. Szeged. 2005, 49, 17–19. [Google Scholar]
- Roossinck, M.J. Lifestyles of plant viruses. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1899–1905. [Google Scholar] [CrossRef]
- Arancibia, R.; Valverde, R.; Can, F. Properties of a cryptic virus from pepper (Capsicum annuum). Plant Pathol. 1995, 44, 164–168. [Google Scholar]
- Valverde, R.A.; Gutierrez, D.L. Molecular and Biological Properties of a Putative Partitivirus from Jalapeño Pepper (Capsicum annuum L.). Rev. Mex. Fitopatol. 2008, 26, 1–6. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Matoušek, J.; Steger, G.; Schubert, J. Complete sequence of a cryptic virus from hemp (Cannabis sativa). Arch. Virol. 2012, 157, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.S.; Antoniw, J.F.; White, R.F.; Jolliffe, T.H. Effects of beet cryptic virus infection on sugar beet in field trials. Ann. Appl. Biol. 1994, 124, 451–459. [Google Scholar] [CrossRef]
- Nakatsukasa-Akune, M.; Yamashita, K.; Shimoda, Y.; Uchiumi, T.; Abe, M.; Aoki, T.; Kamizawa, A.; Ayabe, S.-I.; Higashi, S.; Suzuki, A. Suppression of Root Nodule Formation by Artificial Expression of the TrEnodDR1 (Coat Protein of White clover cryptic virus 1) Gene in Lotus japonicus. Mol. Plant-Microbe Interact.® 2005, 18, 1069–1080. [Google Scholar] [CrossRef]
- Velázquez, K.; Agüero, J.; Vives, M.C.; Aleza, P.; Pina, J.A.; Moreno, P.; Navarro, L.; Guerri, J. Precocious flowering of juvenile citrus induced by a viral vector based on Citrus leaf blotch virus: A new tool for genetics and breeding. Plant Biotechnol. J. 2016, 14, 1976–1985. [Google Scholar] [CrossRef]
- Feng, C.; Guo, X.; Gu, T.; Hua, Y.; Zhuang, X.; Zhang, K. Generation of a Triple-Shuttling Vector and the Application in Plant Plus-Strand RNA Virus Infectious cDNA Clone Construction. Int. J. Mol. Sci. 2023, 24, 5477. [Google Scholar] [CrossRef]
- Wang, S.; Chen, B.; Ni, S.; Liang, Y.; Li, Z. Efficient generation of recombinant eggplant mottled dwarf virus and expression of foreign proteins in solanaceous hosts. Virology 2024, 591, 109980. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer Sequence (5′-3′) | PCR Product Length (bp) |
|---|---|---|
| IHCV-RNA1-F | TGTTATAGACGTTGAGAACGGGT | 1000 |
| IHCV-RNA1-R | GATGTTCGTCCAAGGAAACTGAT | |
| IHCV-RNA2-1F | AACAGCAGACCCGCACAGGAATC | 617 |
| IHCV-RNA2-1R | TCGGCCAGTGTAGCTTGAGGAAA | |
| IHCV-RNA2-2F | TAACAGCAATGAAACACCTCAAAGT | 896 |
| IHCV-RNA2-2R | CTTCCTTAACGAAGATGAACTGTG | |
| 3′RACE-IHCV-RNA1-F | TCCCGAATACCCTGTCGAAAC | 310 |
| 3′RACE-IHCV-RNA2-F | ACAGACATTCACACCCAGTCAGCCT | 200 |
| 5′RACE-IHCV-RNA1-R | ACCTCCTTTAGGTCCTTTCTCG | 500 |
| 5′RACE-IHCV-RNA2-R | CATGTCGAGACGTAGATTCTAGCCG | 330 |
| Universal Short Primer | Manufacturer-provided | — |
| Virus | RdRp (IHCV) | CP (IHCV) | ||
|---|---|---|---|---|
| nt (%) | aa (%) | nt (%) | aa (%) | |
| Citrulluslanatus cryptic virus | 63.10 | 68.10 | 41.20 | 30.20 |
| Dichroapartitivirus 2 | 62.60 | 64.50 | 43.10 | 35.60 |
| Polygonatumpartitivirus 2 | 62.60 | 63.60 | 42.50 | 34.70 |
| Alloteropsis cryptic virus 2 | 60.80 | 58.50 | 32.90 | 11.70 |
| Dactylorhiza cryptic virus 3 | 59.80 | 55.90 | 44.30 | 33.50 |
| Sinapis alba cryptic virus 1 | 59.60 | 59.50 | 42.30 | 29.10 |
| Panax cryptic virus 1 | 59.60 | 59.70 | 40.50 | 30.40 |
| Pepper cryptic virus 1 | 59.10 | 58.80 | 42.00 | 31.40 |
| Vitis cryptic virus | 56.60 | 64.40 | 25.90 | 35.80 |
| Cannabis cryptic virus | 46.00 | 18.70 | 45.80 | 18.10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xu, Y.; Su, X.; Guan, F.; Zheng, K.; Chen, X. The Genomic Characteristics of a Novel Partitivirus Infecting Industrial Hemp in Yunnan, China. Microorganisms 2025, 13, 2682. https://doi.org/10.3390/microorganisms13122682
Liu Y, Xu Y, Su X, Guan F, Zheng K, Chen X. The Genomic Characteristics of a Novel Partitivirus Infecting Industrial Hemp in Yunnan, China. Microorganisms. 2025; 13(12):2682. https://doi.org/10.3390/microorganisms13122682
Chicago/Turabian StyleLiu, Yuying, Yanpin Xu, Xiaoxia Su, Fang Guan, Kuanyu Zheng, and Xuan Chen. 2025. "The Genomic Characteristics of a Novel Partitivirus Infecting Industrial Hemp in Yunnan, China" Microorganisms 13, no. 12: 2682. https://doi.org/10.3390/microorganisms13122682
APA StyleLiu, Y., Xu, Y., Su, X., Guan, F., Zheng, K., & Chen, X. (2025). The Genomic Characteristics of a Novel Partitivirus Infecting Industrial Hemp in Yunnan, China. Microorganisms, 13(12), 2682. https://doi.org/10.3390/microorganisms13122682





