Lacticaseibacillus casei Combats Biofilm Formation and Exhibits Antibacterial Activity Against Clinical Isolates of Staphylococcus aureus, Salmonella enterica, and Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Eukaryotic Cell Cultures
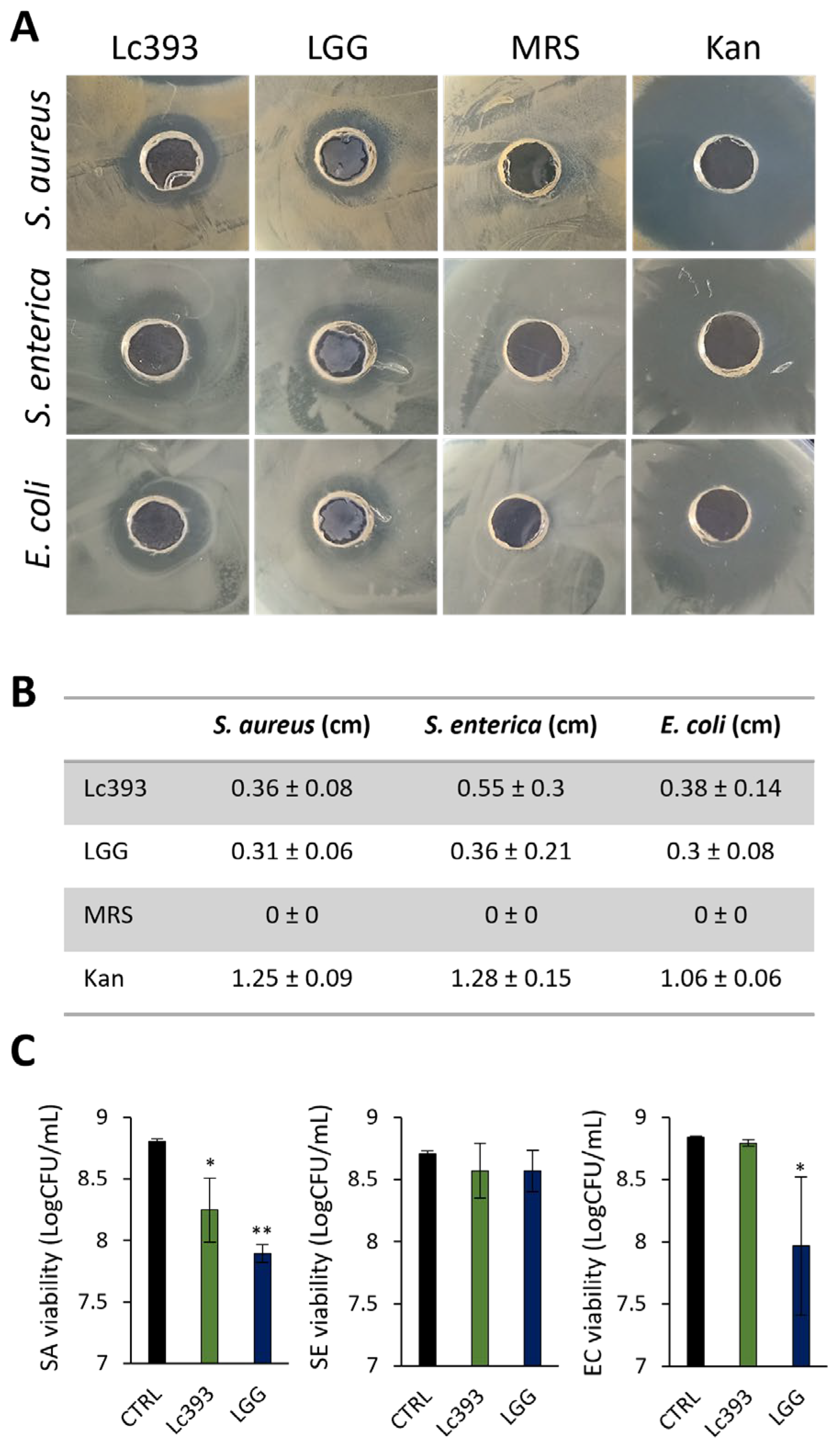
2.3. Well Diffusion Assay
2.4. Pathogen Growth Inhibition Assays
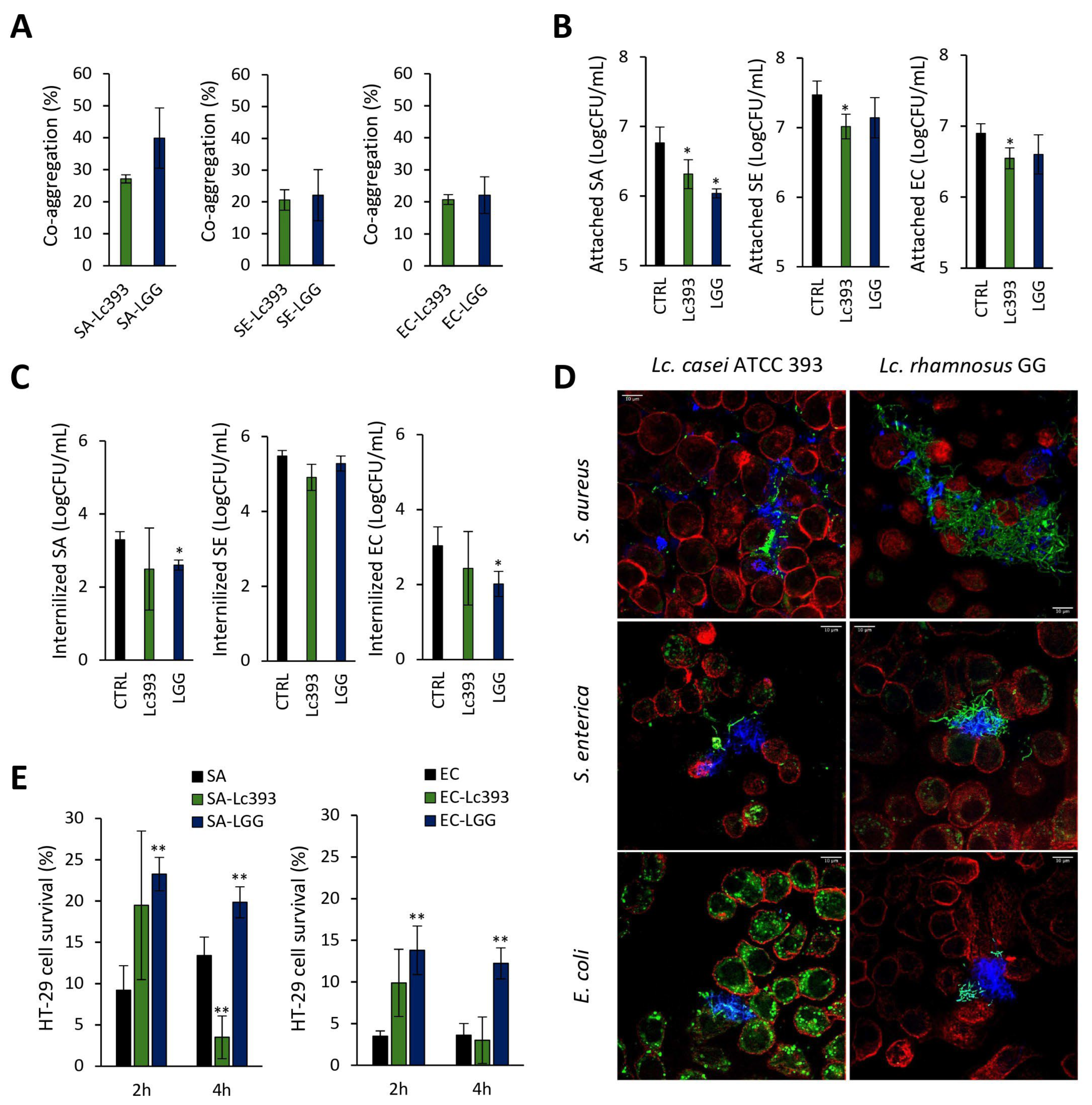
2.5. Aggregation Capacity
2.6. Competitive Exclusion Assay
2.7. Gentamicin Protection Assay
2.8. Confocal Microscopy
2.8.1. Sample Preparation
2.8.2. Image Acquisition
2.9. Eukaryotic Cell Viability Assay
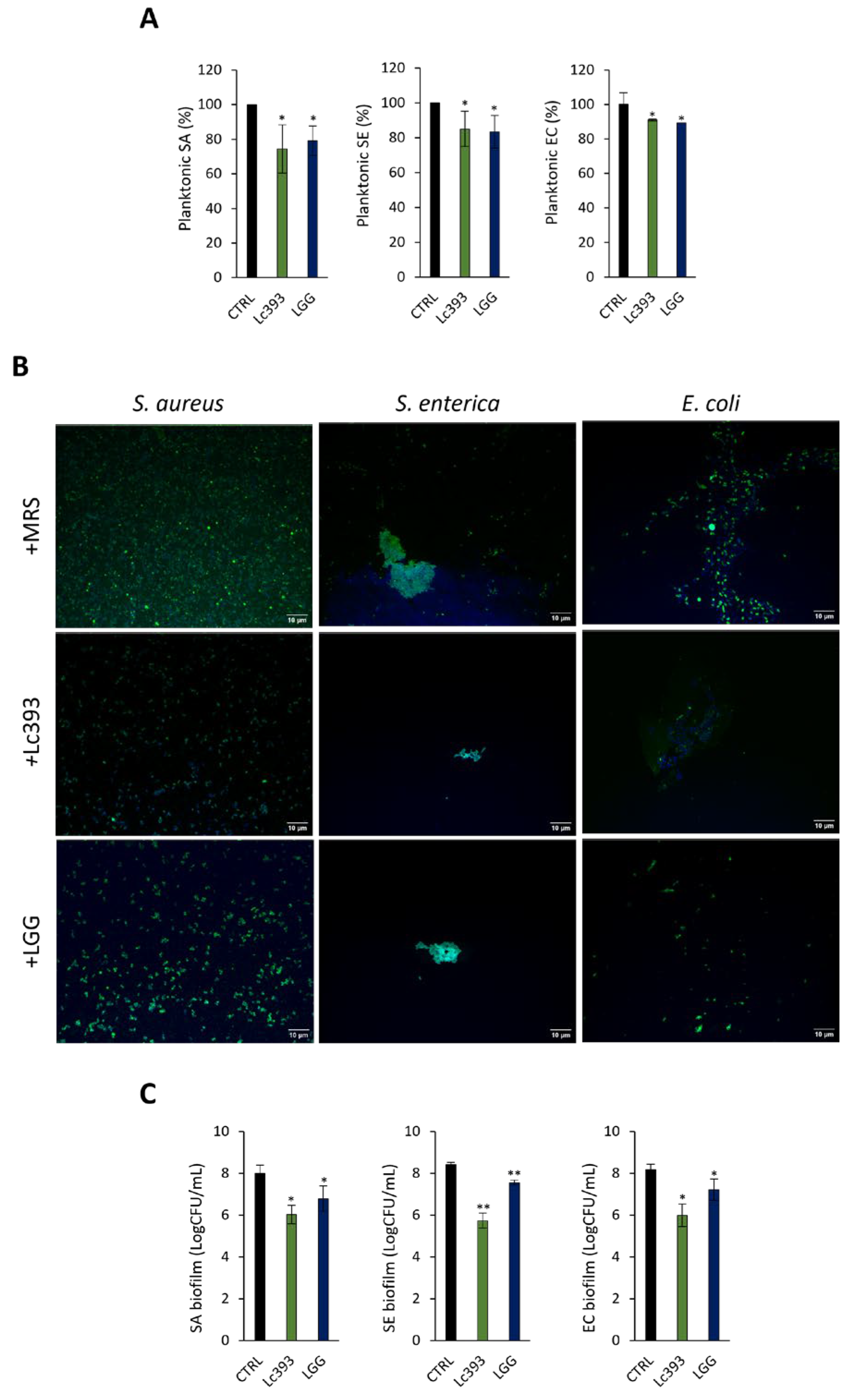
2.10. Antibiofilm Activity of Lactobacilli CFCS
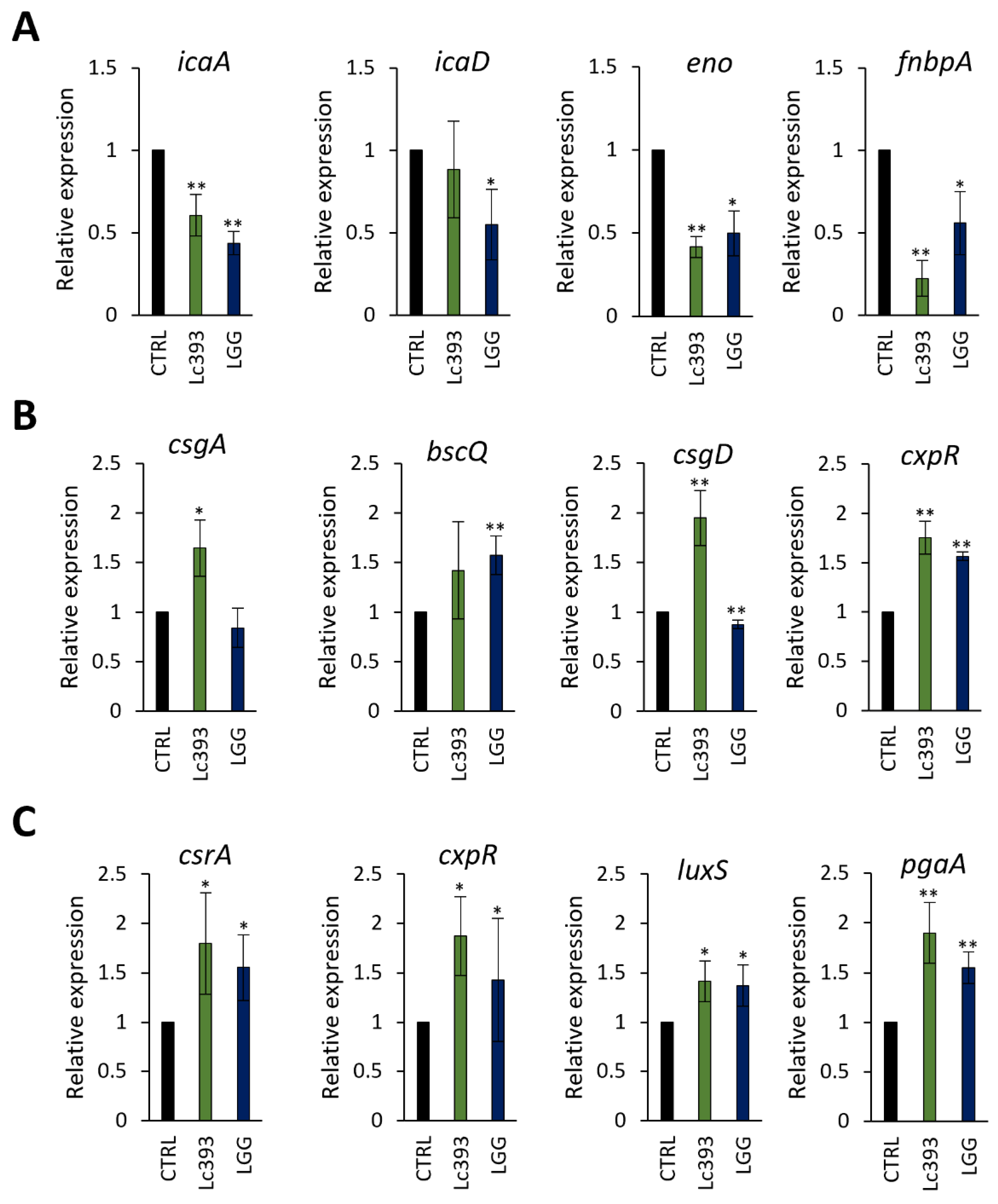
2.11. RNA Extraction, cDNA Synthesis, and qPCR
2.12. In Silico Analysis
2.13. Metabolomics
2.14. Statistical Analysis
3. Results
3.1. Viable Lc393 Exerts Antibacterial Activity
3.2. Investigation of Lc393–Pathogens–Host Interactions
3.3. Lc393 CFCS Inhibits Planktonic Pathogen Viability and Biofilm Formation Capacity
3.4. In Silico Analysis of Genes and Genetic Clusters Coding for Bacteriocins and Antimicrobial Metabolites
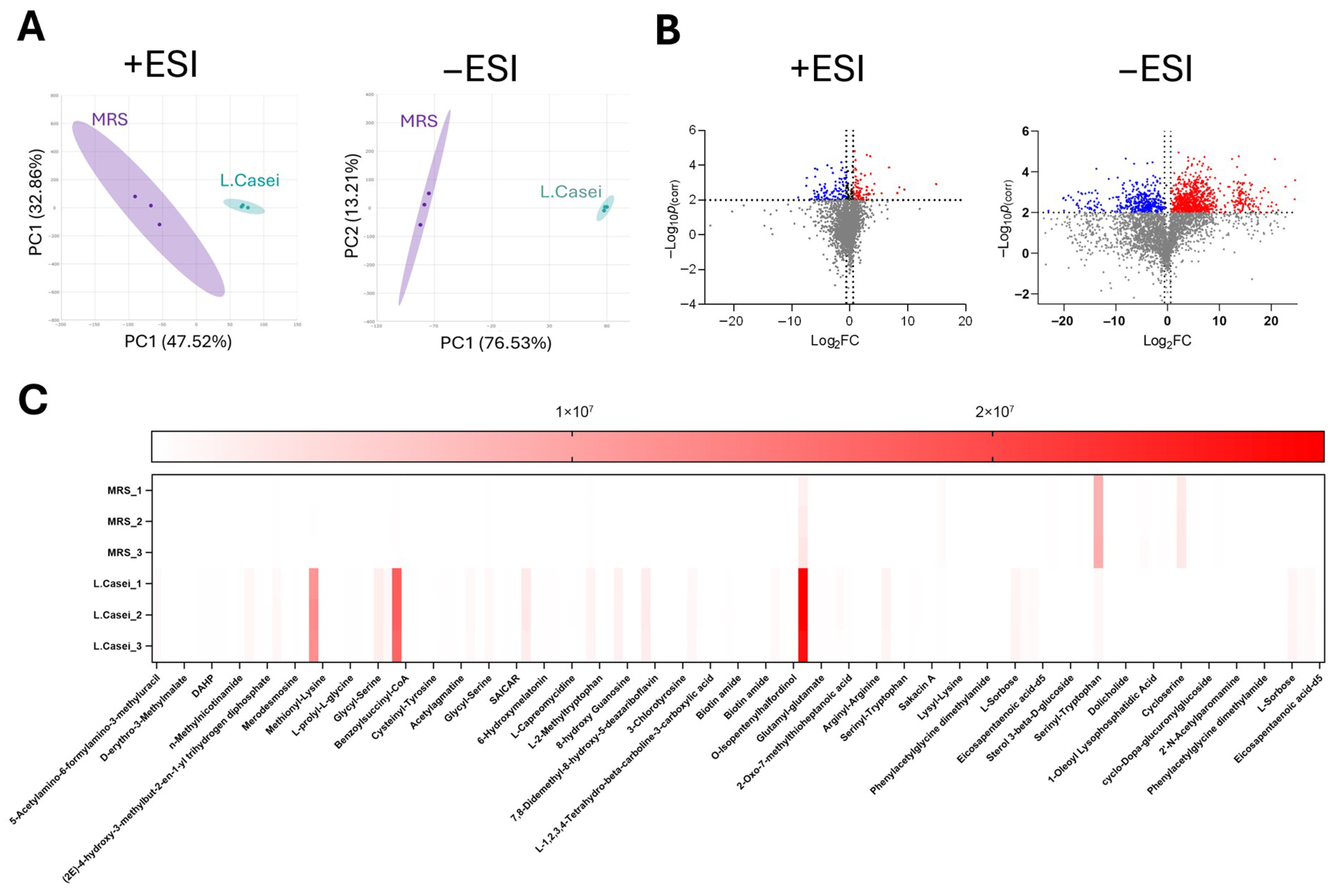
3.5. Untargeted Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grari, O.; Ezrari, S.; El Yandouzi, I.; Benaissa, E.; Lahlou, Y.B.; Lahmer, M.; Saddari, M.; Elouennass, M.; Maleb, A. A comprehensive review on biofilm-associated infections: Mechanisms, diagnostic challenges, and innovative therapeutic strategies. Microbe 2025, 8, 100436. [Google Scholar] [CrossRef]
- Bano, S.; Hassan, N.; Rafiq, M.; Hassan, F.; Rehman, M.; Iqbal, N.; Ali, H.; Hasan, F.; Kang, Y.Q. Biofilms as Battlefield Armor for Bacteria against Antibiotics: Challenges and Combating Strategies. Microorganisms 2023, 11, 2595. [Google Scholar] [CrossRef]
- Elshobary, M.E.; Badawy, N.K.; Ashraf, Y.; Zatioun, A.A.; Masriya, H.H.; Ammar, M.M.; Mohamed, N.A.; Mourad, S.; Assy, A.M. Combating Antibiotic Resistance: Mechanisms, Multidrug-Resistant Pathogens, and Novel Therapeutic Approaches: An Updated Review. Pharmaceuticals 2025, 18, 402. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Tang, M.; Aqib, A.I.; Zhang, X.; Wu, Z.; Wen, Y.; Hou, X.; Xu, J.; Hao, R.; Wang, S.; et al. Dairy farm waste: A potential reservoir of diverse antibiotic resistance and virulence genes in aminoglycoside- and beta-lactam-resistant Escherichia coli in Gansu Province, China. Environ. Res. 2024, 263, 120190. [Google Scholar] [CrossRef] [PubMed]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation, and roles in human disease. Virulence 2024, 2, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Ramírez Castillo, F.Y.; Guerrero Barrera, A.L.; Harel, J.; Avelar González, F.J.; Vogeleer, P.; Arreola Guerra, J.M.; González Gámez, M. Biofilm Formation by Escherichia coli Isolated from Urinary Tract Infections from Aguascalientes, Mexico. Microorganisms 2023, 11, 2858. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial Biofilms in the Food Industry-A Comprehensive Review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef]
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella Biofilm Formation, Chronic Infection, and Immunity Within the Intestine and Hepatobiliary Tract. Front. Cell. Inf. Microbiol. 2021, 10, 624622. [Google Scholar] [CrossRef]
- Fuochi, V.; Coniglio, M.A.; Laghi, L.; Rescifina, A.; Caruso, M.; Stivala, A.; Furneri, P.M. Metabolic Characterization of Supernatants Produced by Lactobacillus spp. with in vitro Anti-Legionella Activity. Front. Microbiol. 2019, 10, 1403. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Efstathiou, C.; Tzampazlis, V.; Plessas, S.; Panopoulou, M.; Koffa, M.; Galanis, A. Genetic and phenotypic assessment of the antimicrobial activity of three potential probiotic lactobacilli against human enteropathogenic bacteria. Front. Cell. Inf. Microbiol. 2023, 13, 1127256. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Panopoulou, M.; Pappa, A.; Galanis, A. Lactobacilli-host interactions inhibit Staphylococcus aureus and Escherichia coli-induced cell death and invasion in a cellular model of infection. Front. Microbiol. 2024, 15, 1501119. [Google Scholar] [CrossRef] [PubMed]
- Walsham, A.D.; MacKenzie, D.A.; Cook, V.; Wemyss-Holden, S.; Hews, C.L.; Juge, N.; Schüller, S. Lactobacillus reuteri Inhibition of Enteropathogenic Escherichia coli Adherence to Human Intestinal Epithelium. Front. Microbiol. 2016, 7, 244. [Google Scholar] [CrossRef] [PubMed]
- Toh, H.; Oshima, K.; Nakano, A.; Takahata, M.; Murakami, M.; Takaki, T.; Nishiyama, H.; Igimi, S.; Hattori, M.; Morita, H. Genomic adaptation of the Lactobacillus casei group. PLoS ONE 2013, 8, e75073. [Google Scholar] [CrossRef] [PubMed]
- Dimitrellou, D.; Sakadani, E.; Kandylis, P. Enhancing Probiotic Viability in Yogurt: The Role of Apple Fibers in Supporting Lacticaseibacillus casei ATCC 393 During Storage and Gastrointestinal Transit. Foods 2025, 14, 376. [Google Scholar] [CrossRef]
- Abdel-Hamid, M.; Romeih, E.; Gamba, R.R.; Nagai, E.; Suzuki, T.; Koyanagi, T.; Enomoto, T. The biological activity of fermented milk produced by Lactobacillus casei ATCC 393 during cold storage. Int. Dairy J. 2019, 91, 1–8. [Google Scholar] [CrossRef]
- Sidira, M.; Karapetsas, A.; Galanis, A.; Kanellaki, M.; Kourkoutas, Y. Effective survival of immobilized Lactobacillus casei during ripening and heat treatment of probiotic dry-fermented sausages and investigation of the microbial dynamics. Meat Sci. 2014, 96, 948–955. [Google Scholar] [CrossRef]
- Saxami, G.; Ypsilantis, P.; Sidira, M.; Simopoulos, C.; Kourkoutas, Y.; Galanis, A. Distinct adhesion of probiotic strain Lactobacillus casei ATCC 393 to rat intestinal mucosa. Anaerobe 2012, 18, 417–420. [Google Scholar] [CrossRef]
- Sidira, M.; Galanis, A.; Ypsilantis, P.; Karapetsas, A.; Progaki, Z.; Simopoulos, C.; Kourkoutas, Y. Effect of probiotic-fermented milk administration on gastrointestinal survival of Lactobacillus casei ATCC 393 and modulation of intestinal microbial flora. J. Mol. Microbiol. Biotechnol. 2010, 19, 224–230. [Google Scholar] [CrossRef]
- Dou, X.; Qiao, L.; Chang, J.; Yan, S.; Song, X.; Chen, Y.; Xu, Q.; Xu, C. Lactobacillus casei ATCC 393 and it’s metabolites alleviate dextran sulphate sodium-induced ulcerative colitis in mice through the NLRP3-(Caspase-1)/IL-1β pathway. Food Funct. 2021, 12, 12022–12035. [Google Scholar] [CrossRef]
- Zhu, L.; Qiao, L.; Dou, X.; Song, X.; Chang, J.; Zeng, X.; Xu, C. Lactobacillus casei ATCC 393 combined with vasoactive intestinal peptide alleviates dextran sodium sulfate-induced ulcerative colitis in C57BL/6 mice via NF-κB and Nrf2 signaling pathways. Biomed. Pharmacother 2023, 165, 115033. [Google Scholar] [CrossRef]
- Tiptiri-Kourpeti, A.; Spyridopoulou, K.; Santarmaki, V.; Aindelis, G.; Tompoulidou, E.; Lamprianidou, E.E.; Saxami, G.; Ypsilantis, P.; Lampri, E.S.; Simopoulos, C.; et al. Lactobacillus casei Exerts Anti-Proliferative Effects Accompanied by Apoptotic Cell Death and Up-Regulation of TRAIL in Colon Carcinoma Cells. PLoS ONE 2016, 11, e0147960. [Google Scholar] [CrossRef]
- Qiao, L.; Chen, Y.; Song, X.; Dou, X.; Xu, C. Selenium Nanoparticles-Enriched Lactobacillus casei ATCC 393 Prevents Cognitive Dysfunction in Mice Through Modulating Microbiota-Gut-Brain Axis. Int. J. Nanomed. 2022, 17, 4807–4827. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Kamiya, S.; Hanawa, T.; Fukuda, M.; Kawakami, H.; Takahashi, H.; Yokota, H. Effect of probiotic bacterial strains of Lactobacillus, Bifidobacterium, and Enterococcus on enteroaggregative Escherichia coli. J. Infect. Chemother. 2010, 16, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, A.; Ataellahi Eshkoor, P.; Saeedi, P.; Halabian, R. Evaluation of antioxidant and antibacterial effects of lactobacilli metabolites- preconditioned bone marrow mesenchymal stem cells in skin lesions amelioration. Bioorg. Chem. 2022, 124, 105797. [Google Scholar] [CrossRef] [PubMed]
- Amat, S.; Subramanian, S.; Timsit, E.; Alexander, T.W. Probiotic bacteria inhibit the bovine respiratory pathogen Mannheimia haemolytica serotype 1 in vitro. Lett. Appl. Microbiol. 2017, 64, 343–349. [Google Scholar] [CrossRef]
- Wu, L.; Luo, Y. Bacterial Quorum-Sensing Systems and Their Role in Intestinal Bacteria-Host Crosstalk. Front. Microbiol. 2021, 12, 611413. [Google Scholar] [CrossRef]
- Prabhurajeshwar, C.; Chandrakanth, R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017, 40, 270. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Vader, L.; Szenei, J.; Reitz, Z.L.; Augustijn, H.E.; Cediel-Becerra, J.D.D.; de Crécy-Lagard, V.; Koetsier, R.A.; Williams, S.E.; et al. antiSMASH 8.0: Extended gene cluster detection capabilities and analyses of chemistry, enzymology, and regulation. Nucleic Acids Res. 2025, 53, W32–W38. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457. [Google Scholar] [CrossRef]
- Jangra, M.; Travin, D.Y.; Aleksandrova, E.V.; Kaur, M.; Darwish, L.; Koteva, K.; Klepacki, D.; Wang, W.; Tiffany, M.; Sokaribo, A.; et al. A broad-spectrum lasso peptide antibiotic targeting the bacterial ribosome. Nature 2025, 640, 1022–1030, Correction in Nature 2025, 645, E11. [Google Scholar] [CrossRef]
- De Keersmaecker, S.C.J.; Verhoeven, T.L.A.; Desair, J.; Marchal, K.; Vanderleyden, J.; Nagy, I. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 2006, 259, 89–96. [Google Scholar] [CrossRef]
- Kathayat, D.; Closs, G., Jr.; Helmy, Y.A.; Deblais, L.; Srivastava, V.; Rajashekara, G. In Vitro and In Vivo Evaluation of Lacticaseibacillus rhamnosus GG and Bifidobacterium lactis Bb12 Against Avian Pathogenic Escherichia coli and Identification of Novel Probiotic-Derived Bioactive Peptides. Probiotics Antimicrob. Proteins 2022, 14, 1012–1028. [Google Scholar] [CrossRef]
- Kang, E.A.; Choi, H.I.; Hong, S.W.; Kang, S.; Jegal, H.Y.; Choi, E.W.; Park, B.S.; Kim, J.S. Extracellular Vesicles Derived from Kefir Grain Lactobacillus Ameliorate Intestinal Inflammation via Regulation of Proinflammatory Pathway and Tight Junction Integrity. Biomedicines 2020, 8, 522. [Google Scholar] [CrossRef]
- Allonsius, C.N.; van den Broek, M.F.L.; De Boeck, I.; Kiekens, S.; Oerlemans, E.F.M.; Kiekens, F.; Foubert, K.; Vandenheuvel, D.; Cos, P.; Delputte, P.; et al. Interplay between Lactobacillus rhamnosus GG and Candida and the involvement of exopolysaccharides. Microb. Biotechnol. 2017, 10, 1753–1763. [Google Scholar] [CrossRef]
- El-Chami, C.; Choudhury, R.; Mohammedsaeed, W.; McBain, A.J.; Kainulainen, V.; Lebeer, S.; Satokari, R.; O’Neil, C.A. Multiple Proteins of Lacticaseibacillus rhamnosus GG Are Involved in the Protection of Keratinocytes From the Toxic Effects of Staphylococcus aureus. Front. Microbiol. 2022, 13, 875542. [Google Scholar] [CrossRef]
- Gatej, S.M.; Marino, V.; Bright, R.; Fitzsimmons, T.R.; Gully, N.; Zilm, P.; Gibson, R.J.; Edwards, S.; Bartold, P.M. Probiotic Lactobacillus rhamnosus GG prevents alveolar bone loss in a mouse model of experimental periodontitis. J. Clin. Periodontol. 2018, 45, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gu, Z.; Song, F.; Zhang, H.; Zhao, J.; Chen, W. Lactobacillus plantarum ZS2058 and Lactobacillus rhamnosus GG Use different mechanisms to prevent Salmonella infection in vivo. Front. Microbiol. 2019, 10, 299. [Google Scholar] [CrossRef] [PubMed]
- Massip, C.; Oswald, E. Siderophore-Microcins in Escherichia coli: Determinants of Digestive Colonization, the First Step Toward Virulence. Front. Cell. Infect. Microbiol. 2020, 10, 381. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.W.L.; Tsolis, R.M.; Bäumler, A.J. Salmonella versus the Microbiome. Microbiol. Mol. Biol. Rev. 2020, 85, e00027-19. [Google Scholar] [CrossRef]
- Spacova, I.; O’Neill, C.; Lebeer, S. Lacticaseibacillus rhamnosus GG inhibits infection of human keratinocytes by Staphylococcus aureus through mechanisms involving cell surface molecules and pH reduction. Benef. Microbes 2020, 11, 703–715. [Google Scholar] [CrossRef]
- Hirano, J.; Yoshida, T.; Sugiyama, T.; Koide, N.; Mori, I.; Yokochi, T. The effect of Lactobacillus rhamnosus on enterohemorrhagic Escherichia coli infection of human intestinal cells in vitro. Microbiol. Immunol. 2003, 47, 405–409. [Google Scholar] [CrossRef]
- Wen Fang Wu Wu, J.; Redondo-Solano, M.; Uribe, L.; WingChing-Jones, R.; Usaga, J.; Barboza, N. First characterization of the probiotic potential of lactic acid bacteria isolated from Costa Rican pineapple silages. PeerJ 2021, 9, e12437. [Google Scholar] [CrossRef]
- Saidi, N.; Saderi, H.; Owlia, P.; Soleimani, M. Anti-Biofilm Potential of Lactobacillus casei and Lactobacillus rhamnosus Cell-Free Supernatant Extracts against Staphylococcus aureus. Adv. Biomed. Res. 2023, 12, 50. [Google Scholar] [CrossRef]
- Singh, N.; Gulhane, R.D.; Singh, A.; Goel, M.; Udelal, P.P.; Sangwan, V.; Sihag, M.K.; Goel, G.; Panwar, H.; Puniya, A.K. Exploring the antimicrobial potential of lactobacilli against early-stage and mature biofilms of Staphylococcus aureus and Pseudomonas aeruginosa. Front. Chem. 2025, 13, 1425666. [Google Scholar] [CrossRef]
- Abou Elez, R.M.M.; Elsohaby, I.; Al-Mohammadi, A.R.; Seliem, M.; Tahoun, A.B.M.B.; Abousaty, A.I.; Algendy, R.M.; Mohamed, E.A.A.; El-Gazzar, N. Antibacterial and anti-biofilm activities of probiotic Lactobacillus plantarum against Listeria monocytogenes isolated from milk, chicken and pregnant women. Front. Microbiol. 2023, 14, 1201201. [Google Scholar] [CrossRef]
- Niranjan, R.; Patil, S.; Dubey, A.; Lochab, B.; Priyadarshini, R. Small cyclic dipeptide produced by Lactobacillus rhamnosus with anti-biofilm properties against Streptococcus mutans biofilm. Biofilm 2024, 8, 100237. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhou, Y.; Tang, G.; Wu, R.; Lin, H. Exploration of the Main Antibiofilm Substance of Lactobacillus plantarum ATCC 14917 and Its Effect against Streptococcus mutans. Int. J. Mol. Sci. 2023, 24, 1986. [Google Scholar] [CrossRef] [PubMed]
- Dalvand, M.; Mirhosseini, S.A.; Amini, K.; Khani, S.; Mahmoodzadeh Hosseini, H.; Mansoori, K. Evaluation of anti-biofilm activity of Lactobacillus rhamnosus GG and Nisin on the expression of aap, ica-A and ica-D as biofilm-associated genes of Staphylococcus epidermidis. Iran. J. Microbiol. 2023, 15, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Pompilio, A.; Kaya, E.; Lupetti, V.; Catelli, E.; Bianchi, M.; Maisetta, G.; Esin, S.; Di Bonaventura, G.; Batoni, G. Cell-free supernatants from Lactobacillus strains exert antibacterial, antibiofilm, and antivirulence activity against Pseudomonas aeruginosa from cystic fibrosis patients. Microb. Inf. 2024, 26, 105301. [Google Scholar] [CrossRef]
- Barzegari, A.; Kheyrolahzadeh, K.; Hosseiniyan Khatibi, S.M.; Sharifi, S.; Memar, M.Y.; Zununi Vahed, S. The Battle of Probiotics and Their Derivatives Against Biofilms. Inf. Drug Resist. 2020, 13, 659–672. [Google Scholar] [CrossRef]
- Arciola, C.R.; Baldassarri, L.; Montanaro, L. Presence of icaA and icaD Genes and Slime Production in a Collection of Staphylococcal Strains from Catheter-Associated Infections. J. Clin. Microbiol. 2001, 39, 2151–2156. [Google Scholar] [CrossRef]
- Jeong, D.; Kim, D.H.; Song, K.Y.; Seo, K.H. Antimicrobial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J. Oral Microbiol. 2018, 10, 1472985. [Google Scholar] [CrossRef]
- Araujo, L.U.; Grabe-Guimaraes, A.; Mosqueira, V.C.; Carneiro, C.M.; Silva-Barcello, N.M. Profile of wound healing process induced by allantoin. Acta Cir. Bras. 2010, 25, 460–466. [Google Scholar] [CrossRef]
- Alferova, V.A.; Maviza, T.P.; Biryukov, M.V.; Zakalyukina, Y.V.; Polshakov, V.I.; Sergiev, P.V.; Korshun, V.A.; Osterman, I.A. Characterization of a novel natural tetracenomycin reveals crucial role of 4-hydroxy group in ribosome binding. Biochimie 2023, 206, 150–153. [Google Scholar] [CrossRef]
- Naegeli, H.U.; Loosli, H.R.; Nussbaumer, A. Clavamycins, new clavam antibiotics from two variants of Streptomyces hygroscopicus. II. Isolation and structures of clavamycins A, B and C from Streptomyces hygroscopicus NRRL 15846, and of clavamycins D, E and F from Streptomyces hygroscopicus NRRL 15879. J. Antibiot. 1986, 39, 516–524. [Google Scholar] [CrossRef]
- Jung, D.; Yum, S.J.; Jeong, H.G. Characterization and evaluation of antimicrobial activity of actinonin against foodborne pathogens. Food Sci. Biotechnol. 2017, 26, 1649–1657. [Google Scholar] [CrossRef]
- Meinnel, T.; Blanquet, S. Enzymatic properties of Escherichia coli peptide deformylase. J. Bacteriol. 1995, 177, 1883–1887. [Google Scholar] [CrossRef]
- Rochat, B. Proposed Confidence Scale and ID Score in the Identification of Known-Unknown Compounds Using High Resolution MS Data. J. Am. Soc. Mass Spectrom. 2017, 28, 709–723. [Google Scholar] [CrossRef]
- Malinowska, J.M.; Viant, M.R. Confidence in Metabolite Identification Dictates the Applicability of Metabolomics to Regulatory Toxicology. Curr. Opin. Toxicol. 2019, 16, 32–38. [Google Scholar] [CrossRef]
- Salek, R.M.; Steinbeck, C.; Viant, M.R.; Goodacre, R.; Dunn, W.B. The Role of Reporting Standards for Metabolite Annotation and Identification in Metabolomic Studies. Gigascience 2013, 2, 13. [Google Scholar] [CrossRef]
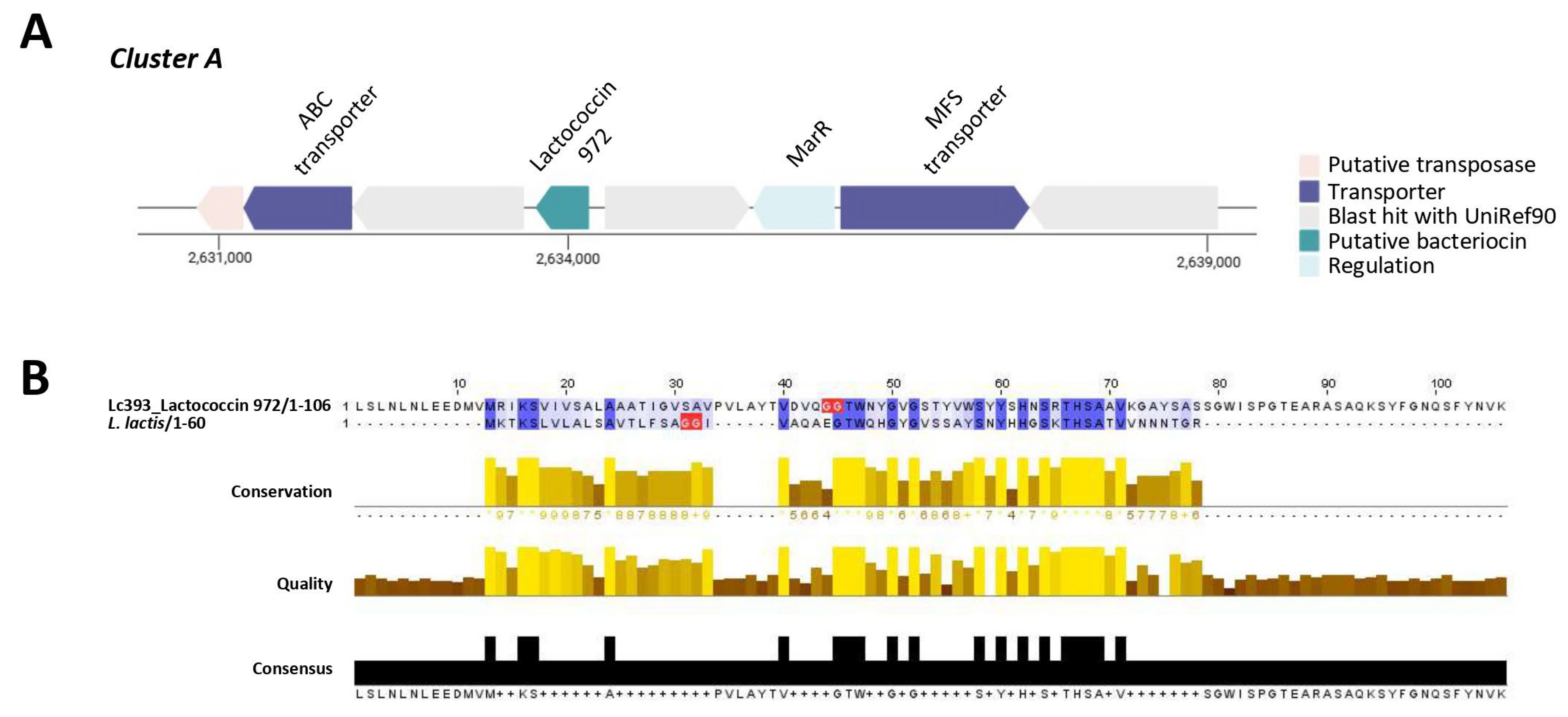
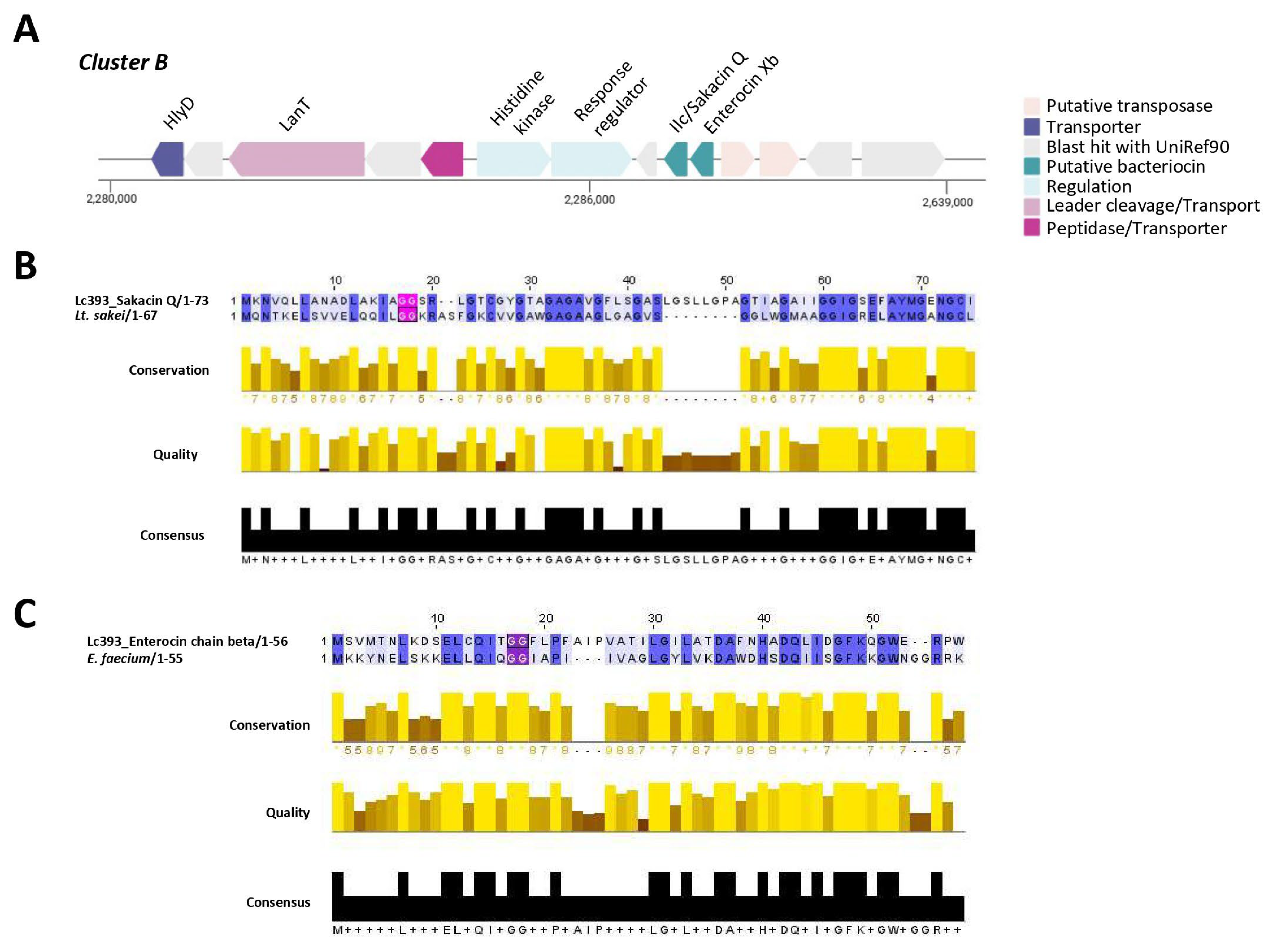
| Putative Bacteriocin | Length (aa) | Theoretical pI | Molecular Weight (kDa) | Protein Family Membership | Signal Seq | Topology |
|---|---|---|---|---|---|---|
| Lactococcin 972 | 105 | 8.97 | 11.2 | Bacteriocin, lactococcin 972 (IPR006540) | Sec/SPI | Outside |
| Bacteriocin II/Sakacin Q | 73 | 5.93 | 6.9 | None predicted | Other | Outside (but with transmembrane regions) |
| Enterocin Xb | 56 | 4.52 | 6.2 | None predicted | Other | Outside (but with transmembrane regions) |
| Metabolite | Theoretical m/z | Actual m/z | Retention Time (min) | Log2Fold of Change | pcorr |
|---|---|---|---|---|---|
| Allantoic acid | 176.0546 | 176.0514 | 15.363 | 7.137223 | 1.86781 × 105 |
| Tetracenomycin A2 | 422.4056 | 422.4053 | 7.154 | 3.459779 | 0.000147944 |
| Isochorismate | 226.0478 | 226.0479 | 7.209 | 5.389228 | 0.000267169 |
| Clavamycin D | 313.1430 | 315.1456 | 3.417 | 14.94609 | 0.003409505 |
| Actinonin | 385.2539 | 385.2537 | 21.841 | 11.01422 | 0.004445146 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiousi, D.E.; Kyriakou, S.; Efstathiou, C.; Didaskalou, S.; Koffa, M.; Pappa, A.; Panopoulou, M.; Panayiotidis, M.I.; Galanis, A. Lacticaseibacillus casei Combats Biofilm Formation and Exhibits Antibacterial Activity Against Clinical Isolates of Staphylococcus aureus, Salmonella enterica, and Escherichia coli. Microorganisms 2025, 13, 2667. https://doi.org/10.3390/microorganisms13122667
Kiousi DE, Kyriakou S, Efstathiou C, Didaskalou S, Koffa M, Pappa A, Panopoulou M, Panayiotidis MI, Galanis A. Lacticaseibacillus casei Combats Biofilm Formation and Exhibits Antibacterial Activity Against Clinical Isolates of Staphylococcus aureus, Salmonella enterica, and Escherichia coli. Microorganisms. 2025; 13(12):2667. https://doi.org/10.3390/microorganisms13122667
Chicago/Turabian StyleKiousi, Despoina Eugenia, Sotiris Kyriakou, Christos Efstathiou, Stylianos Didaskalou, Maria Koffa, Aglaia Pappa, Maria Panopoulou, Mihalis I. Panayiotidis, and Alex Galanis. 2025. "Lacticaseibacillus casei Combats Biofilm Formation and Exhibits Antibacterial Activity Against Clinical Isolates of Staphylococcus aureus, Salmonella enterica, and Escherichia coli" Microorganisms 13, no. 12: 2667. https://doi.org/10.3390/microorganisms13122667
APA StyleKiousi, D. E., Kyriakou, S., Efstathiou, C., Didaskalou, S., Koffa, M., Pappa, A., Panopoulou, M., Panayiotidis, M. I., & Galanis, A. (2025). Lacticaseibacillus casei Combats Biofilm Formation and Exhibits Antibacterial Activity Against Clinical Isolates of Staphylococcus aureus, Salmonella enterica, and Escherichia coli. Microorganisms, 13(12), 2667. https://doi.org/10.3390/microorganisms13122667











